Key Points
High-risk MMs characterized by high-risk cytogenetics were associated with worse OS in EAs, but not in AAs.
Translocation t(4;14) and gain/amplification of 1q were associated with worse OS differently between EAs and AAs.
Visual Abstract
Cytogenetic abnormalities influence the prognosis of multiple myeloma (MM). How these abnormalities associate with overall survival (OS) in European Americans (EAs) and African Americans (AAs) remains unclear. We collected data on fluorescence in situ hybridization targeting 17 cytogenetic abnormalities from 181 patients newly diagnosed with MM between 2010 and 2019. Vital status was ascertained using the National Death Index. Baseline clinical data were retrieved from electronic medical records. Established high-risk cytogenetic abnormalities (HRCAs) include t(4;14), t(14;16), t(14;20), del 17p, and gain/amplification of 1q. In each population, we evaluated the associations between cytogenetic abnormalities and OS. Among 55 AAs and 126 EAs, 65 deaths occurred (median follow-up: 5.8 years). The distribution of the abnormalities was similar between EAs and AAs. High-risk MM, characterized by HRCAs, was associated with worse OS in EAs (hazard ratio [HR], 2.6; 95% confidence interval [CI], 1.3-5.5), but not AAs. Del 13q was associated with worse OS in both populations. Gain/amplification of 1q was associated with poorer OS in EAs (HR, 3.44; 95% CI, 1.3-9.3) but not AAs, whereas t(4;14) was associated with poorer OS in AAs (HR, 14.51; 95% CI, 2.3-92.3) but not EAs. These associations remained after controlling for prognostic factors or other HRCAs, highlighting the potential of population heterogeneity in the prognostic significance of cytogenetic abnormalities.
Introduction
Multiple myeloma (MM) is the second most common of lymphoid malignancies, affecting >34 000 patients annually in the United States.1 The 5-year relative survival for MM has increased to 58% with the use of autologous stem cell transplant therapy and combined therapy of proteasome inhibitor, immunomodulatory imide drug, and dexamethasone.2,3 However, the improvements in outcomes have varied among populations,4,5 and the disparities cannot be fully attributed to variations in access to care.6,7 African Americans (AAs) experience a 2 to 3 times higher incidence of MM compared with European Americans (EAs)8 and typically develop the disease at an average of 5 years younger,4,6 suggesting population-specific disease biology that might also play a role in the survival outcomes of patients with MM.
Primary cytogenetic abnormalities unique to a cell population of the same origin have diagnostic and therapeutic potential for several cancers,9 including hematological malignancies.10-12 In MM, primary cytogenetic abnormalities, including translocations involving the immunoglobulin heavy chain (IgH) gene locus on chromosome 14 and trisomies of odd-numbered chromosomes,13 and secondary cytogenetic abnormalities, such as deletion of chromosome 17p (del 17p) and gain/amplification of chromosome 1q (gain/amplification 1q), can be identified in nearly all patients.14 These chromosomal abnormalities not only carry prognostic significance but also influence clinical behavior.13 For example, the presence of an IgH translocation t(11;14)/IGH::CCND1 is linked to a standard risk, whereas t(4;14)/FGFR3::IGH, t(14;16)/IGH::MAF, and t(14;20)/IGH::MAFB are associated with a poor prognosis.15-17 Furthermore, patients who have del 17p and gain/amplification of 1q were found to have a more rapid rate of disease progression.2,9,18 As a result, cytogenetics has become an integral component of MM management and is included in the International Myeloma Working Group (IMWG) consensus statement.19 The IMWG designates t(4;14), t(14;16), t(14;20), del 17p, and gain/amplification 1q as high-risk cytogenetic abnormalities (HRCAs), and their presence characterizes high-risk MM.19 Notably, some of the HRCAs have been suggested to occur at different frequencies between EAs and AAs with MM. For example, t(4;14) and del 17p have been found to be less prevalent in AAs compared with EAs with MM,20 whereas t(14;16), t(14;20), and t(11;14) are found to be more prevalent in individuals with greater African genetic ancestry in a study of MM and monoclonal gammopathy of unknown significance (MGUS).21 In another study of patients with MM, t(11;14) was found to be more prevalent in AAs.22 However, it remains unknown whether these HRCAs and other cytogenetic abnormalities associate with MM outcomes differently between EAs and AAs.
Therefore, we designed this study to evaluate the associations of HRCAs and other cytogenetic abnormalities with overall survival (OS) in EA and AA patients newly diagnosed with MM. The cytogenetic abnormalities were detected by fluorescence in situ hybridization (FISH) at the University of Chicago Cancer Cytogenetic Laboratory as part of the diagnostic workup.
Methods
Study population
A detailed description of the UChicago Multiple Myeloma Epidemiology Study, from which subjects for this analysis were obtained, has previously been reported.22,23 Briefly, patients aged ≥20 years old with newly diagnosed MM, MGUS, and smoldering multiple myeloma were prospectively and systematically enrolled from 2010 to 2019 at the University of Chicago Medical Center. MM and plasma cell disorders were defined according to the IMWG updated criteria.24 Blood samples and epidemiological questionnaire data were collected at the time of diagnosis from 586 enrolled patients with MM. The study was approved by the University of Chicago Institutional Review Board and conducted in accordance with the Declaration of Helsinki.
Data collection
Of the 586 patients with MM in the cohort, data from FISH probes targeting 17 cytogenetic aberrations within 180 days of diagnosis are available for 181 individuals. These FISH analyses were conducted at the University of Chicago Cancer Cytogenetic Laboratory. Briefly, chromosome analysis using the G-banding method was performed on fresh bone marrow aspirate samples following the standard protocol as a part of routine diagnostic workup. At least 20 metaphase cells were evaluated for each case, and the chromosomal abnormalities were described according to the International System for Human Cytogenetic Nomenclature.25 Interphase reflex FISH analysis of CD138+ plasma cells enriched from bone marrow aspirate samples was performed with a panel of probes designed to detect MM-associated abnormalities following the manufacturer’s recommendations. The following FISH probes (Vysis, Abbott Molecular, Abbott Park, IL) were used: dual-fusion dual-color probes to detect the t(4;14)/FGFR3::IGH, t(11;14)/CCND1::IGH, t(14;16)/IGH::MAF, and t(14;20)/IGH::MAFB; break-apart probe to detect loss of IGH (loss of the entire locus) and IGH rearrangements; chromosome enumeration probes to detect trisomies of chromosomes 3, 7, and 9; 13q14.3 and 13q34 probes set to detect deletion or loss of chromosome 13; locus-specific probes to detect copy number alterations of ATM/11q22.3, TP53/17p13.1, and CDKN2C/1p32.3; and CKS1B/1q21 to detect gain/amplification of 1q. Tetraploidy was detected when all probes revealed 4 signal patterns instead of 2. Certain abnormalities, such as t(11;14) and t(14;20), were evaluated in only a limited number of patients. This may reflect updates in the National Comprehensive Cancer Network guidelines for diagnostic workup, particularly involving FISH, during the study enrollment period. In addition, FISH results were obtained from the UChicago cancer cytogenetic laboratory up to January 2017 and by manual abstraction from electronic health records (EHRs) afterward.
Furthermore, age at diagnosis, sex, and race/ethnicity were obtained from EHRs or a self-administered questionnaire at study enrollment. Vital status was ascertained using the National Death Index. Baseline clinical and laboratory data such as the international staging system (ISS), estimated glomerular filtration rate (eGFR),26 elevated serum lactate dehydrogenase (LDH), serum free light chains, serum albumin, serum β2 microglobulin, and other laboratory results were collected from the EHRs.
Statistical analyses
The study aimed to assess the population-specific associations between cytogenetic abnormalities and OS in MM. Participants were followed up from the date of diagnosis to the date of death, loss to follow-up, or administrative end date of 28 February 2022, whichever occurred first. OS was defined as the time from diagnosis to death from any cause. High-risk MM was defined by the presence of any of the cytogenetic abnormalities associated with poor prognosis according to the IMWG consensus definition of HRCAs, including t(4;14), t(14;16), t(14;20), del 17p, and gain/amplification of 1q.19 Standard-risk MM was defined by the absence of any HRCAs or the presence of any trisomies of odd-numbered chromosomes or t(11;14). Because not all patients were tested for every HRCA, absence of HRCAs was defined as negative for any tested HRCA. We also applied a more restrictive approach, requiring negative test results for all 5 HRCAs. As the results were similar, we present the findings using the former definition with larger sample size.
We used mean (standard deviation) and proportion to compare the distributions of sociodemographic factors and cytogenetic abnormalities between AAs and EAs. The associations between each cytogenetic abnormality and OS were estimated using hazard ratios (HRs) and 95% confidence intervals (CIs) from age-adjusted (years) Cox proportional hazards models. Several clinical indices such as ISS (1/2/3), eGFR (normal: ≥60 mL/min vs abnormal: <60 mL/min), LDH (normal: <240 U/L vs elevated: ≥240 U/L), and serum free light chain (normal: 0.26-1.65 vs low: <0.26 vs high: >1.65) have been linked to the prognosis of MM.27,28 Therefore, we assessed their associations with OS in AAs and EAs, separately. The variables significantly associated with OS (ie, P < .05) in each population were included as covariates in population-specific multivariable Cox proportional hazards models. In EAs, the multivariable models included a priori adjustment of age, sex, ISS, and LDH. In AAs, the multivariable models included age, sex, and ISS, but not LDH because it is not associated with OS in AAs. Last, we used Kaplan-Meier method and the log-rank test to visualize the survival curves of IMWG-defined high- vs standard-risk MMs in EAs and AAs, separately. Data analyses were performed using Stata (version 17.0) and R (version 4.1.2; R Foundation for Statistical Computing, Vienna, Austria; https://cran.r-project.org).
Results
Among the 181 participants included in this analysis, 55 (30.4%) were AAs and 126 (69.6%) were EAs. In a median follow-up of 69.5 months (interquartile range, 36.9-91.5 months), 65 deaths (35.9%) occurred (AA: 23, EA: 42). Table 1 describes the characteristics of participants by vital status. Compared with those at risk, participants who died were older and more likely to have advanced ISS stages and elevated LDH levels at the time of diagnosis. They were similar with respect to education, eGFR, serum free light chain, and other sociodemographic and clinical characteristics evaluated.
Characteristics of AAs and EAs with MM by vital status
| Variables . | At risk, n (%) . | Death, n (%) . | Age-adjusted HRs∗ (95% CI) . |
|---|---|---|---|
| 116 (64.1) | 65 (35.9) | ||
| Age | |||
| Mean (standard deviation) | 59.9 (9.7) | 63.0 (8.1) | 1.0 (1.0-1.1) |
| Race/ethnicity | |||
| EA | 84 (72.4) | 42 (64.6) | 1.0 (Referent)† |
| AA | 32 (27.6) | 23 (35.4) | 1.1 (0.7-1.9) |
| Sex | |||
| Female | 59 (50.9) | 24 (36.9) | 1.0 (Referent)† |
| Male | 57 (49.1) | 41 (63.1) | 1.6 (1.0-2.6) |
| Education | |||
| Below college | 20 (23.3) | 10 (21.2) | 1.0 (Referent)† |
| Some college or completed college | 48 (55.8) | 23 (48.9) | 1.0 (0.5-2.0) |
| Graduate or professional degree | 18 (20.9) | 14 (29.8) | 1.3 (0.6-3.0) |
| ISS | |||
| 1 | 72 (67.3) | 29 (47.5) | 1.0 (Referent)† |
| 2 | 34 (31.8) | 21 (34.4) | 1.5 (0.8-2.6) |
| 3 | 1 (0.9) | 11 (18.0) | 6.8 (3.4-13.9) |
| LDH levels (U/L) | |||
| Normal (<240) | 95 (83.3) | 38 (63.3) | 1.0 (Referent)† |
| Elevated (≥240) | 19 (16.7) | 22 (36.7) | 2.3 (1.4-3.9) |
| eGFR (mL/min) | |||
| ≥60 | 90 (78.3) | 42 (66.7) | 1.0 (Referent)† |
| <60 | 25 (21.7) | 21 (33.3) | 1.4 (0.8-2.4) |
| Serum free light chain | |||
| Low (<0.26) | 31 (27.4) | 14 (22.6) | 0.6 (0.3-1.3) |
| Normal (0.26-1.65) | 18 (15.9) | 12 (19.4) | 1.0 (Referent)† |
| High (>1.65) | 64 (56.6) | 36 (58.1) | 0.7 (0.4-1.4) |
| Variables . | At risk, n (%) . | Death, n (%) . | Age-adjusted HRs∗ (95% CI) . |
|---|---|---|---|
| 116 (64.1) | 65 (35.9) | ||
| Age | |||
| Mean (standard deviation) | 59.9 (9.7) | 63.0 (8.1) | 1.0 (1.0-1.1) |
| Race/ethnicity | |||
| EA | 84 (72.4) | 42 (64.6) | 1.0 (Referent)† |
| AA | 32 (27.6) | 23 (35.4) | 1.1 (0.7-1.9) |
| Sex | |||
| Female | 59 (50.9) | 24 (36.9) | 1.0 (Referent)† |
| Male | 57 (49.1) | 41 (63.1) | 1.6 (1.0-2.6) |
| Education | |||
| Below college | 20 (23.3) | 10 (21.2) | 1.0 (Referent)† |
| Some college or completed college | 48 (55.8) | 23 (48.9) | 1.0 (0.5-2.0) |
| Graduate or professional degree | 18 (20.9) | 14 (29.8) | 1.3 (0.6-3.0) |
| ISS | |||
| 1 | 72 (67.3) | 29 (47.5) | 1.0 (Referent)† |
| 2 | 34 (31.8) | 21 (34.4) | 1.5 (0.8-2.6) |
| 3 | 1 (0.9) | 11 (18.0) | 6.8 (3.4-13.9) |
| LDH levels (U/L) | |||
| Normal (<240) | 95 (83.3) | 38 (63.3) | 1.0 (Referent)† |
| Elevated (≥240) | 19 (16.7) | 22 (36.7) | 2.3 (1.4-3.9) |
| eGFR (mL/min) | |||
| ≥60 | 90 (78.3) | 42 (66.7) | 1.0 (Referent)† |
| <60 | 25 (21.7) | 21 (33.3) | 1.4 (0.8-2.4) |
| Serum free light chain | |||
| Low (<0.26) | 31 (27.4) | 14 (22.6) | 0.6 (0.3-1.3) |
| Normal (0.26-1.65) | 18 (15.9) | 12 (19.4) | 1.0 (Referent)† |
| High (>1.65) | 64 (56.6) | 36 (58.1) | 0.7 (0.4-1.4) |
HRs were calculated using the Cox proportional hazards regression models adjusting for age (years) for all variables except for age.
The referent category, indicated by an HR of 1.0, serves as the baseline for comparison. HRs for other categories are relative to this baseline.
Table 2 shows that the frequency of HRCAs and most cytogenetic abnormalities did not differ significantly between EAs and AAs. However, our data showed that gain/amplification of 1q was slightly more common in EAs than in AAs. We also found that EAs were more likely to have gain of 1q (ie, gain of 1 extra copy of chromosome 1q) than AAs (30.7% vs 8.6%; P = .0), but were less likely to have amplification of 1q (ie, gain of ≥2 extra copies) (data not shown).
Distribution of primary and secondary cytogenetic abnormalities in patients with MM by race/ethnicity
| . | EAs, n (%) . | AAs, n (%) . | P value∗ . |
|---|---|---|---|
| 126 (69.6) | 55 (30.4) | ||
| IMWG HRCA classification† | |||
| Standard risk | 104 (82.5) | 46 (86.8) | .7 |
| High risk | 22 (33.3) | 7 (26.9) | |
| t(4;14)(p16;q32) | |||
| No | 94 (91.3) | 42 (95.5) | .5 |
| Yes | 9 (8.7) | 2 (4.5) | |
| t(11;14)(q13;q32) | |||
| No | 24 (82.8) | 6 (60.0) | .2 |
| Yes | 5 (17.2) | 4 (40.0) | |
| t(14;16)(q32;q23) | |||
| No | 44 (95.7) | 16 (94.1) | 1.0 |
| Yes | 2 (4.3) | 1 (5.9) | |
| t(14;20)(q32;q12) | |||
| No | 36 (100.0) | 16 (94.1) | .3 |
| Yes | 0 (0.0) | 1 (5.9) | |
| Loss of IGH | |||
| No | 49 (72.1) | 16 (61.5) | .3 |
| Yes | 19 (27.9) | 10 (38.5) | |
| IGH rearrangement | |||
| No | 61 (74.4) | 20 (66.7) | .5 |
| Yes | 22 (25.6) | 10 (33.3) | |
| Trisomy of chromosome 3 | |||
| No | 56 (74.7) | 25 (75.8) | 1.0 |
| Yes | 19 (25.3) | 8 (24.2) | |
| Trisomy of chromosome 7 | |||
| No | 48 (80.0) | 14 (70.0) | .4 |
| Yes | 12 (20.0) | 6 (30.0) | |
| Trisomy of chromosome 9 | |||
| No | 51 (64.0) | 20 (58.8) | .7 |
| Yes | 30 (37.0) | 14 (41.2) | |
| Chromosome 13q deletion (del 13q) | |||
| No | 88 (73.3) | 35 (67.3) | .5 |
| Yes | 32 (26.7) | 17 (32.7) | |
| Loss of ATM [11q22.3)] (del 11q) | |||
| No | 107 (98.2) | 42 (100.0) | 1.0 |
| Yes | 2 (1.8) | 0 (0.0) | |
| Gain of ATM | |||
| No | 37 (49.3) | 8 (40.0) | .6 |
| Yes | 38 (50.7) | 12 (60.0) | |
| Loss of TP53 [17p13.1] (del 17p) | |||
| No | 105 (86.1) | 44 (91.7) | .4 |
| Yes | 17 (13.9) | 4 (8.3) | |
| Gain of TP53 | |||
| No | 61 (87.1) | 17 (70.8) | .1 |
| Yes | 9 (12.9) | 7 (29.2) | |
| Loss of CDKN2C [1p32.3] (del 1p) | |||
| No | 71 (97.3) | 30 (90.9) | .2 |
| Yes | 2 (2.7) | 3 (9.1) | |
| Gain/amplification of CKS1B [1q21] (gain/amplification of 1q) | |||
| No | 48 (58.5) | 27 (75.0) | .1 |
| Yes | 34 (41.5) | 9 (25.0) | |
| Tetraploidy | |||
| No | 10 (45.5) | 2 (20.0) | .2 |
| Yes | 12 (54.5) | 8 (80.0) | |
| . | EAs, n (%) . | AAs, n (%) . | P value∗ . |
|---|---|---|---|
| 126 (69.6) | 55 (30.4) | ||
| IMWG HRCA classification† | |||
| Standard risk | 104 (82.5) | 46 (86.8) | .7 |
| High risk | 22 (33.3) | 7 (26.9) | |
| t(4;14)(p16;q32) | |||
| No | 94 (91.3) | 42 (95.5) | .5 |
| Yes | 9 (8.7) | 2 (4.5) | |
| t(11;14)(q13;q32) | |||
| No | 24 (82.8) | 6 (60.0) | .2 |
| Yes | 5 (17.2) | 4 (40.0) | |
| t(14;16)(q32;q23) | |||
| No | 44 (95.7) | 16 (94.1) | 1.0 |
| Yes | 2 (4.3) | 1 (5.9) | |
| t(14;20)(q32;q12) | |||
| No | 36 (100.0) | 16 (94.1) | .3 |
| Yes | 0 (0.0) | 1 (5.9) | |
| Loss of IGH | |||
| No | 49 (72.1) | 16 (61.5) | .3 |
| Yes | 19 (27.9) | 10 (38.5) | |
| IGH rearrangement | |||
| No | 61 (74.4) | 20 (66.7) | .5 |
| Yes | 22 (25.6) | 10 (33.3) | |
| Trisomy of chromosome 3 | |||
| No | 56 (74.7) | 25 (75.8) | 1.0 |
| Yes | 19 (25.3) | 8 (24.2) | |
| Trisomy of chromosome 7 | |||
| No | 48 (80.0) | 14 (70.0) | .4 |
| Yes | 12 (20.0) | 6 (30.0) | |
| Trisomy of chromosome 9 | |||
| No | 51 (64.0) | 20 (58.8) | .7 |
| Yes | 30 (37.0) | 14 (41.2) | |
| Chromosome 13q deletion (del 13q) | |||
| No | 88 (73.3) | 35 (67.3) | .5 |
| Yes | 32 (26.7) | 17 (32.7) | |
| Loss of ATM [11q22.3)] (del 11q) | |||
| No | 107 (98.2) | 42 (100.0) | 1.0 |
| Yes | 2 (1.8) | 0 (0.0) | |
| Gain of ATM | |||
| No | 37 (49.3) | 8 (40.0) | .6 |
| Yes | 38 (50.7) | 12 (60.0) | |
| Loss of TP53 [17p13.1] (del 17p) | |||
| No | 105 (86.1) | 44 (91.7) | .4 |
| Yes | 17 (13.9) | 4 (8.3) | |
| Gain of TP53 | |||
| No | 61 (87.1) | 17 (70.8) | .1 |
| Yes | 9 (12.9) | 7 (29.2) | |
| Loss of CDKN2C [1p32.3] (del 1p) | |||
| No | 71 (97.3) | 30 (90.9) | .2 |
| Yes | 2 (2.7) | 3 (9.1) | |
| Gain/amplification of CKS1B [1q21] (gain/amplification of 1q) | |||
| No | 48 (58.5) | 27 (75.0) | .1 |
| Yes | 34 (41.5) | 9 (25.0) | |
| Tetraploidy | |||
| No | 10 (45.5) | 2 (20.0) | .2 |
| Yes | 12 (54.5) | 8 (80.0) | |
P values of the Fisher exact test.
IMWG molecular classification of MM: High-risk MM was defined by the presence of any HRCAs, including t(4;14), t(14;16), t(14;20), del 17p, and gain/amplification of 1q 19. Standard-risk MM was defined by the absence of HRCAs or the presence of any trisomies of odd-numbered chromosomes or t(11;14).
Table 3 presents the age-adjusted associations between primary and secondary cytogenetic abnormalities and OS. Subjects with high-risk MMs, characterized by the presence of HRCAs, had a suggestive trend toward worse OS compared with those with standard-risk MMs (HR, 1.7; 95% CI, 0.9-3.1; P = .1). We found that most cytogenetic abnormalities were not associated with OS in the age-adjusted univariate analysis, except for t(4;14) and gain/amplification of 1q, which were associated with poorer OS. For gain/amplification of 1q, we further delineated the type of abnormalities according to the number of extra copies of chromosome 1q. We found that only gain of 1q (HR, 2.6; 95% CI, 1.2-5.7; n = 26), but not amplification of 1q (HR, 2.0; 95% CI, 0.6-7.1; n = 9), was significantly associated with worse OS (data not shown).
Presence of cytogenetic abnormalities by vital status and age-adjusted univariate HRs between cytogenetic abnormalities and OS
| . | At risk, n (%) . | Death, n (%) . | Age-adjusted HRs∗ (95% CI) . |
|---|---|---|---|
| 116 (64.1) | 65 (35.9) | ||
| IMWG HRCA classification† | |||
| Standard risk | 100 (86.2) | 50 (79.4) | 1.0 (Referent)‡ |
| High risk | 16 (26.2) | 13 (41.9) | 1.7 (0.9-3.1) |
| t(4;14)(p16;q32) | |||
| No | 93 (95.9) | 43 (86.0) | 1.0 (Referent)‡ |
| Yes | 4 (4.1) | 7 (14.0) | 2.5 (1.1-5.5) |
| t(11;14)(q13;q32) | |||
| No | 16 (72.7) | 14 (82.3) | 1.0 (Referent)‡ |
| Yes | 6 (27.3) | 3 (17.6) | 0.7 (0.2-2.3) |
| t(14;16)(q32;q23) | |||
| No | 42 (97.7) | 18 (90.0) | 1.0 (Referent)‡ |
| Yes | 1 (2.3) | 2 (10.0) | 3.8 (0.8-17.1) |
| t(14;20)(q32;q12) | |||
| No | 36 (97.3) | 16 (100.0) | — |
| Yes | 1 (2.7) | 0 (0.0) | — |
| Loss of IGH | |||
| No | 43 (67.2) | 22 (73.3) | 1.0 (Referent)‡ |
| Yes | 21 (32.8) | 8 (26.7) | 0.7 (0.3-1.7) |
| IGH rearrangement | |||
| No | 58 (76.3) | 23 (63.9) | 1.0 (Referent)‡ |
| Yes | 18 (23.7) | 13 (36.1) | 1.6 (0.8-3.1) |
| Trisomy of chromosome 3 | |||
| No | 59 (73.8) | 22 (78.6) | 1.0 (Referent)‡ |
| Yes | 21 (26.2) | 6 (21.4) | 0.8 (0.3-1.9) |
| Trisomy of chromosome 7 | |||
| No | 47 (79.7) | 15 (71.4) | 1.0 (Referent)‡ |
| Yes | 12 (20.3) | 6 (28.6) | 2.0 (0.8-5.2) |
| Trisomy of chromosome 9 | |||
| No | 52 (61.9) | 19 (61.3) | 1.0 (Referent)‡ |
| Yes | 32 (38.1) | 12 (38.7) | 1.1 (0.5-2.2) |
| Chromosome 13q deletion (del 13q) | |||
| No | 84 (75.7) | 39 (63.9) | 1.0 (Referent)‡ |
| Yes | 27 (24.3) | 22 (36.1) | 1.6 (0.9-2.6) |
| Loss of ATM [11q22.3)] (del 11q) | |||
| No | 97 (98.0) | 52 (100.0) | — |
| Yes | 2 (2.0) | 0 (0.0) | — |
| Gain of ATM | |||
| No | 30 (48.4) | 15 (45.5) | 1.0 (Referent)‡ |
| Yes | 32 (51.6) | 18 (54.5) | 0.9 (0.4-1.9) |
| Loss of TP53 [17p13.1] (del 17p) | |||
| No | 95 (86.4) | 54 (90.0) | 1.0 (Referent)‡ |
| Yes | 15 (13.6) | 6 (10.0) | 0.9 (0.4-2.1) |
| Gain of TP53 | |||
| No | 50 (83.3) | 28 (82.3) | 1.0 (Referent)‡ |
| Yes | 10 (16.7) | 6 (17.6) | 1.2 (0.5-3.0) |
| Loss of CDKN2C [1p32.3] (del 1p) | |||
| No | 74 (93.7) | 27 (100.0) | — |
| Yes | 5 (6.3) | 0 (0.0) | — |
| Gain/amplification of CKS1B [1q21] (gain/amplification of 1q) | |||
| No | 60 (69.0) | 15 (48.4) | 1.0 (Referent)‡ |
| Yes | 27 (31.0) | 16 (51.6) | 2.4 (1.2-4.9) |
| Tetraploidy | |||
| No | 7 (33.3) | 5 (45.5) | 1.0 (Referent)‡ |
| Yes | 14 (66.7) | 6 (54.5) | 0.6 (0.2-2.1) |
| . | At risk, n (%) . | Death, n (%) . | Age-adjusted HRs∗ (95% CI) . |
|---|---|---|---|
| 116 (64.1) | 65 (35.9) | ||
| IMWG HRCA classification† | |||
| Standard risk | 100 (86.2) | 50 (79.4) | 1.0 (Referent)‡ |
| High risk | 16 (26.2) | 13 (41.9) | 1.7 (0.9-3.1) |
| t(4;14)(p16;q32) | |||
| No | 93 (95.9) | 43 (86.0) | 1.0 (Referent)‡ |
| Yes | 4 (4.1) | 7 (14.0) | 2.5 (1.1-5.5) |
| t(11;14)(q13;q32) | |||
| No | 16 (72.7) | 14 (82.3) | 1.0 (Referent)‡ |
| Yes | 6 (27.3) | 3 (17.6) | 0.7 (0.2-2.3) |
| t(14;16)(q32;q23) | |||
| No | 42 (97.7) | 18 (90.0) | 1.0 (Referent)‡ |
| Yes | 1 (2.3) | 2 (10.0) | 3.8 (0.8-17.1) |
| t(14;20)(q32;q12) | |||
| No | 36 (97.3) | 16 (100.0) | — |
| Yes | 1 (2.7) | 0 (0.0) | — |
| Loss of IGH | |||
| No | 43 (67.2) | 22 (73.3) | 1.0 (Referent)‡ |
| Yes | 21 (32.8) | 8 (26.7) | 0.7 (0.3-1.7) |
| IGH rearrangement | |||
| No | 58 (76.3) | 23 (63.9) | 1.0 (Referent)‡ |
| Yes | 18 (23.7) | 13 (36.1) | 1.6 (0.8-3.1) |
| Trisomy of chromosome 3 | |||
| No | 59 (73.8) | 22 (78.6) | 1.0 (Referent)‡ |
| Yes | 21 (26.2) | 6 (21.4) | 0.8 (0.3-1.9) |
| Trisomy of chromosome 7 | |||
| No | 47 (79.7) | 15 (71.4) | 1.0 (Referent)‡ |
| Yes | 12 (20.3) | 6 (28.6) | 2.0 (0.8-5.2) |
| Trisomy of chromosome 9 | |||
| No | 52 (61.9) | 19 (61.3) | 1.0 (Referent)‡ |
| Yes | 32 (38.1) | 12 (38.7) | 1.1 (0.5-2.2) |
| Chromosome 13q deletion (del 13q) | |||
| No | 84 (75.7) | 39 (63.9) | 1.0 (Referent)‡ |
| Yes | 27 (24.3) | 22 (36.1) | 1.6 (0.9-2.6) |
| Loss of ATM [11q22.3)] (del 11q) | |||
| No | 97 (98.0) | 52 (100.0) | — |
| Yes | 2 (2.0) | 0 (0.0) | — |
| Gain of ATM | |||
| No | 30 (48.4) | 15 (45.5) | 1.0 (Referent)‡ |
| Yes | 32 (51.6) | 18 (54.5) | 0.9 (0.4-1.9) |
| Loss of TP53 [17p13.1] (del 17p) | |||
| No | 95 (86.4) | 54 (90.0) | 1.0 (Referent)‡ |
| Yes | 15 (13.6) | 6 (10.0) | 0.9 (0.4-2.1) |
| Gain of TP53 | |||
| No | 50 (83.3) | 28 (82.3) | 1.0 (Referent)‡ |
| Yes | 10 (16.7) | 6 (17.6) | 1.2 (0.5-3.0) |
| Loss of CDKN2C [1p32.3] (del 1p) | |||
| No | 74 (93.7) | 27 (100.0) | — |
| Yes | 5 (6.3) | 0 (0.0) | — |
| Gain/amplification of CKS1B [1q21] (gain/amplification of 1q) | |||
| No | 60 (69.0) | 15 (48.4) | 1.0 (Referent)‡ |
| Yes | 27 (31.0) | 16 (51.6) | 2.4 (1.2-4.9) |
| Tetraploidy | |||
| No | 7 (33.3) | 5 (45.5) | 1.0 (Referent)‡ |
| Yes | 14 (66.7) | 6 (54.5) | 0.6 (0.2-2.1) |
HRs were calculated using the Cox proportional hazards regression models adjusting for age (years).
IMWG molecular classification of MM: High-risk MM was defined by the presence of any HRCAs, including t(4;14), t(14;16), t(14;20), del 17p, and gain/amplification of 1q 19. Standard-risk MM was defined by the absence of HRCAs or the presence of any trisomies of odd-numbered chromosomes or t(11;14).
The referent category, indicated by an HR of 1.0, serves as the baseline for comparison. HRs for other categories are relative to this baseline.
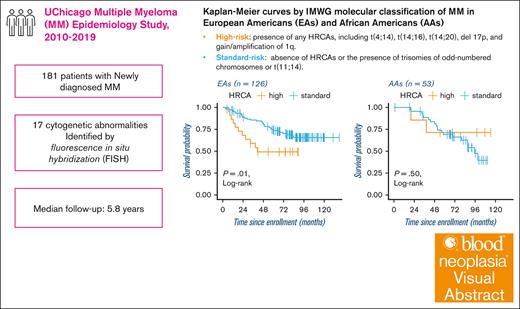
Subsequent population-specific analyses were conducted to examine the associations between cytogenetic aberrations and OS in EAs and AAs, separately (Table 4; Figure 1). Overall, the multivariable analysis adjusting for additional prognostic factors did not materially change the estimates from the age-adjusted univariate analysis. Compared with subjects with standard-risk MM according to the IMWG HRCAs classification, those with high-risk MM were associated with worse OS in EAs only, but not in AAs. For individual cytogenetic abnormalities, t(4;14) was associated with worse OS in AAs, but not in EAs. Furthermore, we observed that gain/amplification of 1q was associated with worse OS in EAs, but not AAs. Additional analyses revealed that gain of 1q was associated with worse OS in EAs (HR, 3.7; 95% CI, 1.2-11.9), but not in AAs, whereas amplification of 1q was not associated with OS in either population (data not shown). However, del 13q was associated with worse OS in both EAs and AAs after adjustment for covariates. Other cytogenetic abnormalities were not associated with OS, regardless of race/ethnicity.
HRs between cytogenetic abnormalities and OS stratified by EAs and AAs with MM
| . | Age-adjusted HR∗ (95% CI) . | Fully adjusted HR† (95% CI) . | ||||||
|---|---|---|---|---|---|---|---|---|
| EA | AA | EA | AA | |||||
| n | n | n | n | |||||
| IMWG HRCAs‡ | 126 | 2.3 (1.1-4.5) | 53 | 0.6 (0.1-2.7) | 120 | 2.6 (1.3-5.5) | 46 | 0.6 (0.1-2.9) |
| t(4;14)(p16;q32) | 103 | 2.1 (0.8-5.4) | 44 | 10.7 (1.9-60.7) | 99 | 1.6 (0.5-4.7) | 38 | 14.5 (2.3-92.3) |
| t(11;14)(q13;q32) | 29 | 0.4 (0.1-3.4) | 10 | 1.6 (0.2-14.0) | 28 | 0.5 (0.0-6.7) | 9 | 1.8 (0.2-14.2) |
| Loss of IGH | 68 | 0.6 (0.2-1.7) | 26 | 1.3 (0.3-5.7) | 65 | 0.6 (0.2-1.9) | 22 | 4.4 (0.7-29.2) |
| IGH rearrangement | 82 | 1.4 (0.6-3.4) | 30 | 1.6 (0.5-5.2) | 80 | 1.4 (0.6-3.3) | 24 | 0.6 (0.1-2.8) |
| Trisomy of chromosome 3 | 75 | 0.5 (0.1-1.7) | 33 | 1.6 (0.4-6.2) | 71 | 0.7 (0.2-2.5) | 29 | 2.1 (0.4-11.9) |
| Trisomy of chromosome 7 | 60 | 1.8 (0.5-6.6) | 20 | 2.0 (0.4-11.0) | 57 | 1.8 (0.5-6.8) | 18 | 3.7 (0.3-46.4) |
| Trisomy of chromosome 9 | 81 | 0.9 (0.4-2.3) | 34 | 1.3 (0.4-4.2) | 77 | 1.4 (0.5-3.5) | 30 | 1.0 (0.2-5.4) |
| Chromosome 13q deletion (del 13p) | 120 | 1.5 (0.8-2.9) | 52 | 2.3 (0.9-6.0) | 114 | 2.0 (1.0-4.2) | 45 | 3.3 (1.0-2.6) |
| Gain of ATM | 75 | 0.8 (0.4-1.8) | 20 | 1.5 (0.3-8.5) | 72 | 0.8 (0.4-1.9) | 15 | — |
| Loss of TP53 [17p13.1] (del 17p) | 122 | 1.0 (0.4-2.5) | 48 | 0.5 (0.1-3.4) | 117 | 1.4 (0.5-3.7) | 42 | 0.5 (0.1-4.0) |
| Gain of TP53 | 70 | 0.6 (0.1-2.7) | 24 | 7.3 (1.5-36.9) | 68 | 0.8 (0.2-3.5) | 22 | — |
| Gain/amplification of CKS1B [1q21](+1q) | 82 | 2.8 (1.1-6.7) | 36 | 1.5 (0.4-5.9) | 77 | 3.4 (1.3-9.3) | 32 | 1.4 (0.3-6.2) |
| Tetraploidy | 22 | 0.4 (0.1-2.4) | 10 | 1.7 (0.2-15.5) | 21 | 0.4 (0.0-3.7) | 7 | — |
| . | Age-adjusted HR∗ (95% CI) . | Fully adjusted HR† (95% CI) . | ||||||
|---|---|---|---|---|---|---|---|---|
| EA | AA | EA | AA | |||||
| n | n | n | n | |||||
| IMWG HRCAs‡ | 126 | 2.3 (1.1-4.5) | 53 | 0.6 (0.1-2.7) | 120 | 2.6 (1.3-5.5) | 46 | 0.6 (0.1-2.9) |
| t(4;14)(p16;q32) | 103 | 2.1 (0.8-5.4) | 44 | 10.7 (1.9-60.7) | 99 | 1.6 (0.5-4.7) | 38 | 14.5 (2.3-92.3) |
| t(11;14)(q13;q32) | 29 | 0.4 (0.1-3.4) | 10 | 1.6 (0.2-14.0) | 28 | 0.5 (0.0-6.7) | 9 | 1.8 (0.2-14.2) |
| Loss of IGH | 68 | 0.6 (0.2-1.7) | 26 | 1.3 (0.3-5.7) | 65 | 0.6 (0.2-1.9) | 22 | 4.4 (0.7-29.2) |
| IGH rearrangement | 82 | 1.4 (0.6-3.4) | 30 | 1.6 (0.5-5.2) | 80 | 1.4 (0.6-3.3) | 24 | 0.6 (0.1-2.8) |
| Trisomy of chromosome 3 | 75 | 0.5 (0.1-1.7) | 33 | 1.6 (0.4-6.2) | 71 | 0.7 (0.2-2.5) | 29 | 2.1 (0.4-11.9) |
| Trisomy of chromosome 7 | 60 | 1.8 (0.5-6.6) | 20 | 2.0 (0.4-11.0) | 57 | 1.8 (0.5-6.8) | 18 | 3.7 (0.3-46.4) |
| Trisomy of chromosome 9 | 81 | 0.9 (0.4-2.3) | 34 | 1.3 (0.4-4.2) | 77 | 1.4 (0.5-3.5) | 30 | 1.0 (0.2-5.4) |
| Chromosome 13q deletion (del 13p) | 120 | 1.5 (0.8-2.9) | 52 | 2.3 (0.9-6.0) | 114 | 2.0 (1.0-4.2) | 45 | 3.3 (1.0-2.6) |
| Gain of ATM | 75 | 0.8 (0.4-1.8) | 20 | 1.5 (0.3-8.5) | 72 | 0.8 (0.4-1.9) | 15 | — |
| Loss of TP53 [17p13.1] (del 17p) | 122 | 1.0 (0.4-2.5) | 48 | 0.5 (0.1-3.4) | 117 | 1.4 (0.5-3.7) | 42 | 0.5 (0.1-4.0) |
| Gain of TP53 | 70 | 0.6 (0.1-2.7) | 24 | 7.3 (1.5-36.9) | 68 | 0.8 (0.2-3.5) | 22 | — |
| Gain/amplification of CKS1B [1q21](+1q) | 82 | 2.8 (1.1-6.7) | 36 | 1.5 (0.4-5.9) | 77 | 3.4 (1.3-9.3) | 32 | 1.4 (0.3-6.2) |
| Tetraploidy | 22 | 0.4 (0.1-2.4) | 10 | 1.7 (0.2-15.5) | 21 | 0.4 (0.0-3.7) | 7 | — |
LDH, elevated lactate dehydrogenase levels.
HRs were calculated using the Cox proportional hazards regression models adjusting for age (years).
In EAs, the Cox model included age (years), sex (female/male), ISS (1/2-3), LDH (normal/elevated). In AAs, the Cox model included age (years), sex (female/male), and (ISS) (1/2-3).
IMWG molecular classification of MM: High-risk MM was defined by the presence of any HRCAs, including t(4;14), t(14;16), t(14;20), del 17p, and gain/amplification of 1q 19. Standard-risk MM was defined by the absence of HRCAs or the presence of any trisomies of odd-numbered chromosomes or t(11;14).
Kaplan-Meier curves by IMWG molecular classification of MM in EAs and AAs. The Kaplan-Meier curves of EAs and AAs are positioned left and right, respectively. HRCA, high or standard, denotes high or standard MMs according to the IMWG molecular classification of MM.
Kaplan-Meier curves by IMWG molecular classification of MM in EAs and AAs. The Kaplan-Meier curves of EAs and AAs are positioned left and right, respectively. HRCA, high or standard, denotes high or standard MMs according to the IMWG molecular classification of MM.
Discussion
In this prospective cohort study of patients newly diagnosed with MM, we found that, despite a similar distribution of HRCAs and individual cytogenetic abnormalities evaluated by interphase FISH between EAs and AAs, high-risk MMs characterized by the presence of any HRCAs were associated with worse OS in EAs, but this may not be true in AAs. We also found that most primary and secondary cytogenetic abnormalities were not associated with OS or population-specific OS, except for del 13q, t(4;14), and gain/amplification of 1q. Specifically, del 13q was associated with worse OS in both EAs and AAs. Nevertheless, gain/amplification of 1q was associated with poorer OS in EAs but not in AAs, whereas t(4;14) was associated with poorer OS in AAs but not in EAs. These population-specific associations were not attenuated with additional adjustment for established clinical prognostic factors or concomitant presence of other HRCAs.
In our patient cohort, individuals with high-risk disease as defined by HRCAs had worse OS, compared with those with standard-risk MMs, aligning with the well-established prognostic value of HRCAs.29 Interestingly, we observed that the positive association between HRCAs and poorer OS was limited to EAs. This population-specific association is consistent with results from a previous study using data from the Multiple Myeloma Research Foundation CoMMpass registry.30 In that study, the authors found a positive association between a greater number of HRCAs and inferior OS in patients of European descent (HR, 1.3; CI, 1.1-1.6; P = .01), but not those of African descent, even after accounting for access to optimal frontline therapies.30 Given the prognostic value of HRCAs, several risk classification schemes have integrated HRCAs to identify high-risk cytogenetic subgroups at diagnosis10,31,32 and guide risk-adapted treatment strategies.33 However, existing HRCA classifications do not adequately consider racial and ethnic variabilities in the influence of HRCAs. Our data revealed that HRCAs might be associated with OS differently between EAs and AAs, highlighting the importance of accounting for racial and ethnic differences in risk classification frameworks in management of MM.
To the best of our knowledge, no study has evaluated the association between cytogenetic abnormalities and OS in MM by race/ethnicity. Our analysis found that although most primary and secondary cytogenetic abnormalities were not associated with OS, the presence of gain/amplification of 1q and t(4;14) had different associations with OS between EAs and AAs. Specifically, the presence of gain of 1q was associated with worse OS in EAs but not in AAs, potentially due to the co-occurrence of other HRCAs. The adverse impact of gain 1q on treatment outcomes of MM may be abrogated by novel drugs when patients do not have other HRCAs.18,34,35 In our patient cohort, none of the AAs with gain of 1q had concurrent presence of t(4;14), t(14;16), t(14;20), or del 17p, whereas 71.4% of EAs with gain of 1q had these additional HRCAs. However, even after adjusting for the presence of these HRCAs, this association in EAs persisted. Furthermore, we found an association between t(4;14) and poorer OS in AAs, but no association in EAs. If confirmed, this difference could be related to variations in MM treatment toxicity, as bortezomib-based treatment can partially alleviate the negative effect of t(4;14).36,37 AAs may be more susceptible to bortezomib-induced peripheral neuropathy, potentially leading to dose reductions or therapy interruptions.38,39
Studies have reported that the simultaneous presence of additional chromosomal abnormalities may modulate the outcomes of patients with high-risk abnormalities.40,41 For example, Hebraud et al showed that the adverse impact of del 13q on OS is linked to the presence of del 17p and t(4;14).41 In our patient populations, the association of del 13q and poor OS in EAs remained irrespective of co-occurrence of del 17p and t(4;14). However, in AAs, this association attenuated after adjusting for t(4;14) (HR, 2.17; 95% CI, 0.47-9.93). We also observed that the association between gain of 1q and worse OS in EAs remained after adjusting for the co-presence of t(4;14), t(14;16), t(14;20), or del 17p. It is possible that EAs and AAs differ in patterns of concomitant presence of multiple HRCAs. Future studies with larger EAs and AAs are needed to disentangle the population-specific impact of the co-occurrence of multiple high-risk abnormalities vs a single abnormality on the prognosis of MM.
In our study, we observed similar distributions of most cytogenetic abnormalities evaluated between AAs and EAs. These findings are consistent with previous studies of cytogenetic changes evaluated by FISH or genomic sequencing.30,42,43 However, it is worth noting that some studies have reported racial differences in the frequency of specific cytogenetic abnormalities between EAs and AAs. For example, Greenberg et al reported a lower prevalence of t(4;14), t(11;14), del(13q), and del(17p) in AAs relative to EAs, based on FISH analysis done on the time of MM diagnosis.20 Furthermore, in a study of patients with MM and MGUS, individuals with a higher percentage of African ancestry calculated by genetic ancestry data were more likely to exhibit the t(14;16), t(14;20), and t(11;14) abnormalities, but not other aberrations such as t(4;14), del(17p), and del(13q).21 A direct comparison across studies is challenging due to the heterogeneity in laboratory practices related to profiling cytogenetic abnormalities.44 This variability encompasses differences in the adoption of CD138 enrichment45 and thresholds for FISH probes,46 both of which can affect the detection and reporting of cytogenetic abnormalities.
Our study has several strengths. First, we used a systematic and prospective enrollment process for patients newly diagnosed with MM with long follow-up. Second, we included a comprehensive panel of 17 cytogenetic abnormalities uniformly evaluated by FISH at a single institution at the onset of MM. Moreover, vital status was determined by linking to the National Death Index, and important clinical prognostic factors were obtained from the EHR, minimizing potential information bias. However, our study also has limitations. Despite having one of the largest representations of AAs with FISH data, this study still lacks sufficient power to detect survival differences in specific cytogenetic abnormalities between AAs and EAs. A sample size of approximately 480 patients per population is needed to achieve 80% power to detect the observed differences in survival rates. Future collaborations across institutions are warranted to validate our novel findings, particularly with the inclusion of sufficient patients with AA with FISH data. Furthermore, similar to other epidemiological studies using EHRs, we were unable to accurately determine treatment modalities, therapeutic responses, and progressive events, which prevented us from accounting for treatment and progression-free survival.
In conclusion, our data showed that the commonly defined HRCAs are associated with worse OS in EAs, but this may not be true for AAs. We also found that, despite similar distributions of most primary and secondary cytogenetic abnormalities between EAs and AAs, gain/amplification of 1q and t(4;14) may have different associations with OS in these populations, which warrants confirmation. These findings suggest that cytogenetic abnormalities may be associated with OS differently in different populations.
Acknowledgments
This work was supported in part by the National Institutes of Health grants R01CA223662, R33 CA269100, R01 CA280637, R56CA282891, and Susan G. Komen TREND21675016.
Authorship
Contribution: B.C.-H.C. designed the study and provided oversight; B.W. conducted the analysis and drafted the manuscript; B.A.D. and A.J. provided clinical advice and helped interpret the results; M.S. collected the cytogenetic data; J.C., D.A., and W.Z. helped interpret the results; and all authors have read and agreed to the published version of the manuscript.
Conflict-of-interest disclosure: B.A.D. consults for Janssen, Sanofi, Canopy, and COTA Healthcare and has received research funding from GlaxoSmithKline and Amgen. A.J. serves on advisory boards with honoraria for AbbVie, Amgen, Bristol Myers Squibb, GlaxoSmithKline, Janssen, and Sanofi. W.Z. consults for Tempus Labs. The remaining authors declare no competing financial interests.
Correspondence: Brian C.-H. Chiu, The University of Chicago, 5841 S. Maryland Ave, MC2000, Chicago, IL 60637; email: bchiu@bsd.uchicago.edu.
References
Author notes
Processed data are available on reasonable request from the corresponding author, Brian C.-H. Chiu (bchiu@bsd.uchicago.edu).