To assess the prognostic relevance of activating mutations of theFLT3 gene in homogeneously treated adults 16 to 60 years of age with acute myeloid leukemia (AML) and normal cytogenetics, pretreatment samples from 224 patients entered into 2 consecutive multicenter treatment trials were analyzed for FLT3internal tandem duplications (ITDs) and Asp835 mutations. Treatment included intensive double-induction therapy and postremission therapy with high cumulative doses of high-dose cytarabine. ITDs were detected in 32% of the patients and were related to de novo AML and to high white blood cell (WBC) counts, percentages of peripheral blood (PB) and bone marrow (BM) blasts, and serum lactate dehydrogenase levels. Asp835 mutations were present in 14% of the patients and were associated with WBC counts and percentages of PB and BM blasts that were higher than those of patients without FLT3mutations. With a median follow-up of 34 months, remission duration and overall survival (OS) were significantly shorter for patients with Asp835 mutations or an ITD than for those without FLT3 mutations (P = .03 and P = .0004, respectively). These results were attributable mainly to the negative prognostic effect of FLT3 ITDs. On multivariate analysis, mutantFLT3 was an independent marker affecting remission duration and OS (hazard ratio, 2.35 and 2.11, respectively). Fluorescence in situ hybridization did not detect monoallelicFLT3 deletions in ITD-positive patients. FLT3mutations identify a subset of young AML patients with normal cytogenetics who do not benefit from intensive chemotherapy, including double-induction and postremission therapy with high-dose cytarabine.
Introduction
Karyotype is widely recognized as one of the most important prognostic factors in acute myeloid leukemia (AML), and cytogenetic data are increasingly used to assign patients to distinct prognostic groups in the context of modern risk-adapted treatment protocols.1-4 By conventional chromosome-banding analysis, approximately 35% to 50% of successfully karyotyped patients lack clonal chromosome aberrations,4 and normal cytogenetics is associated with an intermediate clinical outcome.1,3,5However, several studies indicate that AML patients with normal karyotypes molecularly represent a heterogeneous group and that molecular differences are likely to correlate with prognosis.6-8 Therefore, the identification of new molecular markers and their prospective validation within multicenter treatment trials is critical.
FMS-like tyrosine kinase 3 (FLT3), a member of the class 3 receptor tyrosine kinase (RTK) family, is preferentially expressed on hematopoietic progenitor cells and mediates stem cell differentiation and proliferation.9,10 Interaction of FLT3 with its natural ligand results in activation of the receptor through dimerization and subsequent autophosphorylation of FLT3 proteins,11 followed by induction of multiple intracellular signaling pathways.12 Similarly, ligand-induced FLT3 activation has been shown to enhance the proliferative capacity of AML cells in vitro.13
Like other class 3 RTKs (eg, FMS, KIT, PDGF), FLT3 consists of 5 extracellular immunoglobulinlike domains—a transmembrane domain, a juxtamembrane (JM) domain, 2 intracellular tyrosine kinase (TK) domains separated by a kinase insert domain, and an intracellular C-terminal domain.14 The gene encoding FLT3 maps to chromosome band 13q12 and comprises 24 exons that span a genomic region of approximately 100 kb.15 16
Two types of activating FLT3 mutations have been described in AML. An internal tandem duplication (ITD) of the FLT3gene (FLT3 ITD) can be detected in 20% to 30% of younger adults with AML.17-24 The duplication involves a segment of the JM domain–coding sequence (exons 14 and 15) in direct head-to-tail orientation and is always in-frame.20,21,24,25 In vitro, FLT3 ITDs cause ligand-independent receptor dimerization and constitutive activation of the TK domains, leading to constitutive activation of signaling through the RAS, mitogen-activated protein kinase (MAPK), and signal transducer and activator of transcription 5 (STAT5) pathways. Supporting a potential role in leukemogenesis, FLT3 ITDs from AML patients have been shown to induce autonomous proliferation in cytokine-dependent cell lines.26-29 Point mutations of Asp835 (3-letter amino acid code) within the FLT3 TK domain have recently been described in 7% of adult AML patients.30,31 Like FLT3 ITDs, FLT3Asp835 mutations cause constitutive activation of the receptor and induce autonomous proliferation of 32D cells.31 Of note is that FLT3 Asp835 mutations appear to occur independently ofFLT3 ITDs.
Previous studies have suggested that FLT3 ITD is associated with significantly worse clinical outcomes in younger adults17-19,21 and in pediatric patients25,32with AML. However, studies showing an association of this mutation with poor outcome in adult AML were limited by the fact that patient populations were highly heterogeneous with regard to age at diagnosis, karyotype, and treatment regimens. In particular, none of these studies specifically looked at the subgroup of patients with normal cytogenetics. In a recent study, investigators of the Cancer and Leukemia Group B (CALGB) examined diagnostic bone marrow (BM) samples for FLT3 ITD in 82 adults younger than 60 years of age with de novo AML and normal cytogenetics who had been treated within a single multicenter treatment trial. Within this homogeneous patient population, the mere presence of a FLT3 ITD was not associated with inferior clinical outcome.23 Another important aspect of this study was the identification of 8FLT3 ITD–positive patients who lacked DNA polymerase chain reaction (PCR) evidence of an FLT3 wild-type allele. Multivariate analysis showed that this genotype was the only significant prognostic marker predicting for disease-free survival (DFS) and overall survival (OS). The presence of a FLT3Asp835 mutation did not provide independent prognostic information in the 2 studies published to date.30 31
In this study, we aimed to evaluate the prognostic relevance of activating FLT3 mutations in adult AML patients entered into the German multicenter treatment trials AML HD93 and AML HD98-A of the AML Study Group Ulm (AMLSG ULM). Our primary objective was to determine whether the presence of a FLT3 ITD or a FLT3Asp835 mutation defines a group with poor prognosis within the heterogeneous group of AML patients with normal cytogenetics. Furthermore, we used fluorescence in situ hybridization (FISH) for the detection of monoallelic FLT3 deletions in FLT3ITD–positive patients.
Patients, materials, and methods
Patients
Diagnostic BM or peripheral blood (PB) samples were available from 523 adult patients (aged 16 to 60 years) with AML diagnosed according to French-American-British Cooperative Group criteria33 who had been entered into the German multicenter treatment trials AML HD93 (185 patients; August 1993 to January 1998) and AML HD98-A (338 patients; February 1998 to November 2001). AML HD93 enrolled patients with de novo AML and patients with secondary AML following treatment of a primary malignancy (t-AML). The ongoing AML HD98-A trial also includes patients with refractory anemia with excess blasts in transformation (RAEB-t) and patients with AML following myelodysplastic syndrome (s-AML). Of the 523 patients, 447 (85%) had de novo AML, 37 (7%) had s-AML, 19 (4%) had t-AML, and 20 (4%) had RAEB-t. For this molecular study, the only criterion used to include or exclude patients was the availability of BM or PB samples from diagnosis for mutation analysis of the FLT3 gene. All patients gave informed consent for treatment and cryopreservation of BM and PB. The study was approved by the institutional review boards of the participating AMLSG ULM institutions (see “”). Informed consent was provided according to the Declaration of Helsinki.
Therapy of patients with normal cytogenetics
All patients entered into the AML HD93 and AML HD98-A trials received intensive, response-adapted double-induction and consolidation therapy. Double-induction therapy consisted of a course of ICE (12 mg/m2 idarubicin on days 1 through 3; 100 mg/m2 cytarabine continuously on days 1 through 7; 100 mg/m2 etoposide on days 1 through 3), followed by a second course of ICE started between days 21 and 28 in patients with a response to the first course of induction therapy, or by a course of a HAM-based (3 g/m2 cytarabine every 12 hours on days 1 through 3; 12 mg/m2 mitoxantrone on days 2 and 3) regimen in patients with ICE-refractory disease. First consolidation therapy consisted of a course of HAM.
Second consolidation therapy differed between the 2 trials. In the AML HD93 trial, patients 16 to 54 years of age were assigned to receive a course according to the S-HAM protocol (3 g/m2 cytarabine every 12 hours on days 1, 2, 8, and 9; 10 mg/m2mitoxantrone on days 3, 4, 10, and 11); patients 55 to 60 years of age received the less intensive HAM regimen. In the AML HD98-A trial, patients were randomized to receive either a second course of HAM or myeloablative therapy (total body irradiation/cyclophosphamide or busulfan/cyclophosphamide), followed by autologous stem cell transplantation (SCT). In both trials, patients were assigned to undergo allogeneic SCT if an HLA-compatible donor was available.
Cytogenetic and molecular cytogenetic analysis
Pretreatment samples from all patients were studied centrally by G-banding analysis and FISH. Conventional cytogenetic studies were performed using standard techniques, and chromosomal abnormalities were described according to the International System for Cytogenetic Nomenclature.34 To improve the accuracy of cytogenetic diagnosis, all specimens were also analyzed by FISH using a comprehensive DNA probe set allowing for the detection of the most relevant AML-associated genomic aberrations.35,36 In addition, diagnostic samples from all patients were analyzed for the presence of a partial tandem duplication of the mixed lineage leukemia gene (MLL PTD) by Southern blotting of genomic DNA.7 MLL PTD positivity was confirmed by PCR and sequencing of PCR products.
Analysis of FLT3 ITDs
Genomic DNA was isolated from mononuclear cell preparations stored at −70°C using the DNAzol reagent (Gibco BRL, Eggenstein, Germany) according to the manufacturer's recommendations. Because the location of FLT3 ITDs is restricted to exons 14 and 15,15,18,24,37 PCR amplification of genomic DNA was carried out using primers 11F (5′-GCA ATT TAG GTA TGA AAG CCA GC-3′) and 12R (5′-CTT TCA GCA TTT TGA CGG CAA CC-3′), described previously.18 The total reaction volume of 50 μL contained approximately 500 ng DNA and 10 pmol each primer. Samples were amplified using standard PCR conditions (95°C for 7 minutes; 35 cycles of 95°C for 1 minute, 56°C for 1 minute, 72°C for 2 minutes; 72°C for 10 minutes). PCR products were resolved on a 2% agarose gel stained with ethidium bromide. Repeat analysis was carried out on a 2.5% low-melting–point (LMP) agarose gel if loss of theFLT3 wild-type allele was suspected.
In the 27 patients from the AML HD93 trial with an additional PCR fragment, the amplified product was cloned into the pCR4-TOPO vector (Invitrogen, Groningen, The Netherlands). Ten recombinant colonies were chosen and cultured in Luria-Bertani medium. Plasmid DNA was prepared using the Plasmid Mini Kit (Qiagen, Hilden, Germany). Sequencing was performed using the ABI Ready Reaction Dye Terminator Cycle Sequencing Kit (Applied Biosystems, Weiterstadt, Germany) and the T3 primer on an ABI 310 Prism sequencer (Applied Biosystems).
Analysis of FLT3 Asp835 mutations
Exon 20 of the FLT3 gene was amplified by genomic DNA PCR using primers 20A (5′-CCA GGA ACG TGC TTG TCA-3′) and E20IR (5′-TCA AAA ATG CAC CAC AGT GAG-3′), as previously reported.30Samples were amplified using standard PCR conditions (95°C for 10 minutes; 35 cycles of 95°C for 1 minute, 56°C for 1 minute, 72°C for 1 minute; 72°C for 10 minutes). PCR products were digested withEcoRV and resolved on an agarose gel, as described previously.30 31 Undigested bands, indicative of aFLT3 Asp835 mutation, were cut out for direct nucleotide sequencing using primer 20A to confirm the presence of aFLT3 Asp835 mutation.
Analysis of monoallelic FLT3 deletions by FISH
Sequence analysis using the basic local alignment search tool (BLAST) program (http://www.ncbi.nlm.nih.gov/BLAST/) showed that nucleotides 3 to 2599 of the FLT3 cDNA sequence (GenBank accession no. NM_004119) are contained within bacterial artificial chromosome (BAC) clones RP11-179F17 (GenBank accession no. AL356915) and RP11-9D14 (GenBank accession no. AL445262). These clones were used as probes for the detection of monoallelic deletions of theFLT3 gene in patients with a FLT3 ITD. Dual-color FISH was performed as described.35 36
Criteria for treatment outcomes
Response to induction therapy was assessed at 2 different time points. The first time point was between days 21 and 28 after the first course of induction therapy. Response to the first course of induction therapy was defined as a 50% or greater reduction of BM blasts from pretreatment values. Because the second course of induction therapy was scheduled to start immediately after assessment of the response to the first course of induction therapy, complete hematologic reconstitution was not a requirement. The second time point for assessment of response was after double-induction therapy. In accordance with standard criteria,38 complete remission (CR) required less than 5% BM blasts, an absolute neutrophil count of 1.5 × 109/L or more, a platelet count of 100 × 109/L or more, no blasts in the PB, and no extramedullary leukemia. Therapeutic failure was classified as refractory disease or early/hypoplastic death (death less than 7 days after completion of the first course of induction therapy/death during the remainder of double-induction therapy). Relapse was defined as more than 10% blasts in 2 BM aspirates obtained within 2 weeks or new extramedullary leukemia in patients with previously documented CR. Remission duration end points measured from the date of documented CR were relapse (failure), death in CR (censored), and alive in CR at last follow-up (censored). Overall survival end points measured from the date of study entry were death (failure) and alive at last follow-up (censored).
Statistical analyses
Analyses were based on data available as of November 30, 2001. The median follow-up duration was calculated according to the method of Korn.39 Clinical features were compared across these 3 groups: patients without FLT3 mutation, patients withFLT3 Asp835 mutation, and patients with FLT3 ITD. Binary variables were compared using Fisher exact test. Continuous variables were compared using the Kruskal-Wallis test. For variables reporting significance at P = .05 level, pairwise comparisons were conducted. Pairwise comparisons of binary variables were conducted using Fisher exact test. Pairwise comparisons of continuous variables were conducted using the Wilcoxon rank-sum test. To assess the significance of pairwise comparisons, the α level was adjusted to α = 0.05 ÷ 3 = 0.0167. The Kaplan-Meier method was used to estimate distributions of remission duration and OS.40 Differences between Kaplan-Meier curves were analyzed using the log-rank test.41 The Cox proportional hazards regression model was used to identify differences in remission duration and OS due to prognostic factors.42 As possible prognostic factors, FLT3 mutation status (FLT3Asp835 mutation or FLT3 ITD), a 10-year increment in age, an increment of 50 × 109/L in the diagnostic white blood cell (WBC) count, serum lactate dehydrogenase (LDH) level,MLL PTD status, and response to the first course of induction therapy (for remission duration) were included in the regression model. The variable “study” (AML HD93 or AML HD98-A) was incorporated as a stratification factor to account for study effects on remission duration and OS. Statistical computations were performed using the statistical software packages SAS 6.12 (SAS Institute, Cary, NC) and StatXact (Cytel Software, Cambridge, MA).
Results
Prevalence of FLT3 ITDs in adult AML
The incidence of FLT3 ITDs was determined in all 523 patients with an adequate pretreatment sample: in 27 patients with t(8;21), in 43 patients with inv(16), in 51 patients with t(15;17), in 224 patients with normal karyotypes, in 29 patients with t(11q23), in 45 patients with complex karyotypes (defined as the presence of 3 or more clonal aberrations other than the above balanced translocations), in 78 patients with other chromosome abnormalities, and in 26 patients without evaluable karyotype.
FLT3 ITD was identified in 119 of the 523 (23%) patients. The frequency of FLT3 ITDs differed significantly between cytogenetic groups (P < .0001): of the 224 patients with normal karyotypes, 71 (32%) were FLT3 ITD–positive; of the 51 patients with t(15;17), 20 (39%) exhibited the mutation. In contrast, FLT3 ITDs were rarely observed in patients with t(8;21) (3 of 27 patients; 11%), inv(16) (1 of 43 patients; 2%), or t(11q23) (2 of 29 patients; 7%). Only 1 of the 45 (2%) patients with complex karyotypes was FLT3 ITD– positive. Of the 78 patients with other chromosome abnormalities, 13 (17%) wereFLT3 ITD–positive. Four of these 13 patients had trisomy 8 as the sole cytogenetic abnormality. The remaining 9 patients had the following chromosome aberrations: −Y; t(3;8)(q13.2;q24.1); t(6;9)(p23;q34); der(7)t(7;13)(q32;q12),−13; t(9;15)(q22;q22); add(10)(q26); t(11;15)(p15;q11), t(12;20)(q21;?q12?3); t(11;20)(p15;q11); +20.
There was also a significant difference in the frequency ofFLT3 ITDs between patients with de novo AML (114 of 447 patients; 26%) and secondary AML—that is, s-AML, t-AML, or RAEB-t (7 of 76 patients; 9%; P = .001).
Sequencing analysis of the 27 FLT3 ITD–positive patients with normal cytogenetics from the AML HD93 trial showed FLT3ITDs varying in length from 18 to 122 base pairs (median, 36). Twenty-three patients had simple FLT3 ITDs within exon 14, and 4 patients had FLT3 ITDs involving exon 14 and intron 14. Insertions of 1 to 9 nucleotides (median, 2) of unknown origin between the duplicated regions were observed in 8 patients. As a result, all FLT3 ITDs were readable in-frame.
Prevalence of FLT3 Asp835 mutations in adult AML with normal cytogenetics
The incidence of FLT3 Asp835 mutations was determined in the 224 patients with normal cytogenetics. FLT3 Asp835 mutations were identified in 32 (14%) patients. Sequencing analysis showed 5 kinds of heterozygous FLT3 Asp835 missense mutations: Asp835Tyr (17 patients), Asp835Glu (6 patients), Asp835Val (3 patients), Asp835His (2 patients), and Asp835Ala (1 patient). In 2 patients, sequencing analysis revealed a 1–base pair deletion of Ile836 and a 2–base pair deletion of Met837 (nucleotides 2565 to 2567 of the FLT3 cDNA sequence [GenBank accession no. NM_004119]), resulting in an in-frame deletion of Ile836. One patient had both a 2560G>T transversion, resulting in an Asp835Tyr missense mutation, and a deletion of nucleotides 2565 to 2567. In 1 of the 32 patients with mutations, the presence of an FLT3 Asp835 mutation could not be confirmed because no material was available for sequencing analysis. Four of the 32 (13%) patients withFLT3 Asp835 mutations also had a FLT3ITD.
Analysis of monoallelic FLT3 deletions
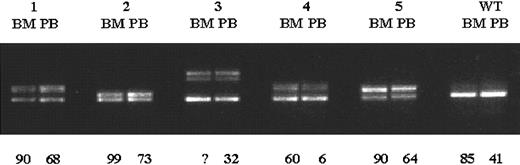
Considerable differences in the intensity of the mutant band were observed in patients with FLT3 ITD (Figure1). Some patients showed strong mutant bands and only faint wild-type bands. On LMP gel electrophoresis, only 1 of the 224 (0.4%) patients with normal cytogenetics displayed a strong mutant band and complete loss of the wild-type allele. Thus, the incidence of loss of the wild-type allele in our study was considerably lower than that found in the CALGB study (8 of 82 patients; 10%).23 The fact that in the latter study only BM samples had been analyzed raised the possibility that the 0.4% value found in our study was an underestimate because the higher proportion of residual normal cells in PB samples would tend to decrease the ratio of the mutant to wild-type FLT3 allele. To investigate whether the frequency of loss of the wild-type allele is related to the material analyzed (BM or PB), we studied diagnostic BM and PB samples from 5 FLT3 ITD–positive patients. Although the percentages of PB blast cells were lower in 4 of 5 patients (68% vs 90%; 73% vs 99%; 32% vs unknown; 6% vs 60%; 64% vs 90%), BM and PB samples were indistinguishable on LMP gel electrophoresis (Figure2).
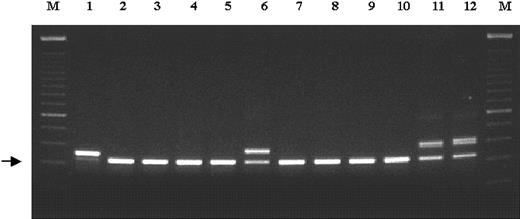
Detection of FLT3 ITD in younger adults with AML.
Exons 14 and 15 of the FLT3 gene were PCR amplified from genomic DNA, and PCR products were resolved on a 2% agarose gel stained with ethidium bromide. Arrow indicates the normalFLT3 gene product (328 bp). High-molecular–weight bands represent FLT3 ITDs. Sample 1 shows a strong mutant band and loss of the wild-type allele. M indicates 100-base pair DNA ladder (Gibco BRL); 1-12, samples from 12 different AML patients.
Detection of FLT3 ITD in younger adults with AML.
Exons 14 and 15 of the FLT3 gene were PCR amplified from genomic DNA, and PCR products were resolved on a 2% agarose gel stained with ethidium bromide. Arrow indicates the normalFLT3 gene product (328 bp). High-molecular–weight bands represent FLT3 ITDs. Sample 1 shows a strong mutant band and loss of the wild-type allele. M indicates 100-base pair DNA ladder (Gibco BRL); 1-12, samples from 12 different AML patients.
Comparison of BM and PB samples from FLT3ITD–positive patients.
Exons 14 and 15 of the FLT3 gene were PCR amplified from diagnostic BM and PB specimens from 5 patients with FLT3ITDs, and PCR products were resolved on a 2.5% LMP agarose gel. Percentages of BM and PB blast cells are shown below the gel. Although the proportion of PB blast cells was considerably lower in 4 of 5 patients, BM and PB samples were indistinguishable on LMP gel electrophoresis. WT indicates samples from a patient withoutFLT3 ITD; 1-5, samples from 5 different AML patients withFLT3 ITDs; and ?, the percentage of BM blasts was unknown in the patient.
Comparison of BM and PB samples from FLT3ITD–positive patients.
Exons 14 and 15 of the FLT3 gene were PCR amplified from diagnostic BM and PB specimens from 5 patients with FLT3ITDs, and PCR products were resolved on a 2.5% LMP agarose gel. Percentages of BM and PB blast cells are shown below the gel. Although the proportion of PB blast cells was considerably lower in 4 of 5 patients, BM and PB samples were indistinguishable on LMP gel electrophoresis. WT indicates samples from a patient withoutFLT3 ITD; 1-5, samples from 5 different AML patients withFLT3 ITDs; and ?, the percentage of BM blasts was unknown in the patient.
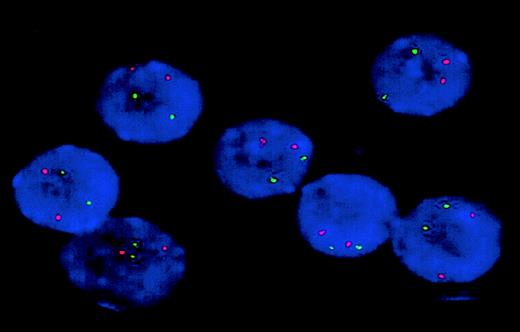
To test the hypothesis that some FLT3 ITDs are associated with monoallelic FLT3 deletions, all 27 FLT3ITD–positive patients with normal cytogenetics from the AML HD93 trial, including the patient with complete loss of the wild-type allele, and 4 additional FLT3 ITD–positive patients with normal cytogenetics from the AML HD98-A trial were studied using FISH with BAC clones RP11-179F17 and RP11-9D14. In all 31 patients, at least 95% of interphase nuclei exhibited 2 strong fluorescence signals with both clones, indicating that no FLT3 allelic deletion had occurred in these patients (Figure 3).
Analysis of monoallelic FLT3 deletions.
Interphase nuclei from the patient with loss of the wild-type allele were studied by FISH using BAC clone RP11-179F17 (green). Cohybridization with a probe recognizing a genomic region at chromosome band 7q35 (red) was performed to monitor hybridization efficiency. All 7 nuclei exhibit 2 green and 2 red fluorescence signals, indicating that no monoallelic FLT3 deletion occurred in this patient. Identical results were obtained with BAC clone RP11-9D14 (not shown). Original magnification × 100.
Analysis of monoallelic FLT3 deletions.
Interphase nuclei from the patient with loss of the wild-type allele were studied by FISH using BAC clone RP11-179F17 (green). Cohybridization with a probe recognizing a genomic region at chromosome band 7q35 (red) was performed to monitor hybridization efficiency. All 7 nuclei exhibit 2 green and 2 red fluorescence signals, indicating that no monoallelic FLT3 deletion occurred in this patient. Identical results were obtained with BAC clone RP11-9D14 (not shown). Original magnification × 100.
Baseline characteristics of patients with normal cytogenetics
Clinical features of the patients with normal cytogenetics were analyzed comparing the following 3 groups (Table1): patients without FLT3mutation, patients with FLT3 Asp835 mutation, and patients with FLT3 ITD. The 4 patients who had both mutations were excluded from this analysis. As noted earlier, comparisons across the 3 groups were followed by relevant pairwise comparisons. Secondary AML was significantly less frequent in patients with FLT3mutations (P = .002 for the comparison across the 3 groups): only 1 of the 28 (4%) patients with a FLT3 Asp835 mutation and only 2 of the 67 (3%) patients with FLT3 ITD had s-AML, t-AML, or RAEB-t. Median WBC counts increased from 12.5 × 109/L in patients without FLT3mutation to 24.8 × 109/L in patients withFLT3 Asp835 mutation and were highest in patients withFLT3 ITD (44.8 × 109/L;P = .0002 for comparison across the 3 groups). Pairwise comparisons showed that the difference between patients withoutFLT3 mutations and patients with FLT3 ITD was statistically significant (P < .0001). In contrast, there were no differences between patients without FLT3 mutations and patients with FLT3 Asp835 mutations (P = .11) or between patients with FLT3 Asp835 mutations and patients with FLT3 ITD (P = .13). Percentages of PB blasts increased from 35% in patients withoutFLT3 mutations to 43% in patients with FLT3Asp835 mutations and were highest in patients with a FLT3ITD (65%; P = .001 for comparison across the 3 groups). Pairwise comparisons showed that the differences between patients without FLT3 mutations and patients with a FLT3ITD and between patients with a FLT3 Asp835 mutation and patients with a FLT3 ITD were statistically significant (P = .0005 and P = .01, respectively). In contrast, there was no difference between patients withoutFLT3 mutations and patients with FLT3 Asp835 mutations (P = .61). Percentages of BM blasts were significantly higher in patients with FLT3 mutations (FLT3 Asp835 mutation, 90%; FLT3 ITD, 90%) than in patients without FLT3 mutations (76%;P = .0002 for comparison across the 3 groups). Serum LDH levels were significantly higher in FLT3 ITD–positive patients (643 U/L) than in patients with FLT3 Asp835 mutations (372 U/L) and in patients without FLT3 mutations (420 U/L; P = .0005 for comparison across the 3 groups). There were no significant differences with respect to other clinical characteristics such as sex, age, hemoglobin level, platelet count, lymphadenopathy, or other extramedullary involvement among the 3 groups. Similar frequencies of MLL PTD-positive patients were found in patients without FLT3 mutations, patients with FLT3 Asp835 mutations, and patients withFLT3 ITD (6%, 8%, and 9%, respectively;P = .64 for comparison across the 3 groups).
Response to double-induction therapy of patients with normal cytogenetics
There was no significant difference in response to double-induction therapy between patients without FLT3mutations, patients with FLT3 Asp835 mutations, and patients with FLT3 ITD (P = .27 for comparison across the 3 groups): 76% of the patients without FLT3 mutations, 82% of the patients with FLT3 Asp835 mutations, and 65% of the FLT3 ITD–positive patients achieved CR.
Remission duration and survival of patients with normal cytogenetics
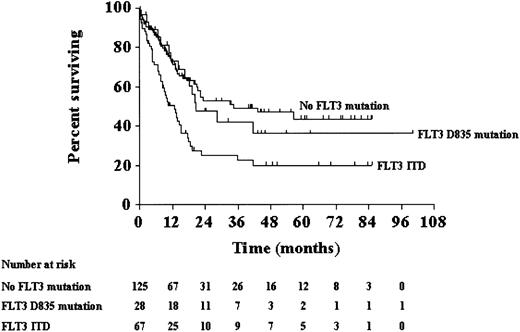
With a median follow-up of 34 months, FLT3 mutation status strongly predicted for remission duration and survival of the patients with normal cytogenetics. Median remission duration was significantly shorter for patients with FLT3 mutations (FLT3 Asp835 mutation, FLT3 ITD, or both) than for those without FLT3 mutations (P = .03 for comparison across the 3 groups; Figure4). Pairwise comparisons showed that the difference in remission duration between patients withoutFLT3 mutations and patients with FLT3 ITD was statistically significant (P = .007). In contrast, there were no differences in remission duration between patients withoutFLT3 mutations and patients with FLT3 Asp835 mutations (P = .18) and between patients withFLT3 Asp835 mutations and patients with FLT3 ITD (P = .42). Median OS was also significantly shorter for patients with FLT3 mutations (FLT3 Asp835 mutation, FLT3 ITD, or both) than for those withoutFLT3 mutations (P = .0004 for comparison across the 3 groups; Figure 5). Pairwise comparisons showed that the difference in OS between patients withoutFLT3 mutations and patients with FLT3 ITD was statistically significant (P = .0001). In contrast, there were no differences in OS between patients without FLT3mutations and patients with FLT3 Asp835 mutations (P = .63) and between patients with FLT3 Asp835 mutations and patients with FLT3 ITD (P = .03).
Remission.
Remission duration for patients with normal cytogenetics according toFLT3 mutation status.
Remission.
Remission duration for patients with normal cytogenetics according toFLT3 mutation status.
Overall survival.
Overall survival for patients with normal cytogenetics according toFLT3 mutation status.
Overall survival.
Overall survival for patients with normal cytogenetics according toFLT3 mutation status.
Cox regression analysis identified response to the first course of induction therapy (hazard ratio, 5.45 [95% confidence interval (CI), 2.26-13.15]), MLL PTD (hazard ratio, 4.47 [95% CI, 2.19-9.11]), and presence of a FLT3 mutation (hazard ratio, 2.35 [95% CI, 1.34-4.12]) as the most significant prognostic factors for remission duration (Table 2). The strongest prognostic markers for OS were presence of a FLT3mutation (hazard ratio, 2.11 [95% CI, 1.39-3.21]), WBC count (hazard ratio, 1.36 [95% CI, 1.16-1.59]), and age (hazard ratio, 1.29 [95% CI, 1.04-1.60]). There was also a significant association between response to the first course of induction therapy and OS (P < .0001).
Discussion
We determined the prognostic significance of activatingFLT3 mutations (ITDs and Asp835 mutations) in a large prospective series of more than 200 young adults with AML and normal cytogenetics. FLT3 ITDs were detected in 32% of the patients and were related to de novo AML and to high WBC counts, high percentages of PB and BM blasts, and elevated serum LDH levels.FLT3 Asp835 mutations were present in 14% of our patients and were also associated with WBC counts and percentages of PB and BM blasts that were higher than those of patients without FLT3mutations. On multivariate analysis, FLT3 mutation status was found to have a major impact on remission duration and OS, which was attributable mainly to the negative prognostic effect ofFLT3 ITDs.
Four previous studies have suggested that the presence ofFLT3 ITD is associated with poor outcomes in younger adults with AML.17-19 21 However, these data were obtained in patient populations that were highly heterogeneous with regard to age at diagnosis, karyotype, and treatment regimens. In contrast, our patients were homogeneous with respect to age (younger adults; 16 to 60 years) and treatment (double-induction therapy, followed by intensive consolidation therapy based on significant cumulative doses of high-dose cytarabine). Furthermore, we specifically looked at the subgroup of patients with normal cytogenetics.
The results from our study allow assessment of the impact ofFLT3 mutations on the outcome of patients receiving consolidation therapy with high-dose cytarabine, which is considered the best conventional treatment option available for patients up to the age of 60 years. The patients reported by Kiyoi et al18were treated according to the AML-87,43AML-89,44 and AML-9245 protocols of the Japan Adult Leukemia Study Group. Within these protocols, the maximum dose of cytarabine per course of postremission therapy was 200 mg/m2 for 5 days as continuous infusion. Furthermore, patients received behenoyl cytarabine instead of cytarabine during postremission therapy or were randomized between these 2 drugs. Patients included in the survival analysis by Rombouts et al21 were treated according to the HOVON-29,46 HOVON-4,47 and HOVON-31 protocols of the Dutch-Belgian Hemato-Oncology Cooperative Group (HOVON), the Swiss Group for Clinical Cancer Research (SAKK), and the AML-11 protocol of the HOVON and EORTC-LCG (European Organisation for the Research and Treatment of Cancer Leukemia Cooperative Group) groups.48 HOVON-31 and AML-11 were designed for elderly patients and did not include high-dose cytarabine. In the HOVON-29 and HOVON-4 trials, patients in CR received further chemotherapy with mitoxantrone and etoposide or underwent autologous or allogeneic SCT. Finally, patients reported by Abu-Duhier et al17 and Kottaridis et al19 had been entered into the recent United Kingdom Medical Research Council (MRC) trials.49 In the MRC AML 10 trial, first consolidation therapy consisted of amsacrine, cytarabine (200 mg/m2 for 5 days as continuous infusion), and etoposide (MACE), followed by a second course of consolidation chemotherapy with mitoxantrone and intermediate-dose cytarabine (5 g/m2). For additional treatment, patients were randomized to allogeneic SCT, autologous SCT, or observation. The current AML 12 trial uses first consolidation therapy with MACE in a similar fashion. Further risk-adapted therapy includes autologous/allogeneic SCT and chemotherapy; the maximum dose of cytarabine administered per cycle is 6 g/m2. In summary, none of these trials used postremission therapy with cytarabine at a dose level of more than 1 g/m2. As a consequence, cumulative doses of cytarabine were significantly lower than those administered within the AML HD93 and AML HD98-A trials (Table3). Intensive consolidation chemotherapy using high-dose cytarabine has been shown to significantly improve DFS and OS of younger adults with AML.50 The effect of cytarabine dose on long-term remission is most marked in patients with t(8;21) or inv(16) and then in patients with normal cytogenetics.1 However, our data suggest that the negative prognostic impact of FLT3 ITDs in patients with normal cytogenetics cannot be overcome by intensive postremission therapy with high-dose cytarabine. These findings contrast the results obtained in a smaller cohort of younger adults with normal cytogenetics who had been entered into a recent treatment trial of the CALGB (CALGB 9621).23 Within this protocol, patients in CR received postremission therapy with cytarabine at a dose level of 2 g/m2, resulting in cumulative doses of cytarabine between 16 and 48 g/m2.51 As noted earlier, CALGB 9621 is the only study published to date in which the mere presence ofFLT3 ITD was not associated with inferior clinical outcome.23
Another way of intensifying antileukemic treatment that has been demonstrated to result in superior DFS of patients with AML is double-induction therapy.52 Of the 9 protocols described above, 4 (HOVON-29, HOVON-4, MRC AML 10, and MRC AML 12) administered 2 courses of induction therapy. In the HOVON trials, the second induction course was scheduled to start within 6 weeks of the first course. In the MRC trials, the second course was administered after a BM sample of adequate cellularity for the assessment of remission status was obtained 18 to 21 days after the end of the first induction course. If the marrow was hypoplastic and assessment of status was not possible, the second course was postponed for another 7 to 10 days. Patients entered into the AML HD93 and AML HD98-A trials received intensive induction treatment consisting of 2 courses of idarubicin and standard-dose cytarabine and etoposide (ICE). Thus, the results from previous studies and our own data indicate that the inferior outcome associated with FLT3 ITDs cannot be overcome by double-induction therapy. Because the timing of induction therapy may be critical, in particular with regard to the effect of prognostic factors on outcome, one may speculate that conventional double-induction approaches do not provide enough dose intensity to overcome the negative prognostic impact of FLT3 ITDs. Data from the French ALFA 9000 trial suggest that the negative prognostic value of FLT3 ITDs can be overcome by reinforced double-induction regimens using mitoxantrone and etoposide on day 22 of a standard 3 + 7 course or on day 8 of a 3 + 3 course.53
In accord with our findings in younger adults, the presence of aFLT3 ITD was an independent risk factor for poor outcome in 91 pediatric patients who had been treated according to a single treatment protocol (Children's Cancer Group-2891).25 This protocol included intensive double-induction and postremission therapy with cytarabine at a dose level of 3 g/m2, resulting in a cumulative cytarabine dose administered during postremission therapy of 12 g/m2.54
Some FLT3 ITD–positive patients lack wild-typeFLT3, and loss of heterozygosity (LOH) for 6 microsatellite markers at chromosome band 13q12 has been demonstrated in such patients.23 Based on these observations, it has been hypothesized that the FLT3 ITD is associated with deletion of the second FLT3 allele. In our study, the frequency of patients with normal cytogenetics with FLT3 ITD who lacked PCR evidence of a FLT3 wild-type allele was 0.4%. This contrasts the data from the CALGB study23 in which the incidence of FLT3 ITD positivity with loss of theFLT3 wild-type allele was 10%. It is possible that this difference is related to the method used to visualize PCR amplification products. In the CALGB study, amplified products were electrophoresed through conventional agarose gels. In our study, repeat analysis on a 2.5% LMP agarose gel revealed a wild-type band in all but one of the patients in whom loss of the FLT3 wild-type allele initially had been suspected. Considering that loss of the wild-type allele has been reported to be associated with significantly inferior clinical outcome in FLT3 ITD–positive patients,23 it appears appropriate to use quantitative approaches to assess the ratio of the mutant to the wild-type FLT3 allele and to determine the prognostic value of this mutation pattern.19 55Agarose gel electrophoresis can definitely not be used to estimate the relative representation of the mutant and wild-type alleles. Even samples with highly different blast percentages—for example, 6% (PB) versus 60% (BM)—were indistinguishable by this technique.
FISH analysis using 2 BAC clones that represent approximately 75% of the FLT3 cDNA sequence failed to identify monoallelicFLT3 deletions in 31 FLT3 ITD–positive patients, including the patient with complete loss of the wild-type allele. Thus, other mechanisms (mitotic nondisjunction, mitotic recombination, gene conversion) resulting in homozygosity rather than hemizygosity56 must be considered to account for the LOH events observed in patients with FLT3 ITD.
Based on the finding that FLT3 Asp835 mutations are gain-of-function mutations that enhance the proliferative capacity of AML cells in vitro to the same extent as FLT3ITDs,31 it has been speculated that FLT3 Asp835 mutations may also affect clinical outcomes of younger adults with AML. In our study, remission duration and OS were shorter for patients with a FLT3 Asp835 mutation than for those without aFLT3 mutation. However, the differences did not reach statistical significance. Considering that FLT3 Asp835 mutations are less frequent than FLT3 ITDs (14% vs 32%), it is possible that larger patient populations must be studied to detect a significant prognostic impact of FLT3 Asp835 mutations.
The discovery of activating FLT3 mutations in AML has important implications for the management of younger adults with this disease. First, assessment of the FLT3 mutation status in patients with normal cytogenetics allows the identification of a subset of patients who do not benefit from intensive chemotherapy, including double-induction and postremission therapy with high-dose cytarabine, and thus may contribute to the stratification of therapy within this heterogeneous subgroup of patients. Second, the high frequency of activating FLT3 mutations may result in their use as molecular targets for monitoring minimal residual disease.57 Finally, the perception that activatingFLT3 mutations play an important role in leukemogenesis has led to the development of biologically targeted therapies with RTK inhibitors that are being studied in phase 1 and 2 trials.58-60
We thank the members of the AML Study Group Ulm (AMLSG ULM) for providing leukemia specimens.
The following AMLSG ULM institutions and investigators participated in this study.
Universitätsklinikum Bonn, Germany, A. Glasmacher; Universitätsklinikum Düsseldorf, Germany, U. Germing; Universitätsklinikum Giessen, Germany, H. Pralle; Universitätsklinikum Göttingen, Germany, D. Haase; Allgemeines Krankenhaus Altona, Hamburg, Germany, H. Salwender; Universitätskliniken des Saarlandes, Homburg, Germany, F. Hartmann; Universitätsklinikum Innsbruck, Austria, G. Gastl; Städtisches Klinikum Karlsruhe, Germany, J. T. Fischer; Universitätsklinikum Kiel, Germany, M. Kneba; Klinikum rechts der Isar der Technischen Universität München, Germany, K. Götze; Städtisches Krankenhaus München-Schwabing, Germany, C. Waterhouse; Städtische Kliniken Oldenburg, Germany, F. del Valle; Caritasklinik St Theresia Saarbrücken, Germany, J. Preiß; Bürgerhospital, Stuttgart, Germany, W. Grimminger; Katharinenhospital Stuttgart, Germany, H. G. Mergenthaler; Krankenhaus der Barmherzigen Brüder, Trier, Germany, W. Weber; and Hanusch-Krankenhaus, Wien, Austria, E. Koller.
Prepublished online as Blood First Edition Paper, August 8, 2002; DOI 10.1182/blood-2002-05-1440.
Supported by grant 01GI9981 from the Bundesministerium für Bildung und Forschung (Kompetenznetz “Akute und chronische Leukämien”).
A complete list of the members of the AML Study Group Ulm appears in “.”
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hartmut Döhner, Department of Internal Medicine III, University Hospital of Ulm Robert-Koch-Str. 8, 89081 Ulm, Germany; e-mail: hartmut.doehner@medizin.uni-ulm.de.