Abstract
Targeting the tyrosine kinase activity ofBCR-ABL represents a very promising therapeutic strategy in chronic myeloid leukemia (CML). Despite strong efficacy of the tyrosine kinase inhibitor STI571, resistance has been observed in a significant proportion of patients in advanced CML stage or in Ph-positive acute lymphoid leukemia (ALL). We investigated in this study the mechanism of resistance to STI571 through point mutations in the tyrosine kinase domain and/or BCR-ABL gene amplification in 24 patients (16 in chronic phase and 8 in accelerated phase of the disease) who obtained no cytogenetic response to STI571 treatment. Screening for the already-described Thr315Ile point mutation in the ABL domain using a reverse transcription polymerase chain reaction restriction fragment length polymorphism (RT-PCR-RFLP) technique, 3 patients showed a proportion of mutated transcript at the time of resistance. The same technique failed to detect mutation at diagnosis, but a specific allele-specific oligonucleotide (ASO)–PCR on DNA for the Thr315Ile mutation and, after sequencing, for 2 newly described Phe311Leu and Met351Thr substitutions, showed the presence of rare mutated cells prior to STI571 therapy. Furthermore, the increased proportion of mutated cells during treatment detected by ASO-PCR strongly suggested clonal selection by the functional inhibiting effect of these mutations. Finally, no BCR-ABL gene amplification was detected by fluorescent in situ hybridization (FISH) in the 24 STI571-resistant patients. Our data support that in CML patients treated with STI571, ABL mutations are not restricted to the accelerated phase of the disease and that, at least in some cases, mutations seem to occur prior to STI571 therapy, probably as second mutational events during the course of CML.
Introduction
Chronic myeloid leukemia (CML) is a hematopoietic stem cell disorder characterized by the reciprocal chromosomal translocation t(9;22)(q34;q11), resulting in a BCR-ABLoncogenic fusion gene on Philadelphia (Ph) 22q- chromosome.1,2BCR-ABL encodes a cytoplasmic fusion protein with constitutive tyrosine kinase activity responsible for transformation and leukemogenic effects.3,4
CML progresses through distinct clinical stages: a chronic phase characterized by expansion of terminally differentiated neutrophils; and an accelerated phase followed by blast crisis with undifferentiated myeloid or lymphoid progenitor cells in maturation arrest. Multiple additional genetic and molecular changes occur at this later disease stage.5
STI571 (Gleevec, Novartis) is a 2-phenylamino pyrimidine derivate that competitively targets the adenosine 5′-triphosphate (ATP)–binding site of the kinase domain of ABL with high specificity6-8 and was recently approved by the Food and Drug Administration9 in the treatment of CML. STI571 induces hematological and cytogenetic remission in phase I and II clinical trial patients in chronic phase as well as in blast crisis10-11CML.
Primary refractoriness or relapse after initial response to STI571 is observed in a significant proportion of patients in advanced CML stage or in Ph-positive acute lymphoid leukemia11 and has been associated with cell-intrinsic changes, including BCR-ABLreactivation of the tyrosine kinase activity by gene amplification12-14 and/or point mutations in the kinase domain of BCR-ABL.9,15,16 Recently, Gorre et al17 reported a Thr315Ile substitution induced by a C to T base change in 9 of 29 acute-phase CML patients showing resistance to STI571. In 2 European reports covering a total of 44 patients also resistant to STI571, no Thr315Ile mutation was detected, but 2 mutations affecting Glu255 amino acid (1 Glu255Lys substitution and 1 Glu255Val substitution, induced by a G to A and A to T base changes, respectively) were described.9,16 Crystallographic studies showed that mutations affecting residues that are in direct contact with ATP or are within the ATP pocket of the kinase domain could have structural effects that influenced inhibition sensitivity.18 The biologic significance of these amino acid changes for STI571 tyrosine kinase inhibiting effect was confirmed by autophosphorylation assays3 or by reconstitution experiments based on Crkl phosphorylation.15
We investigated the mechanism of resistance to STI571 in 24 CML patients who obtained no cytogenetic response to STI571 therapy. Five of them (2 in chronic and 3 in accelerated phase) had mutations in theABL tyrosine kinase domain at the time of resistance that were also demonstrated using sensitive allele-specific oligonucleotide polymerase chain reaction (ASO-PCR) assays, prior to STI571 therapy. No BCR-ABL gene amplification was detected by fluorescent in situ hybridization (FISH) in our study.
Patients, materials, and methods
Patients
Seventy-one CML patients resistant or intolerant to interferon (INF)-α were treated with STI571 in 3 multicenter phase 2 trials (protocols nos. 110, 113, and 114) at our institution between February 2000 and February 2001. After 3 months of therapy, 34 patients showed hematological and cytogenetic response to STI571 (ie, normal blood counts and < 65% of Ph-positive mitoses), whereas 29 showed no cytogenetic response. Twenty-four of them, including 16 in chronic phase and 8 in accelerated phase, were analyzed forABL gene mutation and amplification. No material was available for the 5 remaining patients.
Informed consent was obtained from all patients.
RT-PCR-RFLP assay
RNA extraction.
Total RNA was extracted from frozen aliquots of 107peripheral blood leucocytes with Trizol reagent (Life Technologies, United Kingdom) according to the manufacturer's instructions. RNA pellets were resuspended in 10 μL RNAse-free water, and quantity was estimated by ultraviolet spectrofluorometry.
Reverse transcription.
cDNA was synthesized from 1 μg total RNA in a 20 μL reaction mixture as previously described.19
PCR amplification.
PCR amplification of a 412–base pair (bp) Abl fragment was performed with 2 μL cDNA (corresponding to 100 ng total RNA); 1X TaqGold reaction buffer (Applied Biosystem, Foster City, CA); 1.5 mM MgCl2; 250 μM each dATP, dCTP, dGTP, and dTTP (Pharmacia, Uppsala, Sweden); 0.5U AmpliTaq Gold polymerase (Applied Biosystem); and 50 pmol primer F2: 5′GAG GGC GTG TGG AAG AAA TA 3′ and R2: 5′ GCT GTG TAG GTG TCC CCT GT 3′. Thermocycling conditions used were 12 minutes at 94°C followed by 35 cycles of denaturation at 94°C for 1 minute, annealing at 57°C for 1 minute, extension at 72°C for 1 minute, and a final extension step of 5 minutes at 72°C.
Restriction fragment length polymorphism (RFLP) analysis.
One fifth of PCR product was digested by 5U restriction enzyme Dde I (Roche, Mannheim, Germany) and electrophoresed on 2.5% ethidium-bromide–stained agarose gel.
DNA extraction
Genomic DNA was extracted from 5 × 106 peripheral blood mononuclear cells (PBMCs) using QIAmp DNA minikit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations. Quantity was estimated by ultraviolet spectrofluorometry.
Sequence analysis
The whole kinase and ATP-loop ABL domain (amino acid 242 to 395) was amplified on cDNA in reaction mixture and PCR conditions as described above, using primer F3: 5′CAT CAC CAT GAA GCA CAA GC 3′ and reverse primer R2 at 60°C for annealing.
After purification on QIAquick PCR purification column (Qiagen), 462-bp PCR fragments were sequenced following the ABI protocol for Taq-Dye Terminator Sequencing on an automated ABI377 sequencer. Sequences were analyzed with the Sequence Analysis software V3.3 and the Sequence Navigator software V1.0.1 (Applied Biosystem). Sequencing was performed on both strands.
Detected mutations were always confirmed by sequencing both strands of 207-bp PCR products from DNA. PCR conditions are described above, using forward primer F4: 5′ GTC CTC GTT GTC TTG TTG GC 3′ and reverse primer R4: 5′ CCC CTA CCT GTG GAT GAA GT 3′at 60°C for annealing.
ASO-PCR assays
Mutated or wild-type sequences were specifically amplified in a noncompetitive PCR reaction performed on DNA in 50 μL reaction mixture and PCR conditions as described above, using allele-specific and reverse primers as follows: for the Thr315Ile mutation, F315C: 5′ GCC CCC GTT CTA TAT CAT CAC 3′ or F315T: 5′ CCC GTT CTA TAT CAT CAT 3′and R1: 5′ GGA TGA AGT TTT TCT TCT CCA G 3′ (annealing at 64°C; 158-bp PCR product); for the Phe311Leu mutation, F311T: 5′ CAC CCG GGA GCC CCC GT 3′ or F311C: 5′ CAC CCG GGA GCC CCC GC 3′ and R4 (annealing at 64°C; 174-bp PCR fragment); for the Met351Thr mutation, F351T: 5′CCA CTC AGA TCT CGT CAG CCA T 3′ or F351C: 5′CCA CTC AGA TCT CGT CAG CCA C 3′ and R5: 5′ GCC CTG AGA CCT CCT AGG CT 3′ (annealing at 68 °C; 112-bp PCR fragment).
The sensitivity of this assay was determined for each mutation by amplification of 10-fold limited dilutions of 100 ng patient's DNA at time of resistance in 100 ng healthy control DNA.
FISH analysis
Interphase nuclei were hybridized with fluorescent-labeled probes for BCR-ABL extra signal (ES) (Vysis, Downer Grove, IL) following standard procedure. For each slide, 500 nuclei were analyzed by 2 different observers.
Results
Analysis of Thr315Ile mutation
The recently described STI571-associated resistance mutation Thr315Ile was investigated by studying the loss of Dde I restriction enzyme site induced by C to T base change in our 24 STI571-resistant patients. Analysis was performed after cDNA amplification of a 412-bp PCR fragment at diagnosis and at the time of resistance.
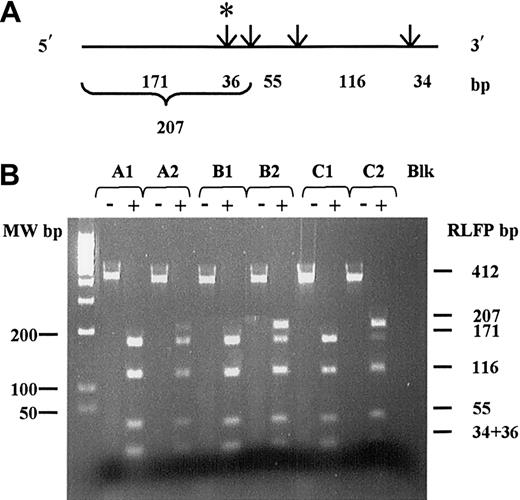
In 3 CML patients with resistance to STI571 (Figure1; patients A, B, and C), the Dde I–restricted pattern showed 2 populations of ABL transcripts, a wild-type sequence characterized by 2 fragments of 171 and 36 bp, respectively, and a mutated sequence characterized by a 207-bp uncut fragment. Differences in band intensities suggested a minor proportion of mutated transcript for patient A and a major proportion of mutated transcript for patients B and C. This RT-PCR-RFLP assay failed to detect Thr315Ile-mutated transcript at diagnosis, as only a 207-bp uncut fragment was detected in those patients.
cDNA restriction map and RT-PCR-RFLP analysis for theAbl Thr315Ile mutation.
The Thr315Ile mutation was investigated by studying the loss of Dde I restriction enzyme site. (A) Dde I restriction map of the F2R2-412 bp cDNA PCR analyzed fragments. Restriction sites are indicated by arrows. The cryptic Dde I site is shown by (*). Length of corresponding fragments obtained after complete digestion are indicated below. (B) Corresponding RFLP pattern on 2.5% ethidium-bromide–stained agarose gel. (-) indicates cDNA PCR fragment prior to Dde I digestion; (+), cDNA PCR fragment after complete Dde I digestion. A1, B1, C1: RFLP pattern obtained from patients A, B, and C, respectively, at diagnosis. A2, B2, C2: RFLP pattern obtained from patients A, B, and C, respectively, at time of resistance. Blk indicates water RT-PCR negative control.
cDNA restriction map and RT-PCR-RFLP analysis for theAbl Thr315Ile mutation.
The Thr315Ile mutation was investigated by studying the loss of Dde I restriction enzyme site. (A) Dde I restriction map of the F2R2-412 bp cDNA PCR analyzed fragments. Restriction sites are indicated by arrows. The cryptic Dde I site is shown by (*). Length of corresponding fragments obtained after complete digestion are indicated below. (B) Corresponding RFLP pattern on 2.5% ethidium-bromide–stained agarose gel. (-) indicates cDNA PCR fragment prior to Dde I digestion; (+), cDNA PCR fragment after complete Dde I digestion. A1, B1, C1: RFLP pattern obtained from patients A, B, and C, respectively, at diagnosis. A2, B2, C2: RFLP pattern obtained from patients A, B, and C, respectively, at time of resistance. Blk indicates water RT-PCR negative control.
Analysis was also performed on 16 patients with complete cytogenetic remission after 3 months of therapy with STI571. None of those patients presented the Thr315Ile mutation (data not shown).
Direct sequence analysis on DNA and RNA
In the 24 STI571-resistant patients, the ABL kinase domain and ATP-loop region were directly sequenced from PCR DNA and cDNA products (a 207-bp F4R4-PCR fragment and a 462-bp F3R2-PCR fragment, respectively) at the time of resistance and prior to STI therapy.
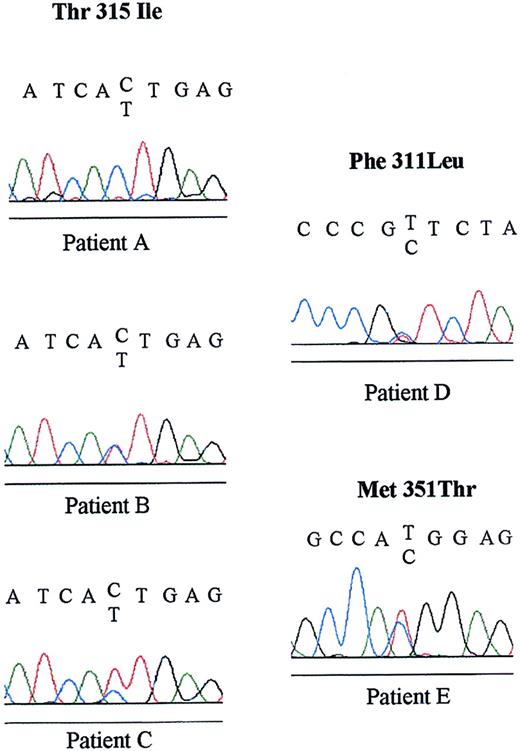
Sequencing data confirmed the Thr315Ile mutation in 2 of the 3 previously RT-PCR-RFLP–detected patients, but failed in the remaining patient (patient A, Figure 2), who presented a lower level of mutated transcript (Figure 1). The heterozygous rate for each patient is presented by comparison of specific C and T signal ranges on chromatographic primary sequence data, according to RT-PCR-RFLP pattern.
Primary sequence data of Thr315Ile, Phe311Leu, and Met351Thr ABL mutations.
Primary sequence data of the 5 mutated patients of our study are presented with the corresponding sequence. Upper reading frame: wild-type sequence. Lower reading frame: mutated sequence. Black indicates guanine (G); blue, cytosine (C); red, thymydine (T); and green, adenosine (A).
Primary sequence data of Thr315Ile, Phe311Leu, and Met351Thr ABL mutations.
Primary sequence data of the 5 mutated patients of our study are presented with the corresponding sequence. Upper reading frame: wild-type sequence. Lower reading frame: mutated sequence. Black indicates guanine (G); blue, cytosine (C); red, thymydine (T); and green, adenosine (A).
Two of the 21 remaining patients showed 2 previously unreported mutations: one patient (patient D) in accelerated phase after 12 months of STI571 therapy had a Phe311Leu substitution induced by a T to C base change, and one patient (patient E) in chronic phase after 18 months of STI571 therapy had a Met351Thr mutation induced by a T to C base change (Figure 2). Clinical and biologic findings of the 5 mutated patients are summarized in Table1.
No mutation affecting the Glu255 amino acid was detected by this direct sequencing method.
ASO-PCR monitoring
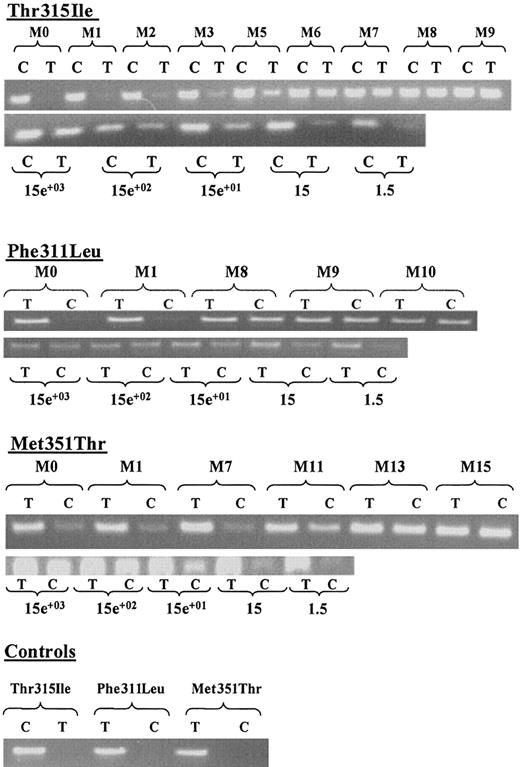
To increase the sensitivity of mutation detection, we developed ASO-PCR strategies. Figure 3 presents monitoring data and corresponding sensitivity for detected Thr315Ile, Phe311Leu, and Met351Thr mutations during STI571 therapy. Results show presence of the 3 mutated sequences on DNA in all patients at M0 prior to STI571 (data from patients A and B are not shown), providing evidence that those point mutations were pre-existent to STI571 treatment. An increased proportion of mutated cells over time is shown by PCR signal intensities on ethidium-bromide–stained agarose gel in the 3 analyzed mutations. This last result strongly suggests clonal selection by functional STI571 resistance of mutated cells during therapy. As specific PCR products of mutated ABL gene were detected even after a 10 000-fold dilution range, we developed a very sensitive ASO-PCR test: assuming that 100 ng DNA represent approximately 15 000 cells, we were able to detect 1.5 ABL-mutated cell in 15 000 wild-type cells. As expected, for each point dilution, signal intensity of nonmutated cells remained constant. The strong specificity of the assay was demonstrated for each mutation by lack detection of ABL sequence from healthy DNA controls.
ASO-PCR monitoring and corresponding sensitivity tests.
For each detected Thr315Ile, Phe311Leu, and Met351Thr mutation, specific ASO-PCR products on ethidium-bromide–stained agarose gel are shown. (C): PCR reaction using Thr315Ile wild-type, Phe311Leu, and Met351Thr mutated primers. (T): PCR reaction using Thr315Ile mutated, Phe311Leu, and Met351Thr wild-type primers. (M): duration of STI571 therapy expressed monthly from diagnosis (M0; ie, before treatment). For sensitivity, corresponding dilution range cell numbers are indicated. Controls: ASO-PCR products on DNA obtained from healthy donor.
ASO-PCR monitoring and corresponding sensitivity tests.
For each detected Thr315Ile, Phe311Leu, and Met351Thr mutation, specific ASO-PCR products on ethidium-bromide–stained agarose gel are shown. (C): PCR reaction using Thr315Ile wild-type, Phe311Leu, and Met351Thr mutated primers. (T): PCR reaction using Thr315Ile mutated, Phe311Leu, and Met351Thr wild-type primers. (M): duration of STI571 therapy expressed monthly from diagnosis (M0; ie, before treatment). For sensitivity, corresponding dilution range cell numbers are indicated. Controls: ASO-PCR products on DNA obtained from healthy donor.
FISH analysis
BCR-ABL gene amplification has been described as a second STI571 mechanism of resistance.12-14 In our study, noBCR-ABL gene amplification was detected by FISH analysis at the time of resistance (data not shown).
Discussion
No BCR-ABL gene amplification has been detected by FISH in any of the 24 CML patients resistant to STI571 studied here.
On the other hand, using 3 different methodological approaches, we found the BCR-ABL Thr315Ile already-described gene mutation15 in 3 of the 24 CML patients, 2 in accelerated phase and one in chronic phase. This incidence of Thr315Ile mutation (ie, 3 of 24 patients, 12.5%) was somewhat lower than that reported by Gorre et al15 17 (ie, 6 of 25 patients, 24%) but higher than that reported in 2 previous European studies (0% of 44 patients).
Direct sequencing failed to detect Thr315Ile mutation in one of our cases. Indeed, this technique can overlook mutations when the proportion of mutated cells is lower than 30% (as would probably be the case for patient A in our study; Figure 1). To maximize sensitivity, sequencing must be done on PCR products amplified with aBCR forward primer (to avoid coamplification of the normalABL allele) or, better, on subcloned PCR products. This latter technical requirement possibly also explains why the Thr315Ile mutation was not found in 2 previous European reports.9 16The lack of a Glu255 detected mutation in our study may also be explained by use of a low sensitivity procedure. Likewise, frequency of the 2 newly described ABL mutations in this report (Phe311Leu substitution in one patient in accelerated phase, and Met351Thr substitution in one patient in chronic phase) may have been underestimated as direct sequencing was performed. Taken together, our data and previous studies suggest that different mutations in the ABL kinase domain may be observed in CML patients resistant to STI571. Our study is also the first report of point mutations in the ABL gene in a chronic-phase CML patient, suggesting that this event is not restricted to patients in the blastic phase of the disease.
Clonal evolution of mutated cells could be monitored over time by ASO-PCR. Our results revealed an increasing proportion of the 3 mutated Thr315Ile, Phe311Leu, and Met351Thr ABL sequences during STI571 therapy. Those results may support a functional effect of newly detected Phe311Leu and Met351Thr mutations on resistance to STI571, as already shown for Thr315Ile substitution.15 Because STI571 interacts with the kinase through hydrogen bonds, any mutation affecting amino acid residues in the active site of ABLcould induce a structural effect, making the drug unable to inhibitABL kinase activity. Nevertheless, further investigations on functional effect of 311 and 351 mutations would be necessary to confirm these points.
Using this very sensitive ASO-PCR technique (sensitivity of 1/10 000), we also demonstrated the presence of rare cells bearing ABL mutations prior to STI571 therapy. This finding shows that, at least in the cases we studied, ABL mutations may be pre-existent to STI571, which only appears to create a clonal selection of the minor population of cells carrying the mutation. Origins of those point mutations of theABL gene are uncertain, but mutations may have been acquired during disease progression through associated genetic instability. Of note is that none of our patients had been exposed to chemotherapy or radiotherapy.
In conclusion, our study shows that among different mechanisms of resistance to STI571, BCR-ABL gene amplification is probably not the most frequent, but ABL mutations affecting binding of STI571 may be more prevalent. Our report seems to confirm that Thr315Ile substitution could be the most frequent mutation in CML.15 20 We report 2 other not previously described mutations, showing that several point mutations can be associated with STI571 resistance and that they are not restricted to the blastic phase of the disease. As we detected resistant mutated cells prior to therapy, at least some ABL mutations do not appear to be induced by STI571 but rather by secondary mutational events during the disease course pre-existent to STI571 treatment. Finally, we suggest the usefulness of detecting already-known STI571-resistant mutations in patient cells prior to therapy or at the early stage of treatment, using strategies such as ASO-PCR, in order to optimize therapeutic decision.
The authors are indebted to Dr Michel Crépin, Sabine Quief, and Institut Federatif de Recherche 22 for expert technical assistance. The authors also thank J. P. Kerckaert for critical reading of the manuscript.
Supported by the Comité Départemental du Nord de la Ligue Nationale contre le Cancer, by the Fondation de France contre le Cancer, by the Programme Hospitalier de Recherche Clinique 1997, and by the CHU de Lille.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Claude Preudhomme, Unité Inserm 524, 1 place de Verdun 59045 Lille Cedex, France; e-mail:cpreudhomme@chru-lille.fr.