Analysis of internal tandem duplications of FLT3(FLT3/ITD) was performed on bone marrow samples obtained at diagnosis and relapse from 108 adult patients with de novo acute myeloid leukemia (AML) to determine the role of this mutation in leukemic relapse. Eighty-three patients had wild-type FLT3at both diagnosis and relapse, 16 had FLT3/ITD at both stages, whereas 8 had acquired the mutation and 1 had lost it at relapse. Using Genescan analysis, we found that FLT3/ITD levels at first relapse were significantly higher than those at diagnosis (mean ± SE, 40.5% ± 4.8% versus 17.9% ± 3.6%,P < .001). The increase in mutation levels at relapse as compared with diagnosis did not correlate with the difference in blast cell percentages at both stages (P = .777). A hemizygous deletion of wild-type FLT3 was found in 4 patients at relapse compared to none at diagnosis. Nine of the 11 patients carrying a single mutation at diagnosis relapsed with an identical mutation. All 6 patients with more than one FLT3/ITD mutation at diagnosis showed changes in mutation patterns and levels at first relapse; however, each patient retained at least one mutation in the relapse sample. The changes of mutation patterns had implications for the monitoring of minimal residual disease. Our results suggest thatFLT3/ITD may contribute as the initial transforming event in AML, and relapse can reflect the selection and outgrowth of a mutant clone or evolution of a new clone harboring this mutation.
Introduction
The fms-like tyrosine kinase 3 (FLT3) gene, located on human chromosome 13q12,1 encodes a class III receptor tyrosine kinase (TK) and plays an important role in hematopoiesis.2 An internal tandem duplication (ITD) in the juxtamembrane (JM) domain of this gene (FLT3/ITD) was found in 20% to 27% of adult acute myeloid leukemia (AML) cases and was associated with a poor prognosis.3-6 Recently, FLT3 was found to be encoded by 24 exons rather than the previously assumed 21 exons.7 Most of the duplicated sequences occurred within exon 14, some also involving intron 14 and the first part of exon 15, with or without insertion.3,5 8
FLT3/ITD has been reported to be associated with leukemic transformation in myelodysplastic syndrome (MDS).9 To date, however, only one study of FLT3/ITD in relapsed AML, which included a small number of patients, has been reported.10 It is of particular interest to know whether the mutation pattern or the level of FLT3/ITD at relapse differs from findings at diagnosis in the same patients. Such information may provide additional insight into the role ofFLT3/ITD in the relapse of AML and may have implications for the monitoring of minimal residual disease (MRD).
In the present study, we analyzed bone marrow (BM) samples collected sequentially at diagnosis and relapse from 108 patients to investigate the role of FLT3/ITD in relapsed AML and to assess the potential usefulness of FLT3/ITD as a marker for detection of MRD. To our knowledge, the present series of adult AML cases is the largest in which FLT3/ITD mutations have been extensively analyzed in paired diagnostic and relapse samples.
Materials and methods
Samples
Beginning in July 1991, BM samples were taken, with informed consent, at diagnosis and during follow-up from 108 consecutively treated adult patients with de novo AML. Each patient had diagnostic and first relapse samples available for analysis, 13 also had second relapse samples, and 2 had third relapse samples. Twenty-five samples from AML patients in complete remission (FLT3/ITD present in 15 and absent in 10) were also studied. All relapse samples contained at least 30% BM blasts. The mononuclear cells from BM samples were enriched by Ficoll-Hypaque (1.077 g/mL, Amersham Pharmacia, Buckinghamshire, England) density gradient centrifugation and cryopreserved in 10% dimethyl sulfoxide and 20% fetal bovine serum at −70°C or in liquid nitrogen. Cytochemical study, immunophenotyping, and cytogenetic analysis were performed at diagnosis. The morphologic subtypes were classified according to French-American-British (FAB) criteria.11 12 All patients were diagnosed and treated at Chang Gung Memorial Hospital, Taipei, Taiwan, where they received induction chemotherapy consisting of daunomycin and cytarabine (3 + 7 regimen). The postremission therapy consisted of high-dose cytarabine plus daunomycin alternating with etoposide for 4 to 6 courses. Since 1994, patients with AML-M3 have received all-trans-retinoic acid in addition to the agents described above. This study was approved by the Human Research Committee of Chang Gung Memorial Hospital.
Cytogenetic analysis and detection of MLLrearrangement and common fusion transcripts
Cytogenetic analysis was performed by a direct method or after short-term unstimulated culture. G-banded metaphases were analyzed. The karyotypes were interpreted according to the International System for Human Cytogenetic Nomenclature.13
All patients, excluding those with t(8;21), inv(16), or t(15;17), were examined for rearrangements of the MLL gene. MLLrearrangement was detected by Southern blot analysis with DNA digested with BamHI and HindIII (New England Biolabs, Beverly, MA) and then hybridized with a 32P-labeled cDNAMLL probe (a kind gift from Dr Behm, St Jude Children's Research Hospital, Memphis, TN) according to the method described by Behm et al.14 Reverse transcription–polymerase chain reaction (RT-PCR) analysis to detect common fusion transcripts ofAML1-ETO, PML-RARα, CBFβ-MYH11, or partial tandem duplication of MLL gene was performed with previously described primers and programs.15-19
Genomic PCR assay for FLT3/ITD
Genomic DNA was extracted from frozen BM cells by use of a DNA extraction kit (Puregene Gentra Systems, Minneapolis, MN) according to the manufacturer's instructions. The PCR reaction was carried out with a 50-μL mixture containing 150 ng gDNA, 200 μM deoxyribonucleotide triphosphate (dNTP), 1 × gold PCR buffer, 1.5 mM MgCl2, 1 U Taq gold polymerase (Applied Biosystems, Foster City, CA), 0.001% gelatin, 50 mM tetramethylammonium chloride, 6% dimethyl sulfoxide, and 30 pmol of the forward primer 5′-CAATTTAGGTATGAAAGCC-3′ and reverse primer 5′-GTACCTTTCAGCATTTTGAC-3′, which completely covered the JM domain through the TK 1 domain,3 on a DNA thermal cycler (ABI 9600, Applied Biosystems), using a program consisting of denaturation at 94°C for 30 seconds, annealing at 59°C for 1 minute, and extension at 72°C for 2 minutes for 35 cycles, with an initial preheating at 95°C for 12 minutes and a final extension at 72°C for 10 minutes. The PCR products were then run on a 3% Nusieve agarose gel (BioWhittaker Molecular Applications, Rockland, ME) or 8% polyacrylamide gel, stained with ethidium bromide, and visualized under a UV lamp. Analysis was repeated on all samples with aberrant band(s).
Detection of FLT3/ITD expression by RT-PCR
Aberrant bands detected by genomic PCR were subjected to RT-PCR. Total RNA was extracted from leukemic cells with Trizol reagent (Gibco, Grand Island, NY) according to the manufacturer's instructions. Then, 2 μg RNA was reverse transcribed with 200 U Moloney murine leukemia virus (MMLV) reverse transcriptase (Life Technologies, Gibco) in 20 μL RT buffer containing 7.5 ng/μL random hexamer primers (Gibco), 125 μM dNTP, 10 μM dithiothreitol (DTT), and RNAsin (20 U; HT Biotechnology, Cambridge, England) at 37°C for 1 hour. cDNA products (1 μL) were amplified with a PCR reaction as described for genomic PCR assay, except that different primer sets were used: R5 (5′-TGTCGAGCAGTACTCTAAACA-3′) and R6 (5′-ATCCTAGTACCTTCCCAAACTC-3′).3
Genescan analysis and determination of FLT3/ITD mutation and transcript levels
The genomic PCR assay was performed again on samples positive for FLT3/ITD, except that the forward primer was labeled at the 5′ end with fluorescein. PCR products (4 μL) were mixed with 5 μL of a solution of formamide (95%) and loading buffer (5% blue dextran, 25 mM EDTA [ethylenediaminetetraacetic acid]) that contained 0.55 μL Rox-1000 (Applied Biosystems). A 1.5-μL sample of this mixture was loaded onto a 5% Long Ranger-6M urea gel (FMC, Rockland, ME; AppliChem, Darmstadt, Germany) with 1 × TBE (Tris 0.089 M, borate 0.089 M, EDTA 0.002 M) running buffer. Electrophoresis was performed at 200 W for 3.5 hours, and then analyzed by an automated ABI PRISM 377 DNA sequencer (Applied Biosystems). The areas under the curves were quantified for FLT3/ITD and the wild-type allele, respectively, by use of Genescan 3.1 software (Applied Biosystems). The level of FLT3/ITD was expressed as a percentage of the area under the curve for FLT3/ITD divided by the sum of the areas under the curves for FLT3/ITD plus wild-typeFLT3. Transcript levels of FLT3/ITD were measured and expressed in the same way as those described for genomic PCR.
Sensitivity of PCR assay
DNA isolated from the leukemic cells of a patient who had a 100% FLT3/ITD level was serially diluted by mixing with the DNA of the HL-60 cell line, which lacks FLT3/ITD, to make 35%, 30%, 25%, 20%, 15%, 10%, 5%, 2%, 1%, and 0.5% positive DNAs, and then analyzed by PCR as described above.
Sequencing of aberrant PCR products
Single abnormal PCR products were directly sequenced in both directions with the BigDye Terminator Cycle Sequencing Ready Reaction kit, which contained AmpliTaq DNA polymerase FS (Applied Biosystems), on an automated DNA sequencing system according to the manufacturer's instructions. Faint aberrant PCR bands or multiple aberrant bands were individually cut from the gel, purified with a MinElute gel extraction kit (Qiagen, Hilden, Germany), cloned into the pCR II-TOPO cloning vector (Invitrogen, Carlsbad, CA), and then sequenced.
Statistical analysis
Frequencies of FLT3/ITD at diagnosis and relapse were compared with McNemar χ2 test. The percentage of leukemic blasts and the level of FLT3/ITD mutations were compared by the nonparametric method with the Wilcoxon signed rank test. The correlation between the percentage of leukemic cells and the level of mutations was analyzed by linear regression.
Results
Frequencies of FLT3/ITD mutation at diagnosis and relapse
The BM samples from 108 adult patients with AML were studied at both diagnosis and relapse. The first relapse occurred at a median of 13 months (range, 3-43 months) following the initial diagnosis.FLT3/ITD was detected in 17 patients at diagnosis compared with 24 at first relapse (P = .298). It was found at both diagnosis and relapse in 16 patients, exclusively at relapse in 8, and exclusively at diagnosis in 1. Eighty-three patients had only wild-typeFLT3 at both diagnosis and relapse. Of the 13 patients with 2 relapses, 3 (nos. 21, 24, and 25) had FLT3/ITD at diagnosis and at both relapses, and 1 (no. 11) who lackedFLT3/ITD at diagnosis acquired it at both relapses. In the remaining 9 patients, the mutation was absent at diagnosis and at both relapses. Neither of the 2 patients with 3 relapses hadFLT3/ITD either at diagnosis or at any of the relapses. Only wild-type FLT3 was detected in samples from the 25 patients in first complete remission.
Characteristics of relapsed AML patients withFLT3/ITD
The frequency of FLT3/ITD according to FAB subtype in relapsed AML was 2 of 5 in M0, 2 of 17 in M1, 15 of 63 in M2, 2 of 6 in M3, 2 of 8 in M4, 2 of 4 in M5, and 0 of 5 in M6. The mutation was found in 14 of the 37 patients who had normal karyotypes and lacked common fusion transcripts or MLLrearrangements, in 2 of the 6 with t(15;17) orPML-RARα, in 1 of the 5 with inv(16) orCBFβ-MYH11, and in 1 with partial tandem duplication of the MLL gene. FLT3/ITD was absent in samples from the 21 patients with t(8;21) or AML1-ETO, the 7 with 11q23 translocations, the 2 with monosomy 7, the 2 with complex anomaly, and the 6 with other abnormalities.
Comparison of mutation patterns and levels found at diagnosis and relapse
The patterns and levels of FLT3/ITD were measured by a quantitative assay based on Genescan analysis and fluorescence detection in 25 patients. Table 1 shows the clinicohematologic features and FLT3/ITD levels in the 17 patients whose samples contained a single mutation. Of the 9 patients (nos. 1-9) with an identical mutation at both diagnosis and relapse, 6 had an increased mutation level at relapse, 1 (no. 7) had a comparable level, and the remaining 2 (nos. 8 and 9) had decreased levels. Patient 10 had a low mutation level (9.8%) at diagnosis with loss of FLT3/ITD at relapse. Seven patients (nos. 11-17) had acquired a single mutation at relapse.
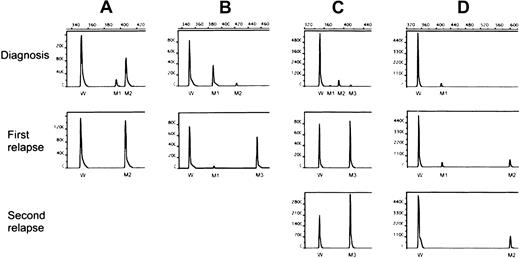
Table 2 shows similar data for the 8 patients harboring more than one FLT3/ITD mutation at diagnosis or first relapse or both. One patient (no. 18) with only wild-type FLT3 at diagnosis had acquired 2 mutations at relapse. Patient 19 with 2 ITD mutations at diagnosis relapsed with the minor mutation but at a higher level. Three (nos. 20-22) of the 5 patients who had 2 mutations at diagnosis lost the minor one while retaining the major mutation at relapse. Patient 23 had 2 mutations at diagnosis, lost the minor one and acquired a new dominant mutation at relapse. Patient 24 had low levels of 3 mutations at diagnosis and lost 2 of them at the first and second relapses. Patient 25 had a single mutation at diagnosis and had acquired an additional mutation at her 2 relapses. The results of Genescan analyses of 4 representative cases (nos. 22-25) are shown in Figure 1. Sequence analysis of PCR products from patients with multiple mutations showed involvement of exon 14 in all 8 patients, intron 14 in 3 (nos. 18, 21 and 25), and exon 15 in 2 (nos. 18 and 25). As demonstrated in Table 2, the multiple mutations, either within a given patient or among different patients, differed from one another with a different initiating end and various duplicated lengths. All altered transcripts were in-frame but were unrelated.
Genescan analysis of FLT3/ITD levels.
The PCR assay was performed with fluorescein-labeled primers and analyzed with an automated DNA sequencer. The area under the curve was calculated for each allele using Genescan software (see “Materials and methods”). M1-M3 indicates FLT3/ITD mutations of different sizes and levels; W, wild-type FLT3. (A) Patient 22 had 2 mutations at diagnosis, with only the dominant mutation (M2) detected at relapse at an increased level.(B) Patient 23 also had 2 mutations at diagnosis, but lost the minor one (M2) while acquiring a new dominant clone (M3) at relapse. (C) Patient 24 had 3 mutations at diagnosis, losing 2 (M1 and M2) and harboring only M3 at a higher level at first relapse, which increased further at second relapse. (D) Patient 25 had a single mutation at diagnosis, acquiring another one (M2) at first relapse, which was the only mutation at second relapse.
Genescan analysis of FLT3/ITD levels.
The PCR assay was performed with fluorescein-labeled primers and analyzed with an automated DNA sequencer. The area under the curve was calculated for each allele using Genescan software (see “Materials and methods”). M1-M3 indicates FLT3/ITD mutations of different sizes and levels; W, wild-type FLT3. (A) Patient 22 had 2 mutations at diagnosis, with only the dominant mutation (M2) detected at relapse at an increased level.(B) Patient 23 also had 2 mutations at diagnosis, but lost the minor one (M2) while acquiring a new dominant clone (M3) at relapse. (C) Patient 24 had 3 mutations at diagnosis, losing 2 (M1 and M2) and harboring only M3 at a higher level at first relapse, which increased further at second relapse. (D) Patient 25 had a single mutation at diagnosis, acquiring another one (M2) at first relapse, which was the only mutation at second relapse.
Among the 25 patients with FLT3/ITD, the levels of mutations measured at diagnosis varied from 0% to 56.5% (mean ± SE, 17.9% ± 3.6%), compared with 0% to 100% (mean ± SE, 40.5% ± 4.8%) at relapse (P < .001). Mutation levels more than 60% were found in 4 relapse samples compared with none in the diagnostic samples. The percentage (mean ± SE) of BM leukemic cells at diagnosis was 74.0% ± 3.7% compared with 70.9% ± 4.1% at relapse (P = .451). Linear regression analysis failed to show a correlation between the levels ofFLT3/ITD and the percentage of leukemic cells, either at diagnosis (P = .492) or at relapse (P = .140). Nor did the difference in mutation levels between diagnostic and relapse samples correlate with the difference in blast percentages at both stages (P = .777).
Sensitivity of the PCR assay for FLT3/ITD detection
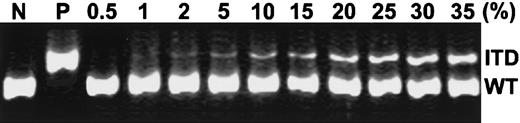
To determine the sensitivity of the PCR assay in detection ofFLT3/ITD in our study, we serially diluted DNA from patient 1 at relapse (mutation level, 100%) with DNA from the HL-60 cell line, which lacks FLT3/ITD, and then analyzed the mixture by PCR assay. As shown in Figure 2, this assay could detect FLT3/ITD levels at 1% to 2% of the positive DNA.
Sensitivity of PCR detection of FLT3/ITD.
gDNA isolated from a patient relapsing with a 100% FLT3/ITD level (patient 1) was diluted by mixing with various amounts of DNA from the HL-60 cell line (FLT3/ITD−) to yield final concentrations of positive DNA at levels of 0.5%, 1%, 2%, 5%, 10%, 15%, 20%, 25%, 30%, and 35%. Undiluted HL-60 DNA served as a negative control (N) and undiluted DNA from patient 1 as a positive control (P). PCR was performed as described in “Materials and methods.” ITD indicates FLT3/ITD allele; WT, wild-typeFLT3 allele.
Sensitivity of PCR detection of FLT3/ITD.
gDNA isolated from a patient relapsing with a 100% FLT3/ITD level (patient 1) was diluted by mixing with various amounts of DNA from the HL-60 cell line (FLT3/ITD−) to yield final concentrations of positive DNA at levels of 0.5%, 1%, 2%, 5%, 10%, 15%, 20%, 25%, 30%, and 35%. Undiluted HL-60 DNA served as a negative control (N) and undiluted DNA from patient 1 as a positive control (P). PCR was performed as described in “Materials and methods.” ITD indicates FLT3/ITD allele; WT, wild-typeFLT3 allele.
RT-PCR for FLT3/ITD expression analysis
The RNAs from the 25 patients carrying FLT3/ITD mutations were subjected to RT-PCR for FLT3/ITD expression analysis. The results for expression of FLT3/ITD mutations showed the same trends as those obtained by DNA PCR (Tables 1 and 2). The FLT3/ITD transcript levels were higher than the mutation levels measured by DNA PCR. Eight patients had transcript levels of more than 60% at relapse.
Discussion
Although FLT3/ITD has been extensively studied in cases of newly diagnosed AML, there has been only one small study of 28 patients on these mutations in relapsed AML.11 We analyzed a considerably larger number of patients and found a higher prevalence of FLT3/ITD at relapse than at diagnosis.FLT3/ITD mutations were present at relapse but were not detected at diagnosis in 8 patients, suggesting that they emerge during the process of relapse in some patients. Alternatively, a smallFLT3/ITD+ subclone below the limits of detection with our assay might have been present at diagnosis and then expanded after chemotherapy, eventually mediating relapse. The results of a dilutional analysis to validate our PCR assay indicate that the putative subclone would have to represent less than 1% to 2% of the DNA sample to escape detection with the PCR method described in this report. Unlike the fusion transcripts generated by chromosomal translocations in acute leukemia, which are almost uniformly present in leukemic cells, the proportion of FLT3/ITD mutations among leukemic blasts in our study ranged widely, from 1% to 100% (median, 35%). These observations are very similar to findings for the N-ras or p53 mutation in AML,20 21suggesting that not all leukemic blasts within a given sample harbor the FLT3/ITD and the evolutional process may vary from patient to patient.
We have also demonstrated a significantly higher mutation level at relapse compared to diagnosis. Because we always enriched for mononuclear cells prior to freezing, the percentages of leukemic cells in relapse and diagnostic samples did not differ appreciably; therefore, the increase in mutation levels at relapse was unlikely to be caused by an increased percentage of leukemic blasts at relapse. An increase in FLT3/ITD levels at relapse suggests that overgrowth of an FLT3/ITD clone was responsible for relapse in a subset of patients. The present results show that the majority of patients harboring FLT3/ITD had a heterozygous mutation with a dominant wild-type FLT3 band in addition to the aberrant bands. Four patients at relapse had FLT3/ITD levels of 60% to 100%, suggesting acquisition of a biallelic mutation or loss of heterozygosity during leukemic relapse. Recently, Whitman et al22 found that 10% of adult patients with AML had a hemizygous deletion of wild-type FLT3 as demonstrated by loss-of-heterozygosity analysis. We used 60% as the cutoff level for hemizygous deletion of one allele, based on data from our previous study at the human androgen receptor gene, in which a PCR assay was used for clonality analysis.23 A single aberrant PCR product without wild-type FLT3 or a PCR product with significantly reduced wild-type FLT3 was also detected in previous studies.5,24,25 Interestingly, transcript levels of FLT3/ITD expression assayed by RT-PCR were even more prominent than the mutation levels assayed by DNA PCR. All but 2 of the positive samples had a higher transcript level at relapse than at diagnosis with 8 samples having a level more than 60%, suggesting suppression of wild-type FLT3 expression. Other investigators have demonstrated that FLT3/ITD causes constitutional activation of the FLT3 receptor tyrosine kinase and induces autonomous cytokine-independent cellular proliferation, leukemic transformation, or outgrowth of AML cases.26-30These observations together with ours indicate thatFLT3/ITD may contribute to the pathogenesis and progression of AML in patients harboring this mutation. The significant increase inFLT3/ITD levels in relapsed AML as observed in our analysis, reinforces the view that an FLT3/ITD subclone present at diagnosis might confer a growth advantage through clonal expansion and become the dominant clone at relapse.
More than one ITD mutation has been detected by other investigators in 23% of AML cases with FLT3/ITD at diagnosis.6However, there are no reports addressing the changes in mutation patterns at relapse. Nine of our 11 patients who had a single ITD mutation at diagnosis relapsed with the identical mutation, although the mutation levels were significantly higher. By contrast, changes in mutation patterns occurred at first relapse in each of the 6 patients who harbored 2 or more mutations at diagnosis; 4 of them lost the minor clone at relapse and 1 patient gained a new clone. We also observed that the expression of minor FLT3/ITD transcripts was frequently suppressed. Among the 4 patients with multiple relapses, comparison of FLT3/ITD mutation patterns between first and second relapse samples showed that 3 patients had identical patterns at both relapses, whereas 1 (no. 25) retained at second relapse the mutation acquired at first relapse but lost the one detected at diagnosis. Although we found that some patients gained or lost mutations in the subsequent relapses, it is noteworthy that none of our patients who exhibited changes of mutation patterns had a totally different mutation in the sequential samples. Thus, the changes of mutation patterns in our patients indicate selection of a previously undetected FLT3/ITD subclone or evolution of a new mutant clone during the process of relapse.
The distribution of FLT3/ITD mutations at relapse did not favor any particular FAB subtype. The frequency of FLT3/ITD was higher in patients with a normal karyotype or t(15;17) than in those with MLL rearrangement or inv(16); none of the patients with t(8;21) had this mutation. None of the 25 patients with an FLT3/ITD mutation had other unfavorable cytogenetic abnormalities. It should be noted in this regard that the distribution of FAB subtypes and cytogenetic or molecular subgroups in our relapsed AML patients with FLT3/ITD was similar to that in previous studies of the newly diagnosed AML.5,6,24 31 Twenty-three patients harboring the FLT3/ITD received reinduction chemotherapy after first relapse, with 10 achieving a second complete remission. However, the small number of patients in this subgroup and their heterogeneous clinical characteristics and treatments preclude any analysis of the prognostic impact of FLT3/ITD at relapse.
The detection of MRD is of growing importance in AML management. As demonstrated here as well as in other studies,6,31 the vast majority of patients with AML with FLT3/ITD lack a previously defined molecular marker of MRD, suggesting that this mutation might serve as a useful target for PCR-based assays; such assays are not without limitations, including the requirement for optimal patient-specific primers,32 and the lack of relevance in cases with a change in the FLT3/ITD mutation pattern from diagnosis to relapse. Also, the very low level of mutations in some cases despite a very high marrow blast percentage may further restrict the use of FLT3/ITD in standard PCR monitoring for MRD. Very recently, Stirewalt et al,33using the quantitative real-time PCR assay, were able to detectFLT3/ITD at levels between 0.01% and 0.001% of positive DNA in 4 patients harboring single mutations of different lengths and locations. The data we report imply that most patients will retain at least one original mutation in their relapse sample, which will have a unique duplicated fragment. Hence, a patient-specific real-time PCR assay using primer sets that cover the junction regions of the different mutations, permitting the mutation but not wild-type alleles to be amplified, would probably overcome the intrinsic technical difficulties.33 For patients with more than oneFLT3/ITD, we suggest that all mutations be monitored during follow-up, although additional study is needed to confirm this strategy. Recently, mutation of Asp835 within the activation loop ofFLT3 was found in 5% to 10% of AML cases.34 35 Use of the Asp835 mutation as a marker of MRD also requires further study.
In conclusion, our study shows that AML patients might acquire or lose FLT3/ITD at relapse and that levels of these mutations are significantly higher at relapse than at diagnosis. Most patients had a heterozygous mutation, whereas a small subset of patients had a hemizygous deletion of the wild-type FLT3 allele. We also demonstrated significant differences in mutation patterns between diagnostic and relapse samples, which have implications for MRD monitoring. Our results suggest that FLT3/ITD mutations could play a dual role in AML pathobiology. They may contribute, first, as the initial transforming event and, second, as a mechanism for drug resistance through clonal evolution or the selection and expansion of a resistant clone.
We thank Mr Ching-Tai Lee for technical assistance, and Ms Yu-Feng Wang for secretarial assistance. The authors are also grateful to Dr Ching-Hon Pui for his advise and encouragement, and to Mr John R. Gilbert for the editorial revision.
Prepublished online as Blood First Edition Paper, May 31, 2002; DOI 10.1182/blood-2002-01-0195.
Supported by grant NSC90-2314-B-182-086 from the National Science Council, Taiwan, grant NHRI-EX90-9011SL from the National Health Research Institutes, Taiwan, and grant CMRP860 from Chang Gung Memorial Hospital, Taiwan.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Lee-Yung Shih, Division of Hematology-Oncology, Department of Internal Medicine, Chang Gung Memorial Hospital, 199 Tung Hwa North Rd, Taipei 105, Taiwan; e-mail: sly7012@adm.cgmh.org.tw.