The clinical progression of chronic myeloid leukemia (CML) from chronic phase to blast crisis is characterized by the increasing failure of myeloid precursors to differentiate into mature granulocytes. This study was undertaken to investigate the influence of Bcr-Abl and of the small molecule Abl tyrosine–kinase inhibitor imatinib mesylate on granulocyte colony-stimulating factor (G-CSF)–induced neutrophilic differentiation. We show that differentiation of 32Dcl3 cells into mature granulocytes is accompanied by the increased expression of the antigens macrophage adhesion molecule–1 (Mac-1) and Gr-1, of the G-CSF receptor (G-CSFR), of myeloid transcription factors (CCAAT/enhancer-binding protein–α [C/EBPα], C/EBPε, and PU.1), and of the cyclin-dependent kinase inhibitor p27Kip1. In 32Dcl3 cells transfected with thebcr-abl gene (32DBcr-Abl), G-CSF did not trigger either granulocytic differentiation or the up-regulation of C/EBPα, C/EBPε, and the G-CSFR. This could be correlated to a defect in c-Myc down-regulation. In contrast, the up-regulation of PU.1 and p27Kip1 by G-CSF was not affected by Bcr-Abl. Importantly, incubation of 32DBcr-Ablwtcells with the kinase inhibitor imatinib mesylate prior to G-CSF stimulation completely neutralized the effects of Bcr-Abl on granulocytic differentiation and on C/EBPα and C/EBPε expression. Taken together, the results suggest that the Bcr-Abl kinase induces a reversible block of the granulocytic differentiation program in myeloid cells by disturbing regulation of hematopoietic transcription factors such as C/EBPα and C/EBPε.
Introduction
Chronic myeloid leukemia (CML) is a clonal malignancy of a hematopoietic stem cell caused in most cases by a reciprocal translocation between chromosomes 9 and 22 (Philadelphia translocation),1 which results in the generation of a fusion protein, Bcr-Abl, with constitutive tyrosine kinase and transforming activity in hematopoietic cells.
At 3 to 5 years after onset, CML often progresses to a fatal blast crisis, characterized by a profound block in differentiation. It is unclear whether this differentiation block is caused by Bcr-Abl or by secondary mutations acquired during disease progression. Indeed, progression from chronic phase to blast crisis has been linked to multiple secondary cytogenetic or molecular alterations. These include trisomy 8, isochromosome i(17q), trisomy 19, and an extra Philadelphia chromosome.2 Among the molecular abnormalities found during blast crisis are alterations in p16INK4A, p53, pRB, Ras, and c-Myc.2 Moreover, fusion genes resulting from reciprocal translocations such as AML/EVI-1 or NUP98/HoxA9 have been reported to be associated with some cases of blast crisis.2 3
Bcr-Abl is the molecular target for imatinib mesylate (STI571), an adenosine triphosphate (ATP)–competitive inhibitor of the Abl tyrosine kinase.4 In several clinical trials, imatinib mesylate was shown to induce hematologic and cytogenetic remissions in up to 98% of patients with chronic phase CML.5,6Surprisingly, therapeutic effects were also seen in patients with myeloid blast crisis or Bcr-Abl+ lymphoid disease, suggesting that even at this late stage and despite the accumulation of numerous secondary and tertiary genetic alterations, the disease was still dependent on Bcr-Abl.7 However, in many cases of late-stage disease, relapses occurred,7 and this could be correlated to the development of direct resistance of Bcr-Abl induced by gene amplifications or point mutations.8
Hematopoietic cell differentiation is regulated by a complex network of growth and differentiation factors.9,10 Granulocyte colony-stimulating factor (G-CSF) and its receptor (G-CSFR) are of pivotal importance for the differentiation of myeloid precursors into mature granulocytes. Mice carrying homozygous deletions of the G-CSF or the G-CSFR genes show reduced levels of morphologically mature neutrophils (about 20% of normal mice levels).11,12 Among the signaling molecules mediating maturation and differentiation of hematopoietic cells induced by G-CSF and other growth factors are transcription factors such as CCAAT/enhancer-binding protein–α (C/EBPα) and C/EBPε, 2 members of the family of CCAAT/enhancer-binding proteins.9,10 C/EBPα-knockout mice display a complete differentiation arrest at the myeloid progenitor level, with no mature neutrophils in the peripheral blood.13 The induced overexpression of C/EBPα in a myeloid cell line was able to induce neutrophilic differentiation without the need for any other signal.14 C/EBPε-knockout mice show impaired granulopoiesis with major functional and morphologic defects of the remaining mature granulocytes. These defects include hyposegmentation of nuclei and a reduced respiratory burst.15 Both C/EBPα and C/EBPε are targets of the activated G-CSF receptor.16 17 The mechanisms leading to C/EBP transcription-factor activation, however, are not fully understood.
Bcr-Abl was recently shown to block G-CSF–induced granulocytic differentiation in a murine hematopoietic progenitor cell line, 32Dcl3.18 We made use of this model (1) to elucidate the effects of Bcr-Abl on the G-CSFR–dependent granulocytic differentiation program and (2) to investigate the reversibility of these effects by imatinib mesylate. We show that blockage of differentiation induced by Bcr-Abl is accompanied by a deficient up-regulation of C/EBPα and C/EBPε, but not of PU.1. Dysregulation of C/EBP transcription factors correlated c-Myc misregulation. All effects on differentiation and C/EBP regulation could be reversed by the addition of the Abl kinase inhibitor imatinib mesylate. Taken together, the results suggest that the block of granulocytic differentiation may occur as a direct interference of the kinase activity of Bcr-Abl with C/EBP transcription-factor regulation.
Materials and methods
Antibodies
Rabbit polyclonal antibodies (abs) to PU.1 (T-21), C/EBPα (14AA), C/EBPε (C-22), c-Jun (H-79), c-Myc (N-262), and G-CSFR (M-20), as well as mouse monoclonal antibodies to p21Waf1/Cip1 (F-5), p27Kip1 (F-8), and phosphotyrosine (PY99), were obtained from Santa Cruz Biotechnology (CA). The monoclonal antibodies to Abl (Ab3) and β-tubulin were purchased from Oncogene Sciences (Uniondale, NJ) and Roche Molecular Biochemical (Mannheim, Germany).
Cell lines and cell culture
L-GM and 32Dcl3 cells were kindly provided by S. Nagata (Osaka, Japan) and U. Just (Munich, Germany). L-GM cells, NFS-60 cells, and 32Dcl3 cells, as well as 32D cells expressing Bcr-Abl and mutants thereof, were grown in Iscove modified Dulbecco medium (IMDM) supplemented with 10% fetal bovine serum (FBS) (Sigma, Deisenhofen, Germany), and 10% WEHI3B-conditioned medium. Jurkat cells were grown in RPMI supplemented with 10% FBS. All media were purchased from Gibco Life Sciences Technologies (Karlsruhe, Germany). For induction of neutrophilic differentiation, cells were counted, washed twice with phosphate-buffered saline (PBS) (Gibco), and seeded at a density of 2 × 105 cells per millliliter into media containing 10% FBS and 10 ng/mL human recombinant (rh) G-CSF (Amgen, Munich, Germany). Fresh media were added at days 2 and 4 to keep cells at a density of around 4 × 105 cells per milliliter throughout culture. For some experiments, imatinib mesylate was added to the cells at a concentration of 1 μM. Morphologic differentiation was assessed by May-Gruenwald/Giemsa staining of cells that had earlier been cytospun onto glass slides. For proliferation assays, cells were washed extensively with cold PBS and seeded into media containing 10% FBS and either 10% WEHI3B-conditioned media or 10 ng/mL human recombinant G-CSF (Amgen). Cell density at day 0 was 3 × 104 cells per milliliter. Imatinib mesylate was added at a concentration of 1 μM where indicated. The number of viable cells was determined daily by trypan blue exclusion.
Plasmids and generation of stably expressing cell lines
Generation of the kinase-inactive mutant of Bcr-Abl was described previously.19 Generation of the imatinib mesylate-resistant mutant Bcr-AblThr315Iso will be described elsewhere. Wild-type (wt) and mutant cDNAs were cloned into pLXSN or pMSCV. Stably expressing cell lines were generated by electroporation. Cells were selected in the presence of 1 mg/mL neomycin (G418). Subclones were generated by limited dilution starting at day 4 after transfection. Two independent clones of each cell type were used for further experiments. The 2 clones had the same phenotype and gave rise to comparable results. Importantly, cells were grown in the presence of interleukin 3 (IL-3) during selection and prior to G-CSF–induced differentiation to avoid secondary alterations due to selection for growth factor independence. New frozen stocks of cells were used every 2 weeks. Generation of retroviral stocks using pMSCV vectors, infection of 32Dcl3 cells, and generation of Bcr-Abl–expressing mass populations are described in the accompanying article by Warmuth et al,20 beginning on page 664.
Cell lysis
For lysis, 32D cells were harvested at the indicated time points after G-CSF stimulation and washed twice in cold PBS. For experiments evaluating the activity profile of imatinib mesylate, cells were incubated with either inhibitor or dimethyl sulfoxide (DMSO) at a density of 5 × 106 cells per milliliter for 1.5 to 2 hours. Then, 107 cells were lysed in 100 μL lysis buffer containing 1% Nonidet P-40 (NP-40), 20 mM Tris (tris(hydroxymethyl)aminomethane) (pH 8.0), 50 mM NaCl, and 10 mM EDTA (ethylenediaminetetraacetic acid), 1 mM phenylmethyl sulfonyl fluoride, 10 μg/mL aprotinin, 10 μg/mL leupeptin, and 2 mM sodium orthovanadate. After resuspension in lysis buffer, cells were incubated for 25 minutes on ice. Unsoluble material was removed by centrifugation at 15 000g. Lysates were checked for protein concentrations by means of a BioRad protein assay (Bio-Rad Laboratories, Muenchen, Germany).
Gel electrophoresis and immunoblotting
Gel electrophoresis and immunoblotting were performed as described previously.19
Flow cytometry
For flow cytometry, cells were washed twice and resuspended in 100 μL PBS containing 2% FBS. Cells were stained with fluorescein isothiocyanate (FITC)–conjugated α–macrophage adhesion molecule–1 (α–Mac-1) (Pharmingen, Heidelberg, Germany) or α–Gr-1 (Caltag, Burlingame, CA) abs for 30 minutes at room temperature. Fluorescence-activated cell sorter (FACS) analysis was done with an EPICS XL 4-color cytometer.
For detection of G-CSFR surface expression, the Fluorokine human G-CSF phycoerythrin conjugate kit from R&D Systems (Minneapolis, MN) was used according to the manufacturer's guidelines. In brief, cells were collected, washed twice, and resuspended in PBS to a final concentration of 4 to 5 × 106 cells per milliliter. Then, 10 μL phycoerythrin (PE)–labeled G-CSF was added to 25 μL washed cell suspension in a 12 × 75 borosilicate tube. As a control, an identical sample of cells was stained with 10 μL PE-conjugated streptavidin. Cells were incubated for 1 hour at 4°C. Thereafter, cells were washed twice with 2 mL 1× rapid dissolution formula–1 (RDF1) buffer and resuspended in 200 μL RDF1 buffer for flow cytometric analysis. To control for specificity of the staining reaction, an aliquot of washed cells was preincubated with a 25-fold molar excess of unconjugated rhG-CSF for 30 minutes at room temperature prior to addition of 10 μL PE-labeled G-CSF.
Detection of apoptosis by flow cytometry
First, 1 × 105 cells per milliliter were incubated with imatinib mesylate at a concentration of 1 μM. Apoptosis was assessed by measuring the binding of FITC-conjugated annexin V to membranes of apoptosing cells. At the indicated time points, aliquots of cells were taken and washed once in PBS. Thereafter, cells were resuspended in 195 μL annexin V binding buffer, and 5 μL annexin V–FITC (Bender MedSystems Diagnostics, Vienna, Austria) was added. After incubation at room temperature for 10 to 20 minutes, cells were washed once and resuspended in 190 μL annexin V binding buffer. Then, 10 μL of a 20 μg/mL propidium iodide stock solution was added, and the ratio of annexin V+ to negative cells was determined by FACS analysis by means of a Coulter EPICS XL 4-color cytometer.
Indirect immunofluorescence
Cells were placed on poly-L-lysine–covered microscope slides for 1 hour in a humidified chamber at 37°C. Then nonadherent cells were washed off with Hanks buffered saline solution, and adherent cells were fixed and immobilized with freshly prepared 2% (wt/vol) paraformaldehyde in PBS for 1 hour at 4°C. Subsequently, cells were permeabilized for 15 minutes with 0.2% (vol/vol) Triton X-100 in PBS, blocked with 2% (wt/vol) glycine in PBS, and incubated with a G-CSFR antibody in PBS for 2 hours at room temperature. Slides were washed with PBS and incubated with FITC-labeled secondary antibody. After the final wash with PBS, slides were mounted on a 9:1 mixture of glycerol and 100 mM Tris/HCl, pH 9.0, containing n-propyl-gallate at 20 mg/mL as antifading reagent. Then samples were examined on a confocal laser scanning apparatus (Leica TCS-NT system; Leica, Bernsheim, Germany) attached to a Leica DM IRB inverted microscope with a PLAPO 63 × 1.32 oil immersion objective.
RNA extraction, cDNA synthesis, and quantitative real-time PCR
For isolation of total RNA and subsequent synthesis of cDNA, the RNeasy Mini kit and the Omniscript Reverse Transcriptase protocol (Qiagen, Hilden, Germany) were used according to the manufacturer's guidelines. Real-time PCR for G-CSFR, C/EBPα, PU.1, and glucose-6 phosphate dehydrgenase (G6PD) was performed by means of light cycler technology (Roche Diagnostics, Mannheim, Germany). Primers for C/EBPα were designed as published previously21: sense, 5′-CCAGCAAGCTGAGGAGCGGCG-3′; antisense, 5′-AACAGCTGAGCCGTGAACTG-3′. For amplification of PU.1, a commercially available light cycler primer set was used (Search LC, Heidelberg, Germany). Primers for G-CSFR and G6PD were as follows: G-CSFR sense, 5′-GCTTGAGCCAACTCCATAGC-3′; G-CSFR antisense, 5′-AAATGCAGGGAAGGACACAG-3′; G6PD sense, 5′-CCGGATCGACCACTACCTGGGCAAG-3′; G6PD antisense, 5′-GTTCCCCACGTACTGGCCCAGGACCA-3′G6PD. All primer sets gave rise to specific DNA fragments of expected sizes. For real-time PCR, 2 μL master mix (Light Cycler FastStart DNA Master SYBR Green I; Roche Diagnostics), 2 μL cDNA, 4 mM MgCl2, 7.5 μM primer, and water to a final concentration of 20 μL were used. For calculation of fold induction or for comparison of DNA levels in different cell types, DNA amounts were normalized to G6PD mRNA expression. For semiquantitative assessment of mRNA expression, real-time PCR reactions were stopped after the indicated number of cycles, and aliquots of the reactions were loaded onto agarose gels. Again, G6PD mRNA expression was used as a control.
Results
The differentiation block of freshly established 32DBcr-Ablwt cells is not due to a loss of G-CSFR expression, but depends on Bcr-Abl kinase activity
To investigate whether Bcr-Abl interferes with differentiation induced by G-CSF in the presence of G-CSFR expression, stable transfectants of 32Dcl3 cells were established expressing either Bcr-Ablwt (32DBcr-Ablwt) or a kinase-inactive mutant of Bcr-Abl (32DBcr-AblK1172R) (Figure1A). In brief, 32Dcl3 cells were transfected by electroporation as described in “Materials and methods.” At 48 hours after transfection, neomycin was added to the media for selection. At 4 days after transfection, cells were plated into 96-well plates by limited dilution to establish individual single cell clones. At 3 weeks after transfection, several subclones were tested for Bcr-Abl expression. Two subclones of each cell type expressing comparable amounts of Bcr-Ablwt or Bcr-AblK1172R were identified (Figure 1A), and aliquots of such cells were cryopreserved to be used for further experiments. Freshly thawed and expanded aliquots were used for each individual experiment. Importantly, 32DBcr-Ablwt cells had been grown in IL-3–containing media continuously during the selection process and prior to analysis of G-CSF–induced differentiation.
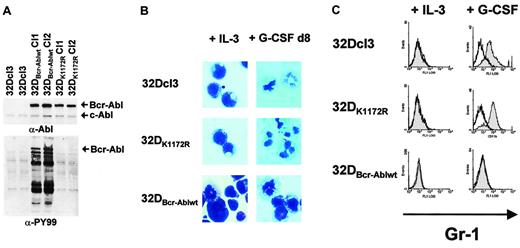
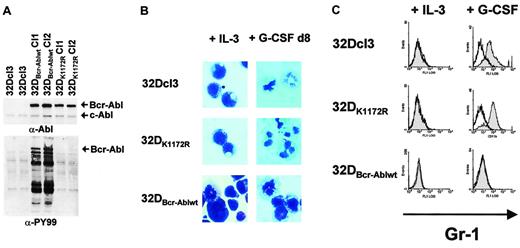
Bcr-Abl inhibition of G-CSF–induced granulocytic differentiation in 32Dcl3 cells by a kinase-dependent mechanism.
(A) Abl (top panel) and phosphotyrosine (bottom panel) immunoblots of lysates from either 32Dcl3, 32DBcr-Ablwt or 32DBcr-AblK1172R cells. (B) Morphological differentiation of 32Dcl3, 32DBcr-Ablwt, or 32DBcr-AblK1172R cells 8 days after stimulation with G-CSF as assessed by May-Gruenwald-Giemsa staining (original magnification, × 400). Cells grown in the presence of IL-3 are shown as a control. (C) Expression of Gr-1 on 32Dcl3, 32DBcr-Ablwt, or 32DBcr-AblK1172R cells upon G-CSF stimulation. Gr-1 expression on cells grown in the presence of IL-3 is shown as a control.
Bcr-Abl inhibition of G-CSF–induced granulocytic differentiation in 32Dcl3 cells by a kinase-dependent mechanism.
(A) Abl (top panel) and phosphotyrosine (bottom panel) immunoblots of lysates from either 32Dcl3, 32DBcr-Ablwt or 32DBcr-AblK1172R cells. (B) Morphological differentiation of 32Dcl3, 32DBcr-Ablwt, or 32DBcr-AblK1172R cells 8 days after stimulation with G-CSF as assessed by May-Gruenwald-Giemsa staining (original magnification, × 400). Cells grown in the presence of IL-3 are shown as a control. (C) Expression of Gr-1 on 32Dcl3, 32DBcr-Ablwt, or 32DBcr-AblK1172R cells upon G-CSF stimulation. Gr-1 expression on cells grown in the presence of IL-3 is shown as a control.
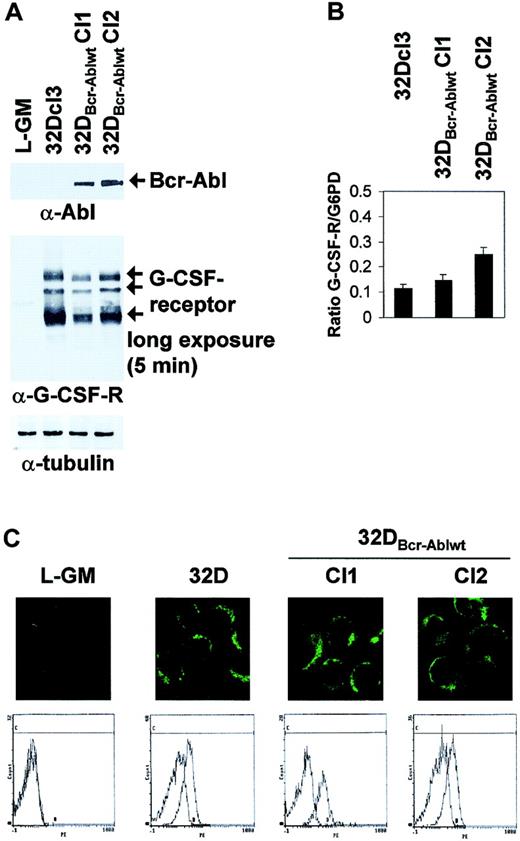
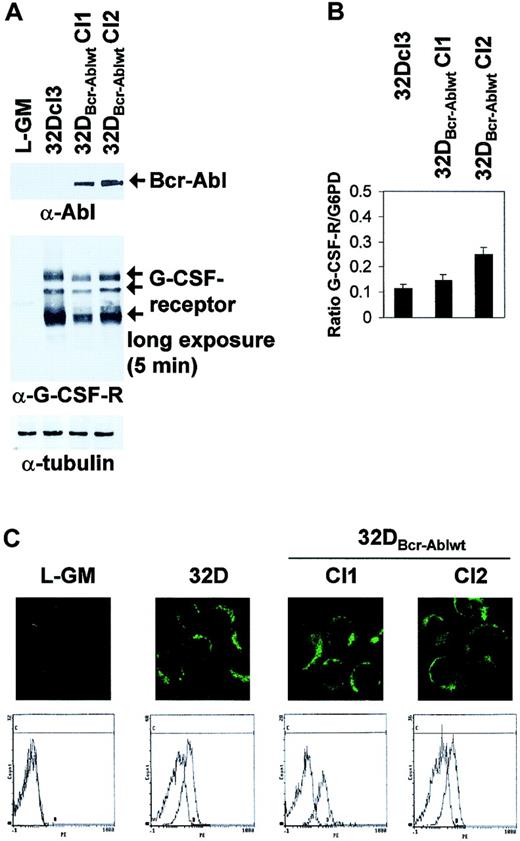
It was previously shown that prolonged culture of Bcr-Abl–transformed 32Dcl3 cells might lead to loss of G-CSF receptor expression.21 However, Western blot analysis and light cycler real-time PCR showed that the 2 clones of 32DBcr-Ablwt cells used for further experiments expressed comparable levels of G-CSFR protein and mRNA (Figure 2A-B). In Western blot analysis, 3 major G-CSFR species of 85 to 90 kDa, 105 to 110 kDa, and 130 to 135 kDa, possibly representing different glycosylation states, were seen (Figure2A, arrows). Moreover, 32Dcl3 cells and 32DBcr-Ablwt cells expressed G-CSFR at comparable levels at the cell surface, as revealed by confocal microscopy (Figure 2C top panels) and FACS analysis (Figure 2C bottom panels). No G-CSFR expression was seen in L-GM cells used as a negative control. FACS controls were performed for every freshly thawed aliquot of 32DBcr-Ablwt cells prior to induction of G-CSF–induced differentiation to confirm appropriate G-CSFR expression.
Effect of Bcr-Abl expression on G-CSF receptor expression in 32DBcr-Ablwt cells.
Expression of Bcr-Abl does not diminish G-CSF receptor expression in 32DBcr-Ablwt cells. The 32Dcl3 or 32DBcr-Ablwt cells grown in the presence of IL-3 were subjected to Western blot (A), real-time PCR (B), confocal microscopy (C, top row), or FACS analysis (C, bottom row) as described in “Materials and methods.” The myeloid, G-CSFR–deficient cell line L-GM was used as a negative control where indicated. Equal expression of G-CSFR in the presence or absence of Bcr-Abl expression was seen in 2 independent subclones of 32DBcr-Ablwt.
Effect of Bcr-Abl expression on G-CSF receptor expression in 32DBcr-Ablwt cells.
Expression of Bcr-Abl does not diminish G-CSF receptor expression in 32DBcr-Ablwt cells. The 32Dcl3 or 32DBcr-Ablwt cells grown in the presence of IL-3 were subjected to Western blot (A), real-time PCR (B), confocal microscopy (C, top row), or FACS analysis (C, bottom row) as described in “Materials and methods.” The myeloid, G-CSFR–deficient cell line L-GM was used as a negative control where indicated. Equal expression of G-CSFR in the presence or absence of Bcr-Abl expression was seen in 2 independent subclones of 32DBcr-Ablwt.
Despite appropriate G-CSFR expression, 32DBcr-Ablwt cells, but not 32DBcr-AblK1172R cells, were defective in G-CSF–induced differentiation as assessed by monitoring morphologic differentiation (Figure 1B) as well as up-regulation of myeloid cell surface markers such as Mac-1 or Gr-1 (Figure 1C and data not shown).
Imatinib mesylate restores G-CSF–induced granulocytic differentiation and C/EBP transcription-factor regulation in 32DBcr-Ablwt cells
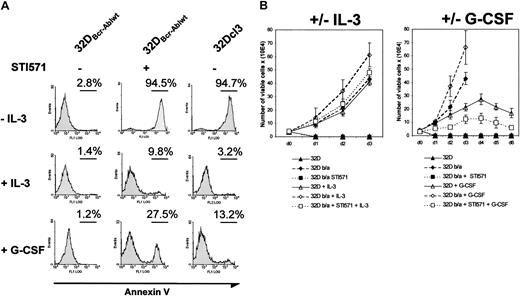
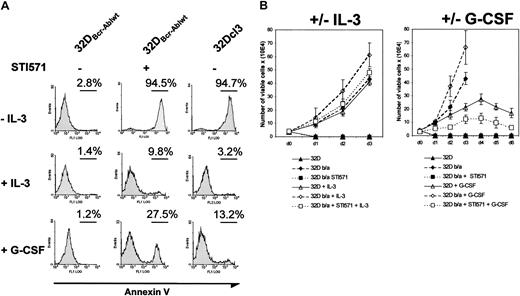
These results suggested that Abl kinase activity was essential for the inhibition of granulocytic differentiation. To further address this issue, we investigated the influence of the Abl kinase inhibitor imatinib mesylate on the differentiation of 32Dcl3 and 32DBcr-Ablwt cells in response to G-CSF. The 32DBcr-Ablwt cells were cultured in the absence of IL-3. These cell clones were readily IL-3 independent for growth and survival on IL-3 withdrawal without any further selection. Incubation of 32DBcr-Ablwt cells with 1 μM imatinib mesylate in the absence of growth factors rapidly decreased cell growth and survival (Figure 3A). Incubation of cells with IL-3 completely rescued 32DBcr-Ablwt cells from imatinib mesylate-induced apoptosis (Figure 3A), and G-CSF decreased the number of apoptotic 32DBcr-Ablwt cells upon imatinib mesylate treatment from 94.5% to 27.5%, as assessed by annexin V staining after 24 hours (Figure 3A). Moreover, proliferation of imatinib mesylate-treated 32DBcr-Ablwt cells in response to IL-3 or G-CSF was similar to the cell growth seen in parental 32Dcl3 cells (Figure 3B). These results demonstrated that 32DBcr-Ablwtcells expressed functional IL-3 and—above all—G-CSF receptors on their surface and regained growth factor responsiveness after inhibition of Bcr-Abl by imatinib mesylate.
Effect of imatinib mesylate on responsiveness of 32DBcr-Ablwt cells to IL-3 and G-CSF stimulation.
Imatinib mesylate restores responsiveness of 32DBcr-Ablwtcells to IL-3 and G-CSF stimulation. (A) The 32Dcl3 and 32DBcr-Ablwt cells were grown in the presence or absence of IL-3, G-CSF, and imatinib mesylate as indicated. The amount of apoptosis in the individual cultures after 48 hours was determined by means of FACS analysis. Both IL-3 and G-CSF led to a rescue of apoptosis induced by 1 μM imatinib mesylate in 32DBcr-Ablwt cells in the absence of cytokines, suggesting that 32DBcr-Ablwt cells express functional IL-3 and G-CSF receptors. (B) Proliferation of 32Dcl3 and 32DBcr-Ablwtcells grown in the presence or absence of IL-3, G-CSF, and imatinib mesylate as indicated. Cell counts were determined by trypan blue exclusion. Imatinib mesylate restored the responsiveness of 32DBcr-Ablwt cells to both IL-3 and G-CSF. Cell counts of 32DBcr-Ablwt cells grown in the presence of 1 μM imatinib mesylate and G-CSF were slightly retarded when compared with the proliferation of 32Dcl3 cells grown in the presence of G-CSF.
Effect of imatinib mesylate on responsiveness of 32DBcr-Ablwt cells to IL-3 and G-CSF stimulation.
Imatinib mesylate restores responsiveness of 32DBcr-Ablwtcells to IL-3 and G-CSF stimulation. (A) The 32Dcl3 and 32DBcr-Ablwt cells were grown in the presence or absence of IL-3, G-CSF, and imatinib mesylate as indicated. The amount of apoptosis in the individual cultures after 48 hours was determined by means of FACS analysis. Both IL-3 and G-CSF led to a rescue of apoptosis induced by 1 μM imatinib mesylate in 32DBcr-Ablwt cells in the absence of cytokines, suggesting that 32DBcr-Ablwt cells express functional IL-3 and G-CSF receptors. (B) Proliferation of 32Dcl3 and 32DBcr-Ablwtcells grown in the presence or absence of IL-3, G-CSF, and imatinib mesylate as indicated. Cell counts were determined by trypan blue exclusion. Imatinib mesylate restored the responsiveness of 32DBcr-Ablwt cells to both IL-3 and G-CSF. Cell counts of 32DBcr-Ablwt cells grown in the presence of 1 μM imatinib mesylate and G-CSF were slightly retarded when compared with the proliferation of 32Dcl3 cells grown in the presence of G-CSF.
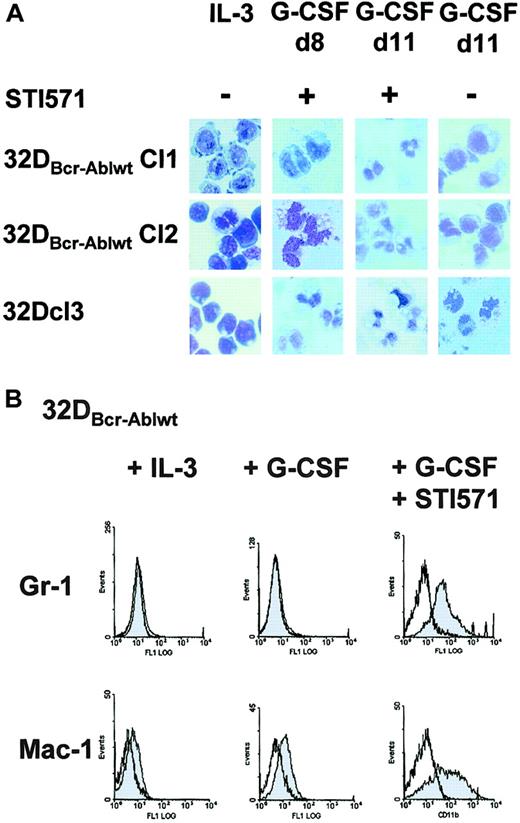
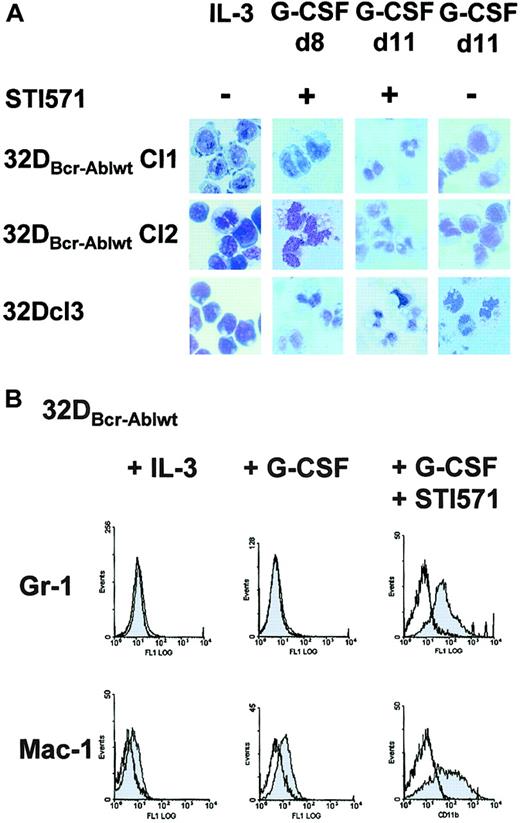
To next investigate whether the block of granulocytic differentiation observed in 32DBcr-Ablwt cells could be reversed by imatinib mesylate, cells were treated with imatinib mesylate and G-CSF for 14 days. In the presence of imatinib mesylate, 32DBcr-Ablwt cells differentiated into morphologically mature neutrophilic granulocytes within 8 to 11 days (Figure4A). Morphologic differentiation was accompanied by up-regulation of myeloid-specific surface markers Mac-1 and Gr-1 (Figure 4B). These data prove that the block of granulocytic differentiation in 32DBcr-Ablwt cells depended on Bcr-Abl kinase activity and could be reversed by the addition of the Abl kinase inhibitor imatinib mesylate.
Effect of imatinib mesylate on G-CSF–induced granulocytic differentiation of 32DBcr-Ablwt cells.
Imatinib mesylate restores G-CSF–induced granulocytic differentiation of 32DBcr-Ablwt cells. (A) 32Dcl3 cells and 2 individual clones of 32DBcr-Ablwt cells were grown in media containing G-CSF. Imatinib mesylate was added at a concentration of 1 μM as indicated. Morphological differentiation of cytospun cells was determined at days 8 and 11 by May-Gruenwald-Giemsa staining (original magnification, × 400). Cells grown in the presence of IL-3 are shown as a control. (B) Expression of Gr-1 and Mac-1 on 32Dcl3 cells and 32DBcr-Ablwt cells upon G-CSF stimulation. Addition of 1 μM imatinib mesylate to the media sufficiently restored up-regulation of both myeloid surface markers.
Effect of imatinib mesylate on G-CSF–induced granulocytic differentiation of 32DBcr-Ablwt cells.
Imatinib mesylate restores G-CSF–induced granulocytic differentiation of 32DBcr-Ablwt cells. (A) 32Dcl3 cells and 2 individual clones of 32DBcr-Ablwt cells were grown in media containing G-CSF. Imatinib mesylate was added at a concentration of 1 μM as indicated. Morphological differentiation of cytospun cells was determined at days 8 and 11 by May-Gruenwald-Giemsa staining (original magnification, × 400). Cells grown in the presence of IL-3 are shown as a control. (B) Expression of Gr-1 and Mac-1 on 32Dcl3 cells and 32DBcr-Ablwt cells upon G-CSF stimulation. Addition of 1 μM imatinib mesylate to the media sufficiently restored up-regulation of both myeloid surface markers.
Bcr-Abl reversibly blocks G-CSF–induced up-regulation of C/EBPα and C/EBPε but not PU.1
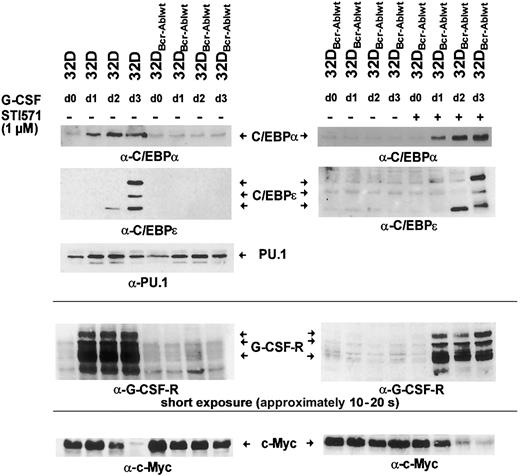
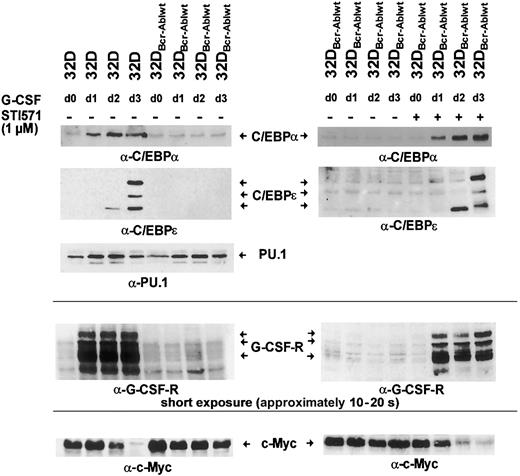
Hematopoietic differentiation is regulated by a network of transcription factors, including C/EBPα, C/EBPε, and PU.1.9 10 To investigate whether Bcr-Abl influences the induction of expression of these transcription factors after G-CSF stimulation, parental 32Dcl3 cells and 32DBcr-Ablwt cells were grown in the presence of G-CSF for 3 days. Moreover, an aliquot of 32DBcr-Ablwt cells was cultured in the presence of both G-CSF and 1 μM imatinib mesylate. Cells grown in the presence of IL-3 were used as a control (Figure 5 left panel, lane 1). Western blot and real-time PCR analysis revealed that in parental 32Dcl3 cells, G-CSF stimulated the expression of these transcription factors in a coordinated manner (Figures 5-6). While considerable amounts of PU.1 were found expressed, only low levels of C/EBPα and no C/EBPε expression were seen in 32Dcl3 cells grown with IL-3 (Figure 5 left panel, lane 1). G-CSF induced a several-fold increase of C/EBPα and a moderate up-regulation of PU.1 protein and mRNA expression (Figure 5 left panel, lanes 1-4, and 6). Moreover, G-CSF stimulated the expression of C/EBPε protein. On day 2, only a faint signal of the 14-kDa C/EBPε isoform missing the transactivation domain was detected (Figure 5 left panel, lane 3). On day 3, 3 different isoforms of C/EBPε were seen (Figure 5 left panel, lane 4).
Effect of Bcr-Abl on G-CSF–induced regulation of C/EBPα, C/EBPε, c-Myc, and PU.1.
Bcr-Abl disturbs G-CSF–induced regulation of C/EBPα, C/EBPε, and c-Myc, but not PU.1. The 32Dcl3 cells and a representative clone of 32DBcr-Ablwt cells were grown in the presence of rhG-CSF (left panel). Lysates of the individual cell types were obtained at day 0 (prior to G-CSF stimulation grown in IL-3) to day 3 and subjected to Western blot analysis by means of rabbit polyclonal antibodies to C/EBPα, C/EBPε, PU.1, G-CSFR, and c-Myc. In addition, aliquots of 32DBcr-Ablwt cells were grown in the presence of rhG-CSF and 1 μM imatinib mesylate (right panel). Lysates were obtained at day 0 (prior to G-CSF stimulation) to day 3; subjected to Western blot analysis by means of rabbit polyclonal antibodies to C/EBPα, C/EBPε, G-CSFR, and c-Myc; and compared with lysates of G-CSF–treated 32DBcr-Ablwt cells grown in the absence of imatinib mesylate. Importantly, lysates of left and right panels are matched; that is, they are from the same subclone and experiments were done at the same time.
Effect of Bcr-Abl on G-CSF–induced regulation of C/EBPα, C/EBPε, c-Myc, and PU.1.
Bcr-Abl disturbs G-CSF–induced regulation of C/EBPα, C/EBPε, and c-Myc, but not PU.1. The 32Dcl3 cells and a representative clone of 32DBcr-Ablwt cells were grown in the presence of rhG-CSF (left panel). Lysates of the individual cell types were obtained at day 0 (prior to G-CSF stimulation grown in IL-3) to day 3 and subjected to Western blot analysis by means of rabbit polyclonal antibodies to C/EBPα, C/EBPε, PU.1, G-CSFR, and c-Myc. In addition, aliquots of 32DBcr-Ablwt cells were grown in the presence of rhG-CSF and 1 μM imatinib mesylate (right panel). Lysates were obtained at day 0 (prior to G-CSF stimulation) to day 3; subjected to Western blot analysis by means of rabbit polyclonal antibodies to C/EBPα, C/EBPε, G-CSFR, and c-Myc; and compared with lysates of G-CSF–treated 32DBcr-Ablwt cells grown in the absence of imatinib mesylate. Importantly, lysates of left and right panels are matched; that is, they are from the same subclone and experiments were done at the same time.
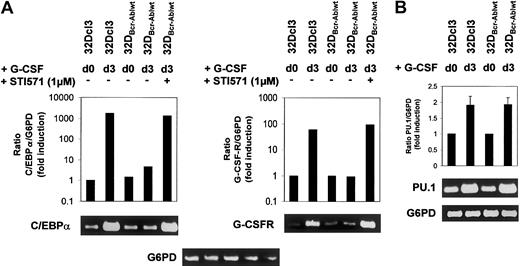
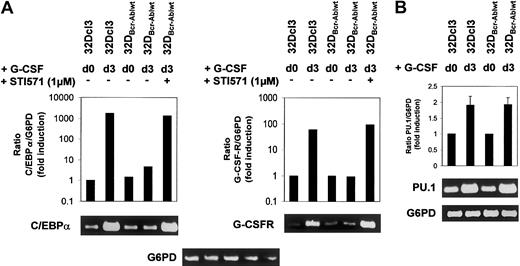
Effect of Bcr-Abl on C/EBPα and G-CSFR mRNA expression.
Bcr-Abl reversibly blocks induction of C/EBPα and G-CSFR mRNA expression. (A-B) Expression of C/EBPα, G-CSFR, and PU.1 mRNA in 32Dcl3 and 32DBcr-Ablwt cells prior to and at day 3 after G-CSF stimulation as assessed by real-time PCR and semiquantitative RT-PCR. Real-time PCR (results shown in graphs) was performed as described in “Materials and methods.” The mRNA expression of C/EBPα, G-CSFR, and PU.1 mRNA was normalized to levels of G6PD mRNA. Induction of mRNA expression is shown as fold induction relative to unstimulated 32Dcl3 cells. Data from 1 of 2 independent experiments are shown. For semiquantitative RT-PCR (results shown in blots), real-time PCR reactions were stopped at appropriate time points (C/EBPα, 24 cycles; G-CSFR, 26 cycles; PU.1, 24 cycles; G6PD, 24 cycles). Aliquots were loaded onto agarose gels for further analyses. G6PD is shown as a control.
Effect of Bcr-Abl on C/EBPα and G-CSFR mRNA expression.
Bcr-Abl reversibly blocks induction of C/EBPα and G-CSFR mRNA expression. (A-B) Expression of C/EBPα, G-CSFR, and PU.1 mRNA in 32Dcl3 and 32DBcr-Ablwt cells prior to and at day 3 after G-CSF stimulation as assessed by real-time PCR and semiquantitative RT-PCR. Real-time PCR (results shown in graphs) was performed as described in “Materials and methods.” The mRNA expression of C/EBPα, G-CSFR, and PU.1 mRNA was normalized to levels of G6PD mRNA. Induction of mRNA expression is shown as fold induction relative to unstimulated 32Dcl3 cells. Data from 1 of 2 independent experiments are shown. For semiquantitative RT-PCR (results shown in blots), real-time PCR reactions were stopped at appropriate time points (C/EBPα, 24 cycles; G-CSFR, 26 cycles; PU.1, 24 cycles; G6PD, 24 cycles). Aliquots were loaded onto agarose gels for further analyses. G6PD is shown as a control.
Analysis of 32DBcr-Ablwt cells revealed that baseline expression of C/EBPα at protein and mRNA levels was equivalent to that of parental 32Dcl3 cells. Up-regulation of C/EBPα and C/EBPε was completely disrupted (Figure 5 left panel, lanes 5-8), but could be restored by the addition of imatinib mesylate (Figure 5 right panel, lanes 5-8). Lack of up-regulation of C/EBPα protein expression was due to inhibition of transcriptional activation of the C/EBPα gene (Figure 6A left panel), whereas considerable induction of C/EBPα mRNA was seen in parental 32DCl3 cells as well as in 32DBcr-Ablwt cells treated with imatinib mesylate. In marked contrast to C/EBPα and C/EBPε, expression of PU.1 was increased by G-CSF in 32DBcr-Ablwt cells in a manner similar to that in 32Dcl3 cells and was correlated to preserved up-regulation of PU.1 mRNA (Figures 5 left panel, and 6B). Taken together, these results suggest that Bcr-Abl directly interferes with parts of the granulocytic differentiation program by inhibiting up-regulation of C/EBPα and C/EBPε. Moreover, the induction of PU.1 by G-CSF demonstrates that the G-CSFR molecules expressed on 32DBcr-Ablwt cells remain functional. With regard to the disturbance of transcription-factor regulation, the effects of Bcr-Abl were essentially similar to the effects of IL-3 (data not shown), which was earlier shown to inhibit G-CSF–induced differentiation in 32Dcl3 cells.23
Bcr-Abl reversibly blocks c-Myc down-regulation upon G-CSF stimulation
C/EBP transcription factors and c-Myc are inversely regulated during differentiation of myeloid cells.17 24 We wished to investigate the influence of Bcr-Abl on the regulation of c-Myc by G-CSF. Therefore, 32Dcl3 and 32DBcr-Ablwt cells treated with G-CSF for 1 to 3 days were analyzed for c-Myc expression. In 32Dcl3 cells, G-CSF led to a decrease of c-Myc expression over a 3-day period (Figure 5 left panel, lanes 1-4). Interestingly, the time kinetics of c-Myc down-regulation correlated with the up-regulation of C/EBPα and C/EBPε (Figure 5 left panel). In contrast, c-Myc expression was only minimally reduced in 32DBcr-Ablwt cells (Figure 5 left panel, lanes 5-8). Appropriate c-Myc regulation could be restored by culturing cells in the presence of 1 μM imatinib mesylate (Figure 5 right panel, lanes 5-8). Kinetics of C/EBPα and C/EBPε up-regulation and c-Myc down-regulation of imatinib mesylate-treated 32DBcr-Ablwtcells were similar to those of 32Dcl3 cells, suggesting that the effects of Bcr-Abl on the regulation of these transcription factors were reversed by imatinib mesylate immediately.
Bcr-Abl inhibits up-regulation of G-CSF receptor expression upon G-CSF stimulation
Rapid up-regulation of the G-CSFR is part of the differentiation program induced by G-CSF,25 and G-CSFR mRNA expression is regulated by both PU.1 and C/EBPα sites in the G-CSFR promotor.25 Western blot analysis showed that baseline expression of the G-CSF receptor was low but equivalent in both 32Dcl3 and 32DBcr-Ablwt cells (Figures 2A, and 5 left panel, lanes 1 and 5). Importantly, the obvious differences between the expression levels of G-CSFR of unstimulated cells seen in Figures 2A and 5 were due to different exposure times of blots. These were necessary as a consequence of the massive up-regulation of G-CSFR protein expression by G-CSF stimulation, but do not reflect a loss of basal level expression of G-CSFR in 32D and 32DBcr-Ablwtcells used for Figure 5 when compared with Figure 2. As expected, G-CSF rapidly induced G-CSFR protein expression in 32Dcl3 cells within 24 hours (Figure 5 left panel, lanes 2-4), which was accompanied by induction of G-CSFR mRNA expression as assessed by real-time PCR and semiquantitative RT-PCR (Figure 6A right panel). In marked contrast, G-CSFR up-regulation was completely disrupted in 32DBcr-Ablwt cells (Figure 5 left panel, lanes 5-8). Block of up-regulation of G-CSFR expression correlated to a block in up-regulation of G-CSFR mRNA (Figure 6). Up-regulation of both G-CSFR protein and mRNA could be restored by growing cells in the presence of 1 μM imatinib mesylate. Identical results were obtained for both clones used in this study. Taken together, these results demonstrate that Bcr-Abl reversibly blocks G-CSF–induced up-regulation of G-CSF receptor expression at the transcriptional level, although basal level of G-CSFR expression was not affected by Bcr-Abl.
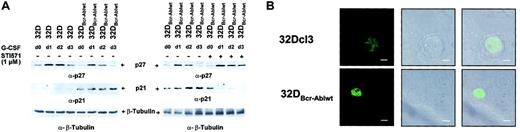
Bcr-Abl disturbs the regulation of cyclin-dependent kinase inhibitors p21Waf1/Cip1 and p27Kip1
Coordinated cell cycle arrest is part of the differentiation program in different cell types. Therefore, we wished to investigate the influence of Bcr-Abl on the expression of cyclin-dependent kinase inhibitors (CDKIs) such as p21Waf1/Cip1 and p27Kip1. Both p27Kip1 and p21Waf1/Cip1 were expressed in 32Dcl3 cells, at least at low levels (Figure 7A left panel, lane 1), but only p27Kip1 was up-regulated transiently upon stimulation with G-CSF (Figure 7A left panel, lanes 1-4). In 32DBcr-Ablwt cells, baseline expression of p27Kip1 was slightly decreased compared with 32Dcl3 cells (Figure 7A left panel, lanes 1 and 5), but p27Kip1 was still up-regulated by G-CSF, albeit less so than in 32Dcl3 cells (Figure 7A left panel, lanes 5-8). Surprisingly, baseline expression of p21Waf1/Cip1 was markedly increased in 32DBcr-Ablwt cells when compared with 32Dcl3 cells (Figure 7A left panel, lanes 5-8). Because p21Waf1/Cip1 is known to be an inhibitor of cell cycle progression, we used immunofluorescence microscopy to investigate whether cell cycle progression in the presence of high levels of p21Waf1/Cip1 could be explained by altered subcellular localization. Surprisingly, no significant influence of Bcr-Abl on subcellular distribution of p21Waf1/Cip1 was seen (Figure7B). As observed for transcription-factor regulation, addition of imatinib mesylate to 32DBcr-Ablwt cells also restored the appropriate regulation of expression of p21Waf1/Cip1 and p27Kip1 (Figure 7A right panel). The p27Kip1 was found transiently upregulated in imatinib mesylate-treated cells as well as in untreated cells in response to G-CSF (Figure 7A right panel). The p27Kip1 expression levels at day 3 after G-CSF stimulation seemed somewhat higher after imatinib mesylate treatment when compared with untreated 32DBcr-Ablwt cells and were approximately equivalent to the levels found in 32Dcl3 cells after G-CSF stimulation. Also, over a 3-day period of imatinib mesylate treatment, p21Waf1/Cip1expression was down-regulated to levels found in parental 32Dcl3 cells (Figure 7A right panel, lane 8).
Effect of Bcr-Abl on regulation of the cyclin-dependent kinase inhibitors p21Waf1/Cip1 and p27Kip1.
Bcr-Abl disturbs regulation of the cyclin-dependent kinase inhibitors p21Waf1/Cip1 and p27Kip1. (A) The 32Dcl3 cells and cells of a representative clone of 32DBcr-Ablwt were stimulated with rhG-CSF (left panel). Lysis of the individual cell types was performed at day 0 (prior to G-CSF stimulation) to day 3. In addition, aliquots of 32DBcr-Ablwt cells were grown in the presence of rhG-CSF and 1 μM imatinib mesylate. Lysates of such cells were compared with lysates of G-CSF–treated 32DBcr-Ablwt cells grown in the absence of imatinib mesylate (right panel). Western blot analysis of the different lysates was performed with the use of antibodies to p27Kip1, and p21Waf1/Cip1. Equal protein loading onto individual lanes was controlled by blotting the stripped membrane with anti–β-tubuline ab. (B) Confocal immunofluorescence microscopy of 32Dcl3 and 32DBcr-Ablwt cells for p21Waf1/Cip1. Representative pictures for each cell type are shown. Scale bar measurements, top row, 4.00 μm; bottom row, 8.00 μm.
Effect of Bcr-Abl on regulation of the cyclin-dependent kinase inhibitors p21Waf1/Cip1 and p27Kip1.
Bcr-Abl disturbs regulation of the cyclin-dependent kinase inhibitors p21Waf1/Cip1 and p27Kip1. (A) The 32Dcl3 cells and cells of a representative clone of 32DBcr-Ablwt were stimulated with rhG-CSF (left panel). Lysis of the individual cell types was performed at day 0 (prior to G-CSF stimulation) to day 3. In addition, aliquots of 32DBcr-Ablwt cells were grown in the presence of rhG-CSF and 1 μM imatinib mesylate. Lysates of such cells were compared with lysates of G-CSF–treated 32DBcr-Ablwt cells grown in the absence of imatinib mesylate (right panel). Western blot analysis of the different lysates was performed with the use of antibodies to p27Kip1, and p21Waf1/Cip1. Equal protein loading onto individual lanes was controlled by blotting the stripped membrane with anti–β-tubuline ab. (B) Confocal immunofluorescence microscopy of 32Dcl3 and 32DBcr-Ablwt cells for p21Waf1/Cip1. Representative pictures for each cell type are shown. Scale bar measurements, top row, 4.00 μm; bottom row, 8.00 μm.
Restoration of G-CSF–induced granulocytic differentiation by imatinib mesylate is blocked by expression of an inhibitor-resistant mutant
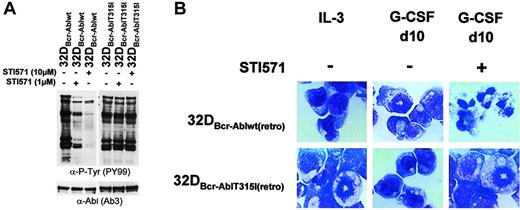
To investigate if restoration of G-CSF–induced differentiation by imatinib mesylate in 32D cells expressing Bcr-Abl was due to the inhibition of Bcr-Abl or instead represented inhibition of other relevant targets, mixed cell populations of 32Dcl3 cells were established by retroviral infection expressing either Bcr-Ablwt or a imatinib mesylate-resistant mutant of Bcr-Abl, Bcr-AblThr315Iso. This mutation has been isolated from patients with clinical imatinib mesylate resistance and has been shown to abolish imatinib mesylate binding to the ATP-binding site of Abl.8 Accordingly, tyrosine phosphorylation of cellular substrates could not be reversed by imatinib mesylate in cells expressing mutant Thr315Iso (Figure8A). At 10 days after retroviral transduction, the 2 cell populations expressed comparable amounts of G-CSFR and C/EBPα as assessed by Western blot analysis and real-time PCR (data not shown). Yet, both Bcr-Ablwt and the imatinib mesylate-resistant mutant Bcr-AblThr315Iso blocked granulocytic differentiation induced by G-CSF as assessed by monitoring morphologic differentiation (Figure 8B) and expression of granulocytic surface markers (data not shown). However, in contrast to 32DBcr-Ablwt cells, a reversal of the block of differentiation by imatinib mesylate was not seen in cells expressing Bcr-AblThr315Iso, proving that granulocytic differentiation of Bcr-Abl+ 32D cells in the presence of imatinib mesylate was dependent on inhibition of Bcr-Abl but not of any other target. As proof of this, we could show that imatinib mesylate did not reverse the block of differentiation induced by IL-3 (data not shown).
Effect of imatinib mesylate on G-CSF–induced granulocytic differentiation in 32DBcr-Abl cells expressing an imatinib mesylate-resistant mutant.
Imatinib mesylate fails to restore G-CSF–induced granulocytic differentiation in 32DBcr-Abl cells expressing an imatinib mesylate-resistant mutant. (A) The 32DBcr-Ablwtand 32DBcr-AblThr315Iso cells established by retroviral infection were either left untreated or incubated with 1 or 10 μM imatinib mesylate for 2 hours prior to lysis. Western blot analysis was performed with antibodies to phosphotyrosines and Abl. (B) The 32Dcl3, 32DBcr-Ablwt, and 32DBcr-AblThr315Iso cells were grown in media containing G-CSF. Imatinib mesylate was added at a concentration of 1 μM as indicated. Morphological differentiation of cytospun cells was determined at day 10 by May-Gruenwald-Giemsa staining (original magnification, × 400). Cells grown in the presence of IL-3 are shown as a control.
Effect of imatinib mesylate on G-CSF–induced granulocytic differentiation in 32DBcr-Abl cells expressing an imatinib mesylate-resistant mutant.
Imatinib mesylate fails to restore G-CSF–induced granulocytic differentiation in 32DBcr-Abl cells expressing an imatinib mesylate-resistant mutant. (A) The 32DBcr-Ablwtand 32DBcr-AblThr315Iso cells established by retroviral infection were either left untreated or incubated with 1 or 10 μM imatinib mesylate for 2 hours prior to lysis. Western blot analysis was performed with antibodies to phosphotyrosines and Abl. (B) The 32Dcl3, 32DBcr-Ablwt, and 32DBcr-AblThr315Iso cells were grown in media containing G-CSF. Imatinib mesylate was added at a concentration of 1 μM as indicated. Morphological differentiation of cytospun cells was determined at day 10 by May-Gruenwald-Giemsa staining (original magnification, × 400). Cells grown in the presence of IL-3 are shown as a control.
Discussion
Although previous reports had suggested that in certain cell types and experimental systems, Bcr-Abl would rather induce than inhibit myeloid differentiation,27,28 we and others could show that in 32Dcl3 cells Bcr-Abl blocks G-CSF–induced neutrophilic differentiation in a kinase-dependent manner.18 Perrotti et al22 recently reported that Bcr-Abl blocks G-CSF–induced differentiation of 32D cells by down-regulating basal level expression of C/EBPα through repression of translation of the C/EBPα mRNA after prolonged time of cells in culture. Loss of C/EBPα expression was associated with decreased G-CSFR expression.22 Using freshly establish Bcr-Abl+ cell clones, we could show that Bcr-Abl blocks G-CSF–induced granulocytic differentiation of 32D cells even prior to the loss of basal levels of C/EBPα and G-CSFR expression, that is, in the presence of normal and functional levels of C/EBPα and G-CSFR protein and mRNA expression. Consequently, inhibition of C/EBPα expression at the translational level is a possible, but not the only, mechanism responsible for blockage of G-CSF–driven granulocytic differentiation in Bcr-Abl–expressing 32DCl3 cells. It should be mentioned that Perrotti et al22 described down-regulation of C/EBPα and G-CSFR expression as developing 15 to 25 days after retroviral infection of parental 32DCl3 cells. In our study, C/EBPα and G-CSFR expression was preserved up to 28 days in cell clones established by electroporation and limited dilution. This obvious discrepancy might be explained by the different transfection and selection methods used, possibly resulting in different onsets and levels of Bcr-Abl protein expression in transfected cells.
Despite preserved basal level expression of C/EBPα and G-CSFR, our data provide evidence that Bcr-Abl specifically interferes with signaling pathways regulating the up-regulation of expression of hematopoietic transcription factors upon G-CSF stimulation. Dysregulation of such transcription factors, above all C/EBPα, has been implicated in the development of acute myeloid leukemias.29,30 Although comparable levels of expression of G-CSFR, C/EBPα, and PU.1 were found in parental 32D and 32DBcr-Ablwt cells, up-regulation of expression of G-CSFR, C/EBPα, and C/EBPε early after G-CSF stimulation was disrupted at the transcriptional level. Interestingly, normal regulation of PU.1 was observed in 32DBcr-Ablwt cells, which is in agreement with recently published data demonstrating that G-CSF regulates PU.1 even in the absence of C/EBP transcription-factor expression.31Interestingly, perturbation of transcription-factor regulation by Bcr-Abl closely resembled the effects seen for IL-3–induced inhibition of G-CSF–driven differentiation.
Our data imply that disturbed regulation of c-Myc expression might be involved in blocking C/EBP transcription-factor up-regulation. It was shown only recently that c-Myc expression is negatively regulated by C/EBPα and that down-regulation of c-Myc is a prerequisite for myeloid progenitors to terminally differentiate.24Consequently, overexpression of c-Myc inhibits C/EBP–dependent transcription and myeloid differentiation.17 32 Our results demonstrate that Bcr-Abl, similarly to IL-3, inhibits the down-modulation of c-Myc expression of cells cultured in G-CSF–containing media, and this might explain the defect of up-regulation of C/EBP transcription factors. Interestingly, this defect was overcome by the addition of imatinib mesylate.
However, disruption of c-Myc regulation might not be the only mechanism by which Bcr-Abl interferes with differentiation. In our study, imatinib mesylate restored up-regulation of G-CSFR expression in Bcr-Abl+ 32Dcl3 cells within 24 hours, clearly preceding restoration of C/EBP transcription-factor up-regulation and down-regulation of c-Myc. Although it is possible that up-regulation of G-CSF receptor expression in this system does not depend on C/EBPα at all,33 this observation might point to a possible involvement of posttranslational modifications, such as protein phosphorylation, in regulating C/EBPα function, a mechanism that has been shown to regulate the function of C/EBPα in adipocytes.34 Inhibition of C/EBPα function by Bcr-Abl–mediated phosphorylation could explain why overexpression of C/EBPα is sufficient to induce differentiation of 32Dcl3 cells in the presence of IL-3 but not if Bcr-Abl is expressed.14 If this is the case, Bcr-Abl could inhibit the initial up-regulation of G-CSFR expression by disrupting functional C/EBPα activation, and this would block up-regulation of other genes, including C/EBPε and C/EBPα itself.
If Bcr-Abl directly interferes with myeloid transcription-factor regulation and differentiation, why do leukemic cells from CML patients in chronic phase show only discrete abnormalities of myeloid differentiation? One possible explanation is that secondary genetic alterations might be necessary to confer a full differentiation block to Bcr-Abl–expressing leukemic cells. The 32Dcl3 cells already harbor such mutations, and these cells have a profound proliferative defect as they grow continuously in culture with just the addition of IL-3 as a single growth factor. Although it is not clear whether 32D cells bear similar secondary genetic alterations such as are found in late-stage CML, this system might reflect blast crisis rather than chronic phase disease.
Moreover, G-CSF is just one of several factors inducing myeloid and, above all, neutrophilic differentiation. G-CSF−/− as well as G-CSFR−/− mice still produce mature granulocytes, although to a lesser degree.11 12 Blockage of C/EBPα and C/EBPε up-regulation by Bcr-Abl might be specific for G-CSF signaling. Other cytokines, such as IL-6 or granulocyte-macrophage CSF (GM-CSF), might provide rescue pathways in vivo. In this context, a defect in G-CSF signaling induced by Bcr-Abl might lead to only subtle alterations in terminal differentiation.
Still, dysregulation of transcription factors such as C/EBPε by Bcr-Abl might explain parts of the leukemic phenotype of CML. C/EBPε−/− mice, for example, have many features that are reminiscent of chronic-phase CML. Bone marrow cellularity of C/EBPε−/− is increased when compared with wild-type mice.35 The fraction of actively proliferating cells in the bone marrow is significantly increased, and these mice display an expansion of the number of CFU-GMs in the marrow.35Usually, these mice die from a myeloproliferative syndrome after 4 to 5 months.15 With regard to this, it is noteworthy that many recent findings suggest a role of C/EBP transcription factors in direct regulation of cell cycle progression by inhibiting cyclin-dependent kinases.36-38 These observations corroborate the potential importance of C/EBP transcription-factor deregulation in Bcr-Abl–induced chronic myeloid leukemia. As C/EBP transcription factors could be a potential target for therapeutic intervention, particularly in CML blast crisis patients who have developed a direct imatinib mesylate resistance, future investigations should focus on the precise mechanisms leading to disturbed regulation of G-CSFR, C/EBPα, and C/EBPε in Bcr-Abl+ cells.
We thank S. Reis for technical assistance, C. Kurzeder for advice on FACS analysis, E. Buchdunger (Novartis, Basel, Switzerland) for providing imatinib mesylate, and S. Nagata and U. Just for kindly providing reagents and cell lines.
Prepublished online as Blood First Edition Paper, September 12, 2002; DOI 10.1182/blood-2002-01-0043.
Supported by grants Ha 1680/2-3 and 1680/2-4 from the Deutsche Forschungsgemeinschaft (M.H.) and by grants from the Novartis Foundation for Therapeutic Research (M.H. and M.W.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Markus Warmuth, KKG Gentherapie, GSF—Forschungszentrum für Umwelt und Gesundheit, Marchioninistrasse 25, D-81377 Munich, Germany; e-mail:mwarmuth@gnf.org.