Abstract
Although many acute myeloid leukemia (AML) colony-forming cells (CFCs) and long-term culture–initiating cells (LTC-ICs) directly isolated from patients are actively cycling, quiescent progenitors are present in most samples. In the current study,3H-thymidine (3H-Tdr) suicide assays demonstrated that most NOD/SCID mouse leukemia-initiating cells (NOD/SL-ICs) are quiescent in 6 of 7 AML samples. AML cells in G0, G1, and S/G2+M were isolated from 4 of these samples using Hoechst 33342/pyroninY staining and cell sorting. The progenitor content of each subpopulation was consistent with the 3H-Tdr suicide results, with NOD/SL-ICs found almost exclusively among G0 cells while the cycling status of AML CFCs and LTC-ICs was more heterogeneous. Interestingly, after 72 hours in serum-free culture with or without Steel factor (SF), Flt-3 ligand (FL), and interleukin-3 (IL-3), most G0 AML cells entered active cell cycle (percentage of AML cells remaining in G0 at 72 hours, 1.2% to 37%, and 0% to 7.6% in cultures without and with growth factors [GFs], respectively) while G0 cells from normal lineage-depleted bone marrow remained quiescent in the absence of GF. All 4 AML samples showed evidence of autocrine production of 2 or more of SF, FL, IL-3, and granulocyte-macrophage colony-stimulating factor (GM-CSF). In addition, 3 of 4 samples contained an internal tandem duplication of theFLT3 gene. In summary, quiescent leukemic cells, including NOD/SL-ICs, are present in most AML patients. Their spontaneous entry into active cell cycle in short-term culture might be explained by the deregulated GF signaling present in many AMLs.
Introduction
Among the malignant cells from most patients with acute myeloid leukemia (AML) are progenitor cells that can be detected in vitro, not only by their ability to form colonies in semisolid media but also by their initiation of malignant hematopoiesis, which will persist over many weeks in long-term culture (LTC). Limiting dilution analysis and cell sorting experiments have demonstrated that both these latter AML LTC-initiating cells (AML LTC-ICs) and the AML progenitors that initiate human leukemia in sublethally irradiated nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice (NOD/SCID leukemia-initiating cells, or NOD/SL-ICs) are rare progenitors that share many of the cell surface phenotypic properties of primitive normal progenitors.1-8 However, NOD/SL-ICs are typically at least 100-fold less frequent than AML LTC-ICs and also have proved more difficult to maintain in short-term culture or transduce with retrovirus.3 9 These latter findings have suggested that the 2 assays detect somewhat different cell populations despite their phenotypic similarity. Nevertheless, it seems likely that such rare progenitors with extensive and long-term proliferative capacity, detected either in vitro or in vivo, are important to the maintenance of leukemia in patients.
A competitive advantage of leukemic over normal hematopoiesis is obvious clinically. One factor that might contribute to this phenomenon is the existence of a larger proliferative fraction among primitive AML progenitors as compared with their normal counterparts. Previous studies have demonstrated that both AML colony-forming cells (AML CFCs) and LTC-ICs are more likely to be found in active cell cycle than similar normal progenitors.10-12 However, in most patient samples at least a small number of quiescent leukemic progenitors could be detected. The current study extends those previous findings by characterizing the proliferative status of NOD/SL-ICs. In addition, Hoechst 33342 (Hst) and pyronin Y (PY) staining and fluorescence-activated cell sorting (FACS) have been used to isolate quiescent AML cells in order to study their functional properties and attempt to manipulate their cell cycle status.13-15 The data demonstrate that many primitive AML progenitors, including most NOD/SL-ICs, are initially quiescent. However, they will spontaneously exit G0 in short-term culture, with only a modest acceleration in this process seen with the addition of growth factors. It is likely that autocrine growth factor production and/or factor-independent activation of signal transduction pathways by mutations such as the Flt-3 internal tandem duplication (ITD) are key determinants of this abnormal proliferative activity in at least some AML samples.
Patients, materials, and methods
Cell preparation
Peripheral blood (PB) samples from newly diagnosed AML patients and normal bone marrow (NBM) samples were obtained with the approval of the Clinical Research Ethics Board of the University of British Columbia and after obtaining informed consent from AML patients. Light-density mononuclear cells were isolated and cryopreserved as previously described.7 Diagnosis and classification of AML were based on the criteria of the French-American-British (FAB) group.16 Cytogenetic analysis was performed on the BM at initial diagnosis. Clinical characteristics of the 7 patients whose cells were studied are listed in Table 1. NBM was obtained from cadaveric donors (North West Tissue Center, Seattle, WA). Ficoll-Hypaque–separated light-density cells were depleted of cells expressing markers of lineage commitment using a Stem-Sep magnetic separation column (StemCell Technologies, Vancouver, BC, Canada). The proportion of CD34+ cells in each of the 2 lineage-depleted (Lin−) NBM samples, A and B, was 28% and 47%, respectively.
Animals
NOD/SCID mice17 were bred and housed in pathogen-free conditions in the British Columbia Cancer Research Center Joint Animal Facility according to protocols approved by the Animal Care Committee of the University of British Columbia.
Hoechst 33342 (Hst) and pyronin Y (PY) staining
Previously described methods were used with minor modifications.14,15 18 Cryopreserved cells were thawed and incubated overnight in serum-free medium (SFM) (StemCell Technologies) to reactivate RNA synthesis and then incubated at 37°C for 45 minutes in Hanks buffered solution with 2% fetal calf serum and 0.02% sodium azide (HFN) and Hst at 5 μg/mL. PY was then added to a final concentration of 1 μg/mL and incubation continued for another 45 minutes. Cells were washed in HFN with the same concentration of Hst and PY and in HFN with Hst, PY, and 2 μg/mL propidium iodide (PI). Finally, cells were resuspended in HFN with Hst and PY and sorted on FACStar Plus (Becton Dickinson Immunocytometry Systems [BDIS], San Jose, CA).
Overnight 3H-Tdr suicide assay
As described in detail previously,12 cells were suspended at fewer than 2 × 106/mL in SFM containing 50 ng/mL human Steel factor (SF) (Terry Fox Laboratory, Vancouver, BC, Canada), 20 ng/mL interleukin-3 (IL-3) (Novartis, Basel, Switzerland), and 20 ng/mL granulocyte colony-stimulating factor (G-CSF) (AMGEN, Thousand Oaks, CA). After 16 hours of incubation at 37°C, in the presence or absence of 20 μCi/mL (740 Bq/mL) of high specific activity 3H-thymidine (3H-Tdr) (25 μCi/mmol [925 Bq/mmol]: Amersham Biosciences, Quebec, QC, Canada), 400 μg/mL cold thymidine (Sigma-Aldrich Canada, Oakville, ON, Canada) was added. Cells were then washed with 40 μg/mL cold thymidine in Iscove medium with 2% fetal calf serum (FCS) and resuspended in Myelocult medium (StemCell Technologies) prior to plating in AML-CFC or LTC-IC assays or injection into NOD/SCID mice.
Detection of AML CFCs and LTC-ICs
AML-CFC and AML LTC-IC assays were performed as previously described.12 Briefly, to detect AML CFCs, cultured or sorted cells were plated at 0.008 × 105 to 2 × 105/mL in methylcellulose-based medium (StemCell Technologies) supplemented with growth factors. Colonies were scored after 10 to 14 days culture at 37°C. To detect AML LTC-ICs, 4 × 106 to 5 × 106 cells from overnight cultures or 0.1 × 106 to 1 × 106 sorted cells in 2.7 mL Myelocult media (StemCell Technologies) with 10−6 M hydrocortisone (Sigma-Aldrich Canada) and 100 ng/mL SF were seeded onto a confluent layer of irradiated (80 cGy) mouse embryonic fibroblasts (Sl/Sl) that had been engineered to produce human IL-3. LTCs were incubated at 37°C in 5% CO2 and received weekly half-media changes and twice-weekly additions of SF to maintain a final concentration of 50 ng/mL. After 5 or 8 weeks, both the adherent and nonadherent cells were harvested, pooled, and plated into methylcellulose assay as described above to detect AML CFCs.
Fluorescence in situ hybridization (FISH)
Among the 7 patient samples studied, numbers 1 and 4 had cytogenetic abnormalities detectable by FISH (Table 1). CD45+ BM cells from mice previously injected with human AML cells were isolated using a FACStar Plus cell sorter (BDIS) and fixed onto glass slides with 3:1 methanol–acetic acid. The yeast artificial clone (YAC 909 g3 or YAC 745 e3) (Research Genetics, Huntsville, AL) for the centromere repeats of chromosome 13 was first amplified by inter-Alu polymerase chain reaction (PCR) using primers and conditions described previously7 19 and then labeled with digoxigenin (DIG, digoxigenin-11-dUTP; Boehringer-Mannheim, Laval, QC, Canada). The 11q23 (MLL) probe labeled with DIG was purchased from Oncor (Gaithersburg, MD).
The slides containing sorted cells were prepared and hybridized overnight with the denatured DNA probes and washed after hybridization as described previously.7
After conterstaining in 0.1 μg/mL PI, slides were visualized on a Zeiss Axioplan (Oberkochen, Germany) fluorescence microscope using double or triple bandpass filters (Omega Optical, Battleboro, NC) to allow simultaneous visualization of fluorescein isothiocyanate (FITC) and PI signals. At least 100 CD45+ cells from mouse BM or Hst/PY-sorted cells were scored for the presence of cytogenetic abnormalities detected by FISH.
Transplantation and detection of AML cells in NOD/SCID mice
Eight- to 10-week-old mice were irradiated with 350 cGy from a137Cs source 16 to 24 hours before 1 × 107 AML cells previously incubated with or without3H-Tdr were injected via the tail vein. In other experiments, NOD/SCID animals were each injected with 1 × 103 to 8 × 105 sorted or cultured AML cells together with 1 × 106 irradiated NBM cells to act as carrier cells. Following irradiation, mice were supplied with acidified drinking water with 100 mg/L ciprofloxacin (Bayer, Leverkusen, Germany). After 4 and/or 8 weeks, femoral BM aspiration was performed on anesthetized mice as described elsewhere.20BM cells were collected in α medium with 50% FCS. Eight or 12 weeks after injection, mice were killed by CO2 inhalation. BM was removed from the 4 long limb bones (femura and humeri) by flushing them with α medium with 50% FCS.
After treatment with 7% ammonium chloride (StemCell Technologies) to lyse red blood cells, cells from mouse tissues were centrifuged and resuspended in HFN and 5% human serum to block human Fc receptors. To block mouse Fc receptors, cells were further incubated with antimouse immunoglobulin G (IgG) Fc receptor monoclonal antibody (2.4 G2; SyStemix, Palo Alto, CA). Half of each sample was incubated with mouse antihuman CD45 conjugated with FITC (Terry Fox Laboratory) and the other half with a fluoresceinated mouse IgG1 isotype control (BDIS). Finally, the cells were stained with 2 μg/mL PI in HFN. Flow cytometry analysis was performed on a FACScan or FACSort flow cytometer (BDIS). A gate was set to exclude at least 99.9% of cells labeled with the isotype control, and the percent of CD45+ cells was then determined using the same gate after excluding nonviable cells. BM cells from mice that had not been injected with human cells were also stained with fluoresceinated anti-CD45 antibody, and the percentage of CD45+ cells was always below 0.1%.
Stimulation of G0 cells with growth factors
AML cells and Lin− NBM were stained with Hst and PY and sorted as described above. Cells with the lowest Hst and PY staining were collected as G0 cells and cultured in SFM with or without 100 ng/mL SF, 100 ng/mL Flt-3 ligand (FL), and 20 ng/mL IL-3 for 72 hours. The cultured cells were restained with Hst and PY for cell cycle analysis, plated into AML-CFC or LTC-IC assays, and injected into NOD/SCID mice.
RT-PCR analyses
To detect growth factor expression by AML blasts, total RNA was extracted with Trizol (Life Technologies, Burlington, ON, Canada) from aliquots of 10 000 light-density mononuclear cells. Complementary DNA was synthesized by reverse transcription using random hexamer priming (Sigma-Aldrich Canada) and reverse transcriptase Superscript II (Gibco, Burlington, ON, Canada). Five microliters of total cDNA was used for a 50 μL total volume primer-specific PCR amplification for IL-3, SF, G-CSF, granulocyte-macrophage colony-stimulating factor (GM-CSF), and FL message. The PCR cocktails also contain 20 mM Tris (tris(hydroxymethyl)aminomethane) HCl (pH 8.4), 50 mM KCl, 2 mM MgCl2, 200 μM of each deoxyribonucleoside triphosphate (dNTP) (Gibco), 5 units of Taq DNA Polymerase (Gibco), and 20 pmol of specific primers for IL-3 (5′-GCTCCCATGACCCAGACAACGTCC-3′ and 5′-CAGATAGAACGTCAGTTTCCTCCG-3′), SF (5′-GGATCTGCAGGAATCGTGTGACTA-3′ and 5′-CTTCAGGAGTAAAGAGCCTGGGTT-3′), G-CSF (5′-CTGTGGACAGTGCAGGAAGCCACC-3′ and 5′-GCTGGGCAAGGTGGCGTAGAACGC-3′), GM-CSF (5′-CAGGAGGCCCGGCGTCTCCTGAAC-3′ and 5′-ACAAGCAGAAAGTCCTTCAGGTTC-3′), FL (5′-AACAACCTATCTCCTCCTGCT-3′ and 5′-GGCACATTTGGTGACAAAGTG-3′), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (5′-GTCTTCACCACCATGGAGAAGG-3′ and 5′-GCCTGCTTCACCACCTTCTTGA-3′) and to give DNA fragments of 345 bp (IL-3), 335 bp (SF), 547 bp (G-CSF), 290 bp (GM-CSF), 306 bp (FL), and 493 bp (GAPDH). Forty cycles (94°C for 30 seconds, 62°C for 1 minute, and 72°C for 1 minute) were performed after the initial denaturation (94°C, 5 minutes). PCR products were detected first in 1.2% agarose gel and then by Southern blotting.
To detect the ITD in the FLT3 gene, cDNA was synthesized as described above from 1 μg total RNA extracted from 1 × 106 to 5 × 106 cells. A total of 0.8 μL cDNA was used in a 50-μL PCR reaction containing 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 2.5 units of Taq DNA Polymerase, and 40 pmol of specific primers for FLT-3 (5′-TGTCGAGCAGTACTCTAAACA-3′ and 5′-ATCCTAGTACCTTCCCAAACTC-3′). After an initial denaturation (94°C, 5 minutes), 35 cycles (94°C for 30 seconds, 62°C for 45 seconds, and 72°C for 1 minute) were performed and followed by an extension step (72°C, 10 minutes). PCR products were electrophoresed in 3% agarose gel, 90 V, 4.5 hours.
Southern blotting
Ten microliters of PCR products was electrophoresed in 1.2% agarose gel, transferred onto nylon membranes (Zeta-probe, Bio-Rad Canada, Mississauga, ON), and hybridized with [32P]deoxycytidine triphosphate ([32P]dCTP) (Amersham Biosciences)–labeled probes overnight at 60°C in 4.6 × saline sodium citrate, 7.6% formamide, 1.5 mM EDTA (ethylenediaminetetraacetic acid), 0.86% skim milk, 0.8% sodium dodecyl sulfate (SDS), 380 μg/mL salmon sperm DNA, and 7.5% dextran sulfate. Probes were labeled by incubating the denatured fragments in the presence of hexamers and the Klenow fragment of polymerase I using a random priming labeling kit (Invitrogen, Burlington, ON, Canada), and unbound [32P]dCTP was removed with a Sephadex G-50 column. Hybridized membranes were washed twice at 60°C for 1 hour in a solution of 0.3 × SSC and 0.1% SDS and exposed to Kodak XAR film (Eastman Kodak, Rochester, NY) at −70°C for 0.5 to 3 hours.
Probes
Complementary DNA probes for human IL-3, SF, G-CSF, GM-CSF, and FL were excised from plasmid vectors by restriction endonuclease digestion. The inserts were separated from the vector backbone by agarose gel electrophoresis. DNA was recovered by electroelution and purified with phenol/chloroform. The amount of DNA recovered was estimated by comparison of band densities in ethidium bromide–stained gels. For GAPDH, products of GAPDH-specific PCR were used as the hybridization probe.
Detection of growth factor protein and bioactivity
SFM was collected from AML PB cells cultured at 2 × 106/mL for 72 hours in the absence of growth factors and concentrated 4-fold with Centricon-10 concentrators (Amicon, Beverly, MA). The presence of IL-3, SF, and GM-CSF was evaluated using a sandwich enzyme immunoassay technique with Quantikine kits (R&D Systems, Minneapolis, MN) according to the manufacturer's protocols. The lower limits of growth factor detection for these assays are 7.8 pg/mL, 31.2 pg/mL, and 7.8 pg/mL for IL-3, SF, and GM-CSF, respectively. To detect FL activity, 1-, 2-, and 4-fold dilutions of concentrated conditioned media were incubated with 4 × 104 BaF3–Flt-3 cells (Immunex, Seattle, WA) plated in round-bottom 96-well plates. After 3 days at 37°C, 1 μCi3H-Tdr was added to each well. After 6 hours of additional incubation, cells were harvested onto a glass fiber filter using LKB Betaplate cell harvester (Wallac, Turku, Finland), and3H-Tdr incorporation was counted in an LKB Betaplate liquid scintillation counter (Wallac). The conditioned media dilutions were tested in duplicate and the results compared with a standard curve created by testing known concentrations of human recombinant FL (Terry Fox Lab) in triplicate. The limit of sensitivity of this assay is 40 pg/mL.
Results
Cycling status of NOD/SL-ICs
Prior investigations had shown that a variable proportion of AML CFCs and LTC-ICs from different patient samples could be quiescent. The results in that previous report are confirmed here for 7 additional AML patients as shown in Table 2. The percent kill in 3H-Tdr suicide assay of AML CFCs and LTC-ICs ranged from 19% to 99% and 0% to 100%, respectively. Five of 7 patient samples showed less than 90% kill of progenitors detected in one or both assays, demonstrating the presence of a quiescent subpopulation. To investigate the possibility that some NOD/SL-ICs might also be in G0, AML cells from the same 7 patients were cultured overnight with or without exposure to 3H-Tdr and then injected intravenously into NOD/SCID mice. Because the level of AML cell engraftment in mouse BM is determined by the number of NOD/SL-ICs initially injected into mice, a comparison of the proportion of human cells detected in mouse BM between animals receiving3H-Tdr–treated and untreated cells allows an estimation of the proportion of cycling NOD/SL-ICs.3,5 8 As shown in Table 3, for 6 of the 7 AML samples there was a modest (less than 50%) change in the level of engraftment of AML cells 8 weeks after exposure to 3H-Tdr, indicating that most of these primitive progenitors were quiescent. In contrast, the 82% reduction in engraftment of 3H-Tdr–treated cells from patient 7 indicates that most NOD/SL-ICs from this patient sample were actively cycling. There was a close correlation between the percent CD45+ cells detected in mouse BM at 4 and 8 weeks after injection of human AML cells (r = 0.75) and no significant difference in the percentage of kill after3H-Tdr exposure detected at the 2 time points (P = .50, t test). FISH was performed on CD45+ human cells sorted from the BM of mice injected with cells from patients 1 and 4. A total of 98 of 112 (88%) versus 290 of 340 (84%) and 46 of 203 (23%) versus 57 of 243 (24%) untreated versus 3H-Tdr–treated cells were cytogenetically abnormal from mice injected with cells from each of these patients, respectively. There was thus no significant change in the proportion of abnormal cells after 3H-Tdr exposure. In addition, the proportion of abnormal cells detected was similar to that reported in the diagnostic BM cytogenetic report on the 2 patients (Table1).
Isolation of quiescent AML progenitors using Hst/PY staining
To isolate and further characterize viable AML cells in different phases of the cell cycle, cells from patients 1, 2, 4, and 5 were sorted into 3 different populations according to their staining characteristics with Hst/PY. Cells in G0 have diploid DNA and low RNA content and hence the lowest Hst and PY staining. Although there is no change in DNA content in G1, RNA synthesis and PY staining increase significantly. S/G2+M cells are synthesizing both DNA and RNA and stain brightly with both Hst and PY (Figure 1). The proportion of cells in the 3 different subpopulations was similar in the 4 different patient samples (Table 4). More than 90% of the AML cells were in G1 in each case. In contrast, the mean ± SD percentage of G0 cells was 3.4% ± 0.8%. Although this latter value was much lower than the percentage of G0cells in Lin− NBM (39.6% and 44.6% for Lin−NBM A and B, respectively), because the analysis of NBM was performed on the small fraction of total BM cells that are Lin−, the proportions are not directly comparable. FISH demonstrated percentages of cytogenetically abnormal cells in the 3 sorted cell subpopulations from patients 1 and 4 similar to that found in the patients' diagnostic BM (Table 1).
FACS profiles of Hst/PY-stained AML cells.
AML cells or NBM Lin− were stained with Hst/PY and sorted into subpopulations. A representative Hst/PY dot profile of AML cells from patient 1 is shown.
FACS profiles of Hst/PY-stained AML cells.
AML cells or NBM Lin− were stained with Hst/PY and sorted into subpopulations. A representative Hst/PY dot profile of AML cells from patient 1 is shown.
Aliquots of these Hst/PY-stained, FACS-sorted subpopulations of AML cells from the same 4 AML patients were plated in AML-CFC and LTC-IC assays and injected into NOD/SCID mice. As shown in Figure2, in patients 2, 4, and 5 where most of the AML CFCs were found to be actively cycling with the3H-Tdr suicide assay, more than 99% of these progenitors were in the G1 and S/G2+M subpopulations isolated by Hst/PY staining. Most AML LTC-ICs from patients 2 and 4 were determined to be actively cycling by3H-Tdr suicide while those from patients 1 and 5 were entirely quiescent (Table 2). Consistent with this result, Hst/PY staining revealed nearly all the LTC-ICs from patients 2 and 4 to be detected in cultures initiated with G1 and S/G2+M cells while LTC-ICs from patients 1 and 5 were only detected among G0 cells (Figure 2).
Distribution of AML CFCs and LTC-ICs in different phases of the cell cycle as determined by Hst/PY staining.
AML cells were stained with Hst/PY and sorted into 3 different subpopulations based on their DNA and RNA content as described in Figure 1, following which they were then plated into CFC and LTC-IC assays. The percentage of total CFCs or LTC-derived colonies in each sorted subpopulation was calculated by multiplying the number of progenitors detected per 106 sorted cells from that population by the percentage of the total cell population represented by that subpopulation and then dividing that number by the total number of progenitors found in all 3 sorted subpopulations (× 100).
Distribution of AML CFCs and LTC-ICs in different phases of the cell cycle as determined by Hst/PY staining.
AML cells were stained with Hst/PY and sorted into 3 different subpopulations based on their DNA and RNA content as described in Figure 1, following which they were then plated into CFC and LTC-IC assays. The percentage of total CFCs or LTC-derived colonies in each sorted subpopulation was calculated by multiplying the number of progenitors detected per 106 sorted cells from that population by the percentage of the total cell population represented by that subpopulation and then dividing that number by the total number of progenitors found in all 3 sorted subpopulations (× 100).
Table 5 shows the engraftment obtained in NOD/SCID mice injected 8 to 12 weeks previously with different doses of Hst/PY-stained and -sorted cells. As expected, engraftment was reliably obtained when 1 × 106 to 10 × 106unsorted AML cells from each of these 4 patient samples were injected. Consistent with the results of 3H-Tdr suicide assays shown in Table 3, similar levels of engraftment were obtained with 5- to 25-fold fewer sorted G0 cells from each of the 4 samples. In contrast, no engraftment was seen with similar numbers of S/G2+M cells. G1 cells, injected at the same or higher doses than those tested for G0 cells, resulted in no detectable engraftment with cells from patients 1 and 5 and low-level engraftment in 1 of 6 mice and 1 of 4 mice only for cells from patients 2 and 4, respectively.
In summary, the 3H-Tdr suicide and Hst/PY staining and sorting results are consistent in demonstrating that NOD/SL-ICs from all 4 AML patient samples are largely quiescent while AML LTC-ICs and CFCs are more heterogeneous in their cycling status.
Spontaneous entry of G0 cells into cell cycle after in vitro growth factor stimulation
To determine if these primitive quiescent AML progenitors could be stimulated to enter active cell cycle, G0 cells from patients 1, 2, 4, and 5 were isolated by Hst/PY fractionation and cultured in SFM with or without 100 ng/mL SF, 100 ng/mL FL, and 20 ng/mL IL-3. Cells were harvested after 24 and 72 hours and reanalyzed by Hst/PY staining for cell cycle distribution and placed in AML CFC, LTC-IC, and NOD/SL-IC assays. Interestingly, when cultured in the absence of growth factors, a significant proportion of quiescent AML cells from each of the 4 patients spontaneously left the resting state and entered G1 or S/G2+M (Table6) so that a mean of 16.7% (range, 1.2%-37%) were in G0 after 72 hours. At the same time point in growth factor–supplemented cultures, the mean proportion of AML cells in G0 was 2.6% (range, 0%-7.6%). In contrast, when cultured for 72 hours in the absence of growth factors in SFM, most G0 cells from 2 Lin− NBM samples remained quiescent (Table 6).
AML CFCs and LTC-ICs, including those from the 2 samples where LTC-IC progenitors were initially quiescent, were easily detected among cells that were in active cell cycle after 72 hours in culture (Table7). In contrast, exit from G0reduced the ability of AML cells from patients 2 and 5 to engraft NOD/SCID mice (Table 8). Compared with mice injected with the same or fewer numbers of uncultured G0 cells where engraftment was easily detectable (range of human CD45+ cells in mouse bone marrow, 1%-97.6%) at 8 to 12 weeks after transplantation, cultured G0 cells resulted in less than 0.1% to 3.7% and less than 0.1% to 0.5% engraftment with cells from patients 2 and 5, respectively.
Autocrine production of growth factors and/or FLT3 ITD in AML cells
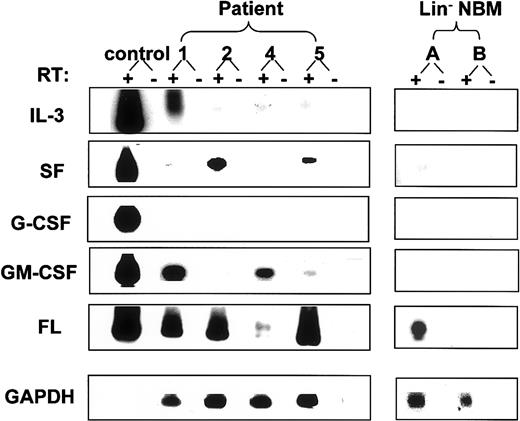
To investigate possible mechanisms whereby initially quiescent AML cells could spontaneously enter active cell cycle, cells from patients 1, 2, 4, and 5 were evaluated for the presence of the FLT-3 ITD and autocrine production of growth factors. Expression of mRNA for 3 or 4 of the 5 growth factor genes studied was detected by reverse transcriptase (RT)–PCR in all 4 patient samples. All 4 samples showed expression of FL and IL-3 while none showed any expression of G-CSF mRNA. In addition, samples 1 and 4 showed expression of GM-CSF, sample 2 showed expression of SF, and sample 5 showed expression of both SF and GM-CSF message. Interestingly, 1 of the 2 Lin− NBM samples tested also showed FL mRNA and a low level of SF transcript expression (Figure3). Enzyme-linked immunosorbent assay (ELISA) of growth medium from cultured AML cells confirmed the presence of IL-3 for AML samples 1, 2, and 4 and GM-CSF for samples 1, 4, and 5 (7.8 or less and 40 to 760 pg/mL, respectively) but was unable to detect soluble SF protein. Using stimulation of 3H-Tdr incorporation in the FL-responsive BaF3 Flt-3 cell line as an indicator, small amounts of bioactive FL were detected in the same growth media (32 to 65 pg/mL) from samples 1, 2, and 5. Interestingly, the FLT-3 ITD was detected in 3 of the 4 AML blast samples (patients 2, 4, and 5) (Figure 4).
Growth factor mRNA expression in AML blasts and normal bone marrow cells.
RT-PCR was performed with total cDNA synthesized from 10 000 AML blasts or NBM cells using a specific primer set for each growth factor as described in “Patients, materials, and methods.” Southern blots of PCR products were hybridized with the corresponding cytokine-specific probes. For each cytokine, the positive control was a cDNA from a murine cell line transfected with the corresponding human cytokine cDNA.
Growth factor mRNA expression in AML blasts and normal bone marrow cells.
RT-PCR was performed with total cDNA synthesized from 10 000 AML blasts or NBM cells using a specific primer set for each growth factor as described in “Patients, materials, and methods.” Southern blots of PCR products were hybridized with the corresponding cytokine-specific probes. For each cytokine, the positive control was a cDNA from a murine cell line transfected with the corresponding human cytokine cDNA.
Demonstration of the FLT3 ITD in AML blasts.
Total RNA was extracted from 1 × 106 to 5 × 106 blast cells. PCR was performed on total cDNA with specific primers flanking exon 11 and 12 of the FLT3gene where the ITD usually occurs. *The positive control (+ve) was an AML sample previously shown to contain this mutation, and the negative control (-ve) was cDNA from cells from a patient with acute lymphoblastic leukemia that did not show the Flt-3 ITD.
Demonstration of the FLT3 ITD in AML blasts.
Total RNA was extracted from 1 × 106 to 5 × 106 blast cells. PCR was performed on total cDNA with specific primers flanking exon 11 and 12 of the FLT3gene where the ITD usually occurs. *The positive control (+ve) was an AML sample previously shown to contain this mutation, and the negative control (-ve) was cDNA from cells from a patient with acute lymphoblastic leukemia that did not show the Flt-3 ITD.
Discussion
In previous studies using 3H-Tdr suicide assays, a substantial proportion of AML CFCs and LTC-ICs from a variety of different patient samples were found to be in active cell cycle.10,12 The current data, using the same technique, confirm these previous findings by demonstrating that 5 of 7 different AML samples contained AML CFCs and/or LTC-ICs that were primarily actively cycling. Determination of the progenitor content of Hst/PY-stained subpopulations of AML cells from 4 of these samples confirmed these findings. In contrast, previous studies have demonstrated that CFCs and LTC-ICs in normal steady-state peripheral blood are uniformly quiescent.11,12 The presence of a larger proportion of actively cycling blasts and their progenitors in AML, as compared with normal hematopoietic cell populations, is consistent with the competitive advantage that AML cells show clinically. Somewhat similar results have been described for progenitors from patients with chronic myeloid leukemia (CML) where an apparent defect in the number and self-renewal potential of leukemic stem cells is more than compensated for by the increased proliferative activity of more mature CML progenitors.18,21 22
Despite the generally active cell cycle status of AML cells detected in vitro, the leukemic progenitors that engraft NOD/SCID mice were largely resistant to killing by 3H-Tdr in 6 of the 7 samples tested. The quiescent status of NOD/SL-ICs in each of 4 of these samples was subsequently confirmed by using Hst/PY staining and cell sorting. These results are also consistent with those from Terpstra et al, who found that 5-fluorouracil selectively spares AML cells with long-term growth potential in SCID mice and in culture.23Interestingly, Jordan and colleagues found that most CD34+CD38− AML cells, which would be expected to contain most NOD/SL-ICs, are also in G0.24
Comparison of the 3H-Tdr suicide results obtained with AML CFCs and LTC-ICs (Table 2) and those obtained on NOD/SL-ICs from the same patient samples (Tables 3) shows that a larger proportion of the former progenitors than the latter were in active cell cycle in samples from patients 2, 4, and 6. To determine if the different duration of the LTC-IC and NOD/SL-IC assays might have influenced these results, the AML LTC-IC assay was extended from 5 to 8 weeks (the usual time point for terminating the NOD/SL-IC assay). However, as shown in Table 2, the percentage of AML LTC-ICs detected after 5 or 8 weeks in culture that were killed by 3H-Tdr was very similar. Previous publications have demonstrated that the same cell surface phenotype that enriches AML cells for the presence of LTC-ICs also partially purifies NOD/SL-ICs.3,25-27 These data had suggested that the 2 assays might detect the same primitive leukemic cell population. Thus, the difference in cycling status observed here between AML LTC-ICs and NOD/SL-ICs is somewhat surprising. However, the low frequency of AML LTC-ICs or NOD/SL-ICs detected in FACS-purified subpopulations in these former experiments allows the possibility of substantial functional heterogeneity among AML cells with the same cell surface phenotype. The possibility that LTCs and immunodeficient mice detect different populations of primitive leukemic cells, at least in some patient samples, is also suggested by the lower frequency of NOD/SL-ICs than AML LTC-ICs and the relative ease with which the latter but not the former progenitors can be transduced with retrovirus.5,9 On the other hand, the functional requirements of the LTC and NOD/SCID mouse assays may place limitations on the detection of certain cells regardless of their usual place in the hematopoietic hierarchy. For examples, the NOD/SL-IC assay requires that cells home to the murine bone marrow microenvironment and engraft as well as proliferate to produce detectable daughter cells over 8 or more weeks. Entry into cell cycle has been shown to inhibit the ability of primitive normal hematopoietic cells to be detected in the NOD/SCID mouse assay while return to the quiescent state rescues this function.15 28 Similar constraints may exist for many primitive AML cells.
The small but significant population of quiescent AML cells identified in all the samples analyzed in these and previous studies are of interest because they appear to be engraftable and presumably relatively resistant to cell cycle active chemotherapeutic drugs. Their existence in AML patients during induction and consolidation therapy and as potential contaminants in stem cell harvests may contribute to disease relapse following standard chemotherapy regimens or autologous transplantation protocols. Thus, experiments within the current studies were designed to study the functional properties of these quiescent leukemic cells in some detail and to attempt to trigger their entry into active cell cycle by ex vivo manipulations.
Interestingly, most of the quiescent AML cells that we isolated from all 4 samples tested exited G0 within 24 to 72 hours of serum-free culture initiation even in the absence of growth factor stimulation, a phenomenon that was not seen with G0 cells isolated from Lin− NBM cells (Table 6). In normal hematopoiesis a variety of factors, including chemokines, inhibitory growth factors, and direct cell-cell interactions, are thought to facilitate the maintenance of primitive progenitors in the quiescent state.28-30 It seems likely that primitive AML cells retain a degree of responsiveness to such stimuli. However, when unrestrained by the in vivo microenvironment, quiescent AML cells tend to spontaneously exit G0. This finding suggests that intrinsic mechanisms that would normally maintain the quiescent state are perturbed in AML.
A variety of abnormalities in growth factor–mediated signal transduction have been described in AML cells. Among these are autocrine production of a variety of stimulatory growth factors and activating mutations in growth factor receptors. One or both of these phenomena were detected in each of the 4 samples tested here. All 4 showed autocrine production of growth factor mRNA with protein detected for at least 2 of the 5 factors evaluated in each case. This demonstration of a very high frequency of detection of growth factor message and protein in AML cells is consistent with our previous results in which 9 of 19 AML samples surveyed showed expression of at least 1 of the 4 growth factors evaluated5 and those of other investigators.31-33 Autocrine growth factor production in AML samples predicts for autonomous proliferation in colony assays and high-level engraftment in NOD/SCID mice while autonomous AML blast proliferation has been associated with a poor clinical prognosis.5,34-36 Expression of mRNA for FL was found in all 4 AML samples tested as well as 1 Lin− NBM sample. These data are consistent with published observations33 37 and suggest that this finding is not specific to cells that have undergone malignant transformation but rather a consequence of the relatively undifferentiated state of AML blasts.
Three of the 4 AML samples studied here contained the FLT-3 ITD. This abnormality causes constitutive activation of the FLT-3 tyrosine kinase, activates intracellular signal transduction pathways, and causes autonomous cell growth when introduced into hematopoietic cell lines.38,39 Primary AML samples containing this mutation show enhanced growth in NOD/SCID mice and a poor clinical prognosis.40 41
It is conceivable that quiescent AML cells would display a different gene expression pattern than the total population of leukemic blasts studied here. However, in addition to mRNA all 4 AML samples tested had FL, IL-3, and/or GM-CSF detected in growth medium, suggesting that the entire leukemic cell population would have been exposed to this stimulatory activity. Thus, it seems likely that autocrine growth factor stimulation and/or factor-independent activation of FLT-3–mediated signal transduction pathways contributed to the observed tendency for quiescent AML cells to spontaneously exit G0. Like autocrine growth factor production, the FLT-3 ITD is found in a substantial proportion of AML samples—that is, 20% to 30% in most large series.41 42 Therefore, although the current study examined only 4 patient samples in depth for their cycling properties, it seems likely that these findings are relevant to many AMLs.
Although quiescent AML cells had a tendency to spontaneously enter active cell cycle when incubated for several days in SFM, this process could be accelerated by the addition of a cocktail of growth factors selected for their ability to stimulate the proliferation of AML blasts.7 However, even after 72 hours of stimulation under these conditions, a small proportion (0.6% to 7.6%) of quiescent cells remained in cultures from 3 of the 4 patient samples (Table 6). Interestingly, other investigators have found subpopulations of quiescent AML cells that failed to enter cell cycle after growth factor stimulation.43 The presence of a small but nevertheless deeply quiescent population of primitive leukemic cells could provide a reservoir of relatively drug-resistant cells from which clinical relapse could ultimately arise.24,44,45 The fact that combining growth factor treatment with conventional chemotherapy has generally produced disappointing clinical results suggests either that the growth factors used were not completely effective in triggering the active cycling of quiescent AML progenitors or that other mechanisms of drug resistance rescued the proliferating leukemic cells.46-49 In the latter case, strategies to induce progenitor cycling must be combined with those to overcome specific molecular mechanisms of drug resistance if AML stem cells are to be eradicated.
Prepublished online as Blood First Edition Paper, December 5, 2002; DOI 10.1182/blood-2002-10-3062.
Supported by a grant from the National Cancer Institute of Canada with funds from the Terry Fox Run (D.E.H.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
D. E. Hogge, Terry Fox Laboratory, 601 West 10th Ave, Vancouver, BC, V5Z 1L3, Canada; e-mail:dhogge@bccancer.bc.ca.