Promyelocytic leukemia protein PML acts as a tumor suppressor, whereas its chimeric mutant promyelocytic leukemia/retinoic acid receptorα (PML/RARα) causes acute promyelocytic leukemia (APL). Because PML has been shown to form transcription-regulatory complexes with various molecules, we speculated that PML and/or PML/RARα might affect signal transducer and activator of transcription 3 (STAT3) activity, which plays a crucial role in granulocyte colony-stimulating factor (G-CSF)–induced growth and survival of myeloid cells. In luciferase assays, PML inhibited STAT3 activity in NIH3T3, 293T, HepG2, and 32D cells. PML formed a complex with STAT3 through B-box and COOH terminal regions in vitro and in vivo, thereby inhibiting its DNA binding activity. Although PML/RARα did not interact with STAT3, it dissociated PML from STAT3 and restored its activity suppressed by PML. To assess the biologic significance of these findings, we introduced PML and PML/RARα into interleukin-3 (IL-3)–dependent Ba/F3 cells expressing the chimeric receptor composed of extracellular domain of G-CSF-R and cytoplasmic domain of gp130, in which gp130-mediated growth is essentially dependent on STAT3 activity. Neither PML nor PML/RARα affected IL-3–dependent growth of these clones. By contrast, gp130-mediated growth was abrogated by PML, whereas it was enhanced by PML/RARα. These results reveal new functions of PML and PML/RARα and suggest that dysregulated STAT3 activity by PML/RARα may participate in the pathogenesis of APL.
Introduction
Acute promyelocytic leukemia (APL) is characterized by a clonal expansion of myeloid precursor cells blocked at the promyelocyte. In more than 95% of APL patients, reciprocal chromosomal translocation t(15; 17) fuses the promyelocytic leukemia (PML) gene to the retinoic acid receptor (RAR) α gene, yielding the fusion protein PML/RARα.1 2
PML is a ubiquitously expressed nuclear protein containing the RING domain, B1 and B2 boxes, and α-helical coiled-coil domain.3,4 PML forms a homodimer and localizes in the speckled subnuclear structure referred to as nuclear bodies (NBs),5 where it interacts with various proteins, including p53 and retinoblastoma protein, and modifies their functions.6-9 It was more feasible to use PML−/− mice to develop lymphomas than PML+/+mice when challenged with mutagens, suggesting a possible role for PML as a tumor suppressor.10,11 In APL cells, PML/RARα delocalizes PML from NBs through direct interaction and inhibits tumor-suppressive activity of PML, which is thought to play some role in leukemogenesis of APL.12-14
The growth, differentiation, and survival of myeloid cells are pivotally regulated by granulocyte colony-stimulating factor (G-CSF). After binding to the receptor, G-CSF activates Janus family tyrosine kinases (JAKs). The activated JAKs, in turn, induce phosphorylation and dimerization of signal transducers and activators of transcription (STATs), which translocate into the nucleus and initiate gene transcription.15-18 Although G-CSF activates STAT1, STAT3, and STAT5, STAT3 was shown to be essential for G-CSF–dependent proliferation and differentiation of myeloid cells.19 20
Considering the fact that STAT3 is a crucial regulator of development of myeloid cells, we speculated that STAT3 activity might be dysregulated in APL cells. Here, we found that PML bound to STAT3 and inhibited its activity, which was canceled by PML/RARα. Furthermore, PML abrogated STAT3-dependent growth of Ba/F3 cells, whereas it was enhanced by PML/RARα. These results suggest that PML/RARα could participate in the pathogenesis of APL through the dysregulation of STAT3 activity.
Materials and methods
Reagents and antibodies
A rabbit anti-STAT3 antibody (Ab) (K-15), a goat anti-PML Ab (N-19), and normal rabbit IgG (control IgG) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). A rhodamine-conjugated murine anti–goat IgG Ab and an FITC-conjugated murine anti–rabbit IgG Ab were purchased from Chemicon International (Temecula, CA).
Plasmid constructs and cDNAs
Expression vectors for PML3 and PML/RARα were provided by Dr M. Alcalay, Department of Experimental Oncology, European Institute of Oncology, Milan, Italy.6 Flag-tagged PML mutants were generated by polymerase chain reaction (PCR) and subcloned into pcDNA3 (Invitrogen, De Schelp, The Netherlands). Glutathione S-transferase (GST)–STAT3 constructs were generated by subcloning PCR products into pGEX-4T-1 (Pharmacia, Uppsala, Sweden). An expression vector for constitutively active STAT5A (1*6-STAT5A) was provided by Dr T. Kitamura (Tokyo University, Japan). pEF-BOS-G-CSF-R/gp130 is an expression vector for G-CSF-R/gp130, a chimeric receptor composed of the extracellular domain of G-CSF receptor and the cytoplasmic domain of gp130.21
Cell lines and cultures
293T, HepG2, and NIH3T3 cells were maintained in Dulbecco modified Eagle medium containing 10% fetal bovine serum (FBS). Murine interleukin-3 (IL-3)–dependent cell lines Ba/F3 and 32D were cultured with RPMI 1640 containing 10% FBS and 1 ng/mL IL-3. To stably express PML and PML/RARα in Ba/F3, we transfected each expression vector by electroporation (250 V, 960 μF) (Bio-Rad Laboratories, Richmond, CA). The transfected cells were screened by the culture with G418 (1.5 mg/mL; Sigma, St Louis, MO). After the selection, we examined the expression levels of the transgene among the stable transformants by Western blot analysis. According to this result, 5 clones, in which each transgene was efficiently expressed, were selected and subjected to further analyses.
Cell proliferation assays
To quantitate the growth of cultured cells, an MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazoloim bromide] (Sigma) rapid colorimetric assay was used as previously reported.22
Northern blot analysis
Northern blot analysis was performed as described previously.23
Immunoprecipitation and immunoblotting
293T cells were transfected with various expression vectors and pEF-BOS-G-CSF-R/gp130 by calcium phosphate precipitation. After 32 hours, the cells were stimulated with 30 ng/mL G-CSF for 30 minutes, and total cellular lysates were prepared as described previously.23 The procedures for immunoprecipitation and immunoblotting were described previously.23
Electrophoretic mobility shift assay
The isolation of nuclear extracts and electrophoretic mobility shift assay (EMSA) were performed as described previously.23 The sequence of the probe to detect DNA binding of STAT3 is based on the IL-6/interferon (IFN) response element in the murine interferon regulatory factor-1 gene promoter as follows: 5′-GCGTGATTT CCCCGAAATGATGAGGCA-3′.24
Luciferase assays
Luciferase assays were performed in NIH3T3, 293T, and HepG2 cells according to the methods described previously.23 The details of the reporter genes for STAT3 (4 × acute phase response elements [APRE]-Luc containing the APRE sequence from the rat α2-microglobulin promoter), STAT5 (3 × β-Cas-Luc containing the γ activation site [GAS] sequence from the bovine β-casein promoter), and STAT1 (4 × interferon α stimulated response element [ISRE]-Luc containing the ISRE sequence from the murine guanylate binding protein [GBP] promoter) were described in previous papers.24,25 The cells were transfected with various expression vectors along with 1 μg of an appropriate reporter gene and 10 ng of pRL-TK, an expression vector for Renillaluciferase. After 8 hours, the cells were washed, incubated without serum for 24 hours, and then cultured with or without an appropriate cytokine indicated (30 ng/mL IL-6, 30 ng/mL IFNα, 5 U/mL erythropoietin (EPO), or 30 ng/mL G-CSF) for 6 hours. We performed luciferase assays in 32D cells by electroporation as described previously.23 Relative luciferase activity was calculated by normalizing transfection efficiency according toRenilla luciferase activity. The experiments were performed in triplicate, and similar results were obtained from at least 4 independent experiments.
GST pulldown assays
GST-STAT3 fusion proteins expressed in Escherichia coli were purified on glutathione-sepharose 4B beads (Pharmacia).35S-labeled PML was prepared with TNT Quick Coupled Transcription/Translation System (Promega, Madison, WI). For each binding reaction, 20 μg GST fusion protein bound to glutathione-sepharose beads was incubated with 50 μL TNT (Promega) reaction solution for 1 hour at 4°C. The resulting complexes were separated by gel electrophoresis and subjected to autoradiography.
Immunostaining
293T cells grown on BioCoat Collagen I CultureSlides (Nippon Becton Dickinson, Tokyo, Japan) were transfected with the indicated expression vectors and pEF-BOS-G-CSF-R/gp130, and then cultured with 30 ng/mL G-CSF for 30 minutes. After fixation and permeabilization, cells were incubated with an anti-STAT3 Ab for 1 hour and then with an FITC-conjugated anti–rabbit IgG Ab. After rinsing with PBS containing 0.2 μg/mL 4′, 6-diamidino-2-phenylindole dihydrochloride (DAPI), the cells were incubated with an anti-PML Ab and then with a rhodamine-conjugated anti–goat IgG Ab. The stained cells were observed under a confocal laser microscope (Zeiss LSM410, Obercochen, Germany).
Results
PML repressed STAT3 activity through the complex formation
At first, we examined the effect of PML on STAT3 activity in NIH3T3 and 293T cells with luciferase assays using 4 × APRE-Luc. In NIH3T3 cells, IL-6–induced STAT activity was suppressed by PML in a dose-dependent manner (Figure1A). Similarly, PML inhibited gp130-mediated or G-CSF-R–mediated STAT3 activity in 293T and HepG2 cells (Figure 1B-C). In addition, PML was found to suppress G-CSF-R–mediated STAT3 activity in a hematopoietic myeloid cell line, 32D (Figure 1D). By contrast, PML did not affect IFNα-induced STAT1 activity or G-CSF–induced STAT1 activity in HepG2 cells (Figure 1E; data not shown). Also, it did not influence 1*6-STAT5 (constitutively active STAT5) activity in NIH3T3 cells (Figure 1F) and did not repress EPO-induced or G-CSF–induced STAT5 activity in HepG2 cells (Figure 1G; data not shown). These results indicate that PML specifically represses STAT3 activity.
PML binds to STAT3 and inhibits its activity.
NIH3T3 (A,F), 293T (B), HepG2 (C,E,G), and 32D (D) cells were transfected with an appropriate reporter gene (4 × APRE-Luc [A-D], 4 × ISRE-Luc [E], or 3 × β-Cas-Luc [F-G]) along with the indicated expression vectors. Also, 293T cells were transfected with pEF-BOS-G-CSF-R/gp130 and HepG2 cells with G-CSF-R. Relative luciferase activities induced by IL-6, G-CSF, EPO, IFNα, or 1*6-STAT5 were quantitated. The results are shown as the means ± standard deviations (SDs) of triplicate cultures. Similar results were obtained from at least 4 independent experiments. (H-I) 293T cells were transfected with pEF-BOS-G-CSF-R/gp130 and the indicated expression vectors. After G-CSF stimulation, total cell lysates were isolated and subjected to immunoprecipitation (IP) and immunoblotting (IB) with the indicated antibodies.
PML binds to STAT3 and inhibits its activity.
NIH3T3 (A,F), 293T (B), HepG2 (C,E,G), and 32D (D) cells were transfected with an appropriate reporter gene (4 × APRE-Luc [A-D], 4 × ISRE-Luc [E], or 3 × β-Cas-Luc [F-G]) along with the indicated expression vectors. Also, 293T cells were transfected with pEF-BOS-G-CSF-R/gp130 and HepG2 cells with G-CSF-R. Relative luciferase activities induced by IL-6, G-CSF, EPO, IFNα, or 1*6-STAT5 were quantitated. The results are shown as the means ± standard deviations (SDs) of triplicate cultures. Similar results were obtained from at least 4 independent experiments. (H-I) 293T cells were transfected with pEF-BOS-G-CSF-R/gp130 and the indicated expression vectors. After G-CSF stimulation, total cell lysates were isolated and subjected to immunoprecipitation (IP) and immunoblotting (IB) with the indicated antibodies.
To explore the mechanism by which PML suppressed STAT3 activity, we examined the in vivo association by coimmunoprecipitation experiments in 293T cells. As shown in Figure 1H, PML was coimmunoprecipitated with STAT3 only when PML and STAT3 were cotransfected (upper panel, lane 8) and vice versa (lower panel, lane 7). Because a negative control Ab (a normal rabbit IgG) did not immunoprecipitate PML (Figure 1I, lane 4), we considered that these reactions were specifically performed by anti-PML and anti-STAT3 Abs.
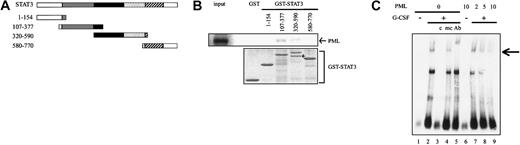
Next, we examined the in vitro binding between PML and several GST-STAT3 fusion proteins (Figure 2A). Because we could not obtain the GST fusion protein containing full-length STAT3 as a soluble protein due to the formation of the inclusion body, we performed GST pulldown experiments using GST-STAT3, each containing the truncated fragment of STAT3. At first, we examined the quality and quantity of fusion proteins by Coomassie brilliant blue staining (Figure 2B, lower panel) and found that purified GST-STAT3 (320-590) partially includes the degraded product (indicated by an arrow). However, we could not prevent this degradation in spite of the repeated experiments. Next, we examined which fusion protein bound to35S-labeled PML. As shown in Figure 2B, upper panel, PML bound to GST-STAT3 (107-377) and also, to a lesser degree, to GST-STAT3 (320-590). In contrast, PML scarcely bound to GST alone, GST-STAT3 (1-154), or GST-STAT3 (580-770). These results imply that STAT3 may bind to PML through the domain spanning amino acids 320-377. However, given that GST-STAT3 (107-377) bound to PML with stronger affinity than GST-STAT3 (320-590), the other domain included by GST-STAT3 (103-377), such as the coiled-coil domain, might enhance and/or stabilize their binding. Because amino acids 320-377 reside in the DNA binding domain, we examined the effect of PML on the DNA binding activity of STAT3 with EMSA. As shown in Figure 2C, the nuclear extract prepared from G-CSF–treated 293T cells bound to the probe containing the STAT3 binding sequence (Figure 2C, lane 2), which was canceled by the wild-type competitor (Figure 2C, lane 3) but not by the mutant competitor (Figure 2C, lane 4) and supershifted by an anti-STAT3 Ab (Figure 2C, lane 5), indicating that this DNA binding complex was formed from STAT3. When PML was cotransfected, PML reduced this DNA binding complex in a dose-dependent manner (Figure 2C, lanes 7-9). In contrast, PML showed no effect on the DNA binding activity of IFNα-activated STAT1 or EPO-activated STAT5 (data not shown), again suggesting that PML would specifically inhibit STAT3 activity.
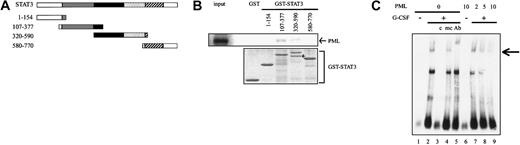
PML inhibits DNA binding activity of STAT3.
(A) Functional domains of STAT3 and the structure of GST-STAT3. Gray indicates coiled-coil domain; black, DNA binding domain; dotted, linker domain; and hatched, SH2 domain. (B) In vitro binding between PML and GST-STAT3 was examined by GST pulldown assays (upper panel). Coomassie brilliant blue staining of each GST-STAT3 protein (lower panel). An arrow indicates the partially degraded product. (C) 293T cells expressing STAT3 and G-CSF-R/gp130 with or without PML were cultured in the presence or absence of 30 ng/mL G-CSF for 30 minutes. Then, nuclear extracts were isolated and subjected to EMSA. An arrow indicates the DNA binding complex including STAT3. c indicates wild-type competitor; mc, mutant competitor; Ab, anti-STAT3 Ab.
PML inhibits DNA binding activity of STAT3.
(A) Functional domains of STAT3 and the structure of GST-STAT3. Gray indicates coiled-coil domain; black, DNA binding domain; dotted, linker domain; and hatched, SH2 domain. (B) In vitro binding between PML and GST-STAT3 was examined by GST pulldown assays (upper panel). Coomassie brilliant blue staining of each GST-STAT3 protein (lower panel). An arrow indicates the partially degraded product. (C) 293T cells expressing STAT3 and G-CSF-R/gp130 with or without PML were cultured in the presence or absence of 30 ng/mL G-CSF for 30 minutes. Then, nuclear extracts were isolated and subjected to EMSA. An arrow indicates the DNA binding complex including STAT3. c indicates wild-type competitor; mc, mutant competitor; Ab, anti-STAT3 Ab.
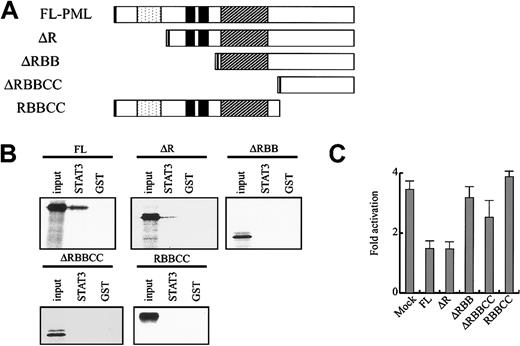
Next, we examined the in vitro binding between GST-STAT3 (107-377) and several PML mutants (Figure 3A). A mutant lacking the RING domain (ΔR) still bound to GST-STAT3 (107-377), whereas its affinity was lower than that of full-length (FL)–PML. In contrast, Δ RING-B-box (RBB), ΔRBB coiled coil (CC), and RBBCC scarcely bound to GST-STAT3 (107-377) (Figure 3B). The similar results were obtained from GST-STAT3 (320-590) (data not shown). Consistent with the data on the in vitro binding, ΔR repressed STAT3 activity as efficiently as FL-PML, whereas ΔRBB, ΔRBBCC, and RBBCC showed little effect on STAT3 activity (Figure 3C). These results suggest that PML directly binds to STAT3 and inhibits its activity through B-box zinc finger and carboxyl-terminal domains.
PML binds to STAT3 and inhibits its activity through B-box and carboxy-terminal domains.
(A) The structure of PML mutants. Dotted indicates RING domain; filled, B1 and B2 boxes; and hatched, coiled-coil domain. (B) In vitro binding between GST-STAT3 (107-377) and PML mutants was examined by GST pulldown assays. (C) NIH3T3 cells were transfected with 4 × APRE-Luc and the effector gene indicated. IL-6–induced fold activation is indicated. The results are shown as the means ± SDs of triplicate cultures. Similar results were obtained from at least 4 independent experiments.
PML binds to STAT3 and inhibits its activity through B-box and carboxy-terminal domains.
(A) The structure of PML mutants. Dotted indicates RING domain; filled, B1 and B2 boxes; and hatched, coiled-coil domain. (B) In vitro binding between GST-STAT3 (107-377) and PML mutants was examined by GST pulldown assays. (C) NIH3T3 cells were transfected with 4 × APRE-Luc and the effector gene indicated. IL-6–induced fold activation is indicated. The results are shown as the means ± SDs of triplicate cultures. Similar results were obtained from at least 4 independent experiments.
PML/RARα restored STAT3 activity inhibited by PML
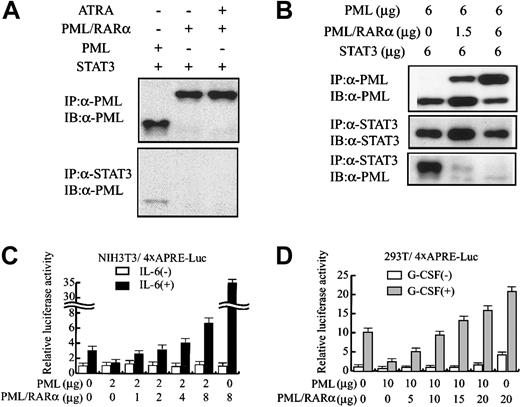
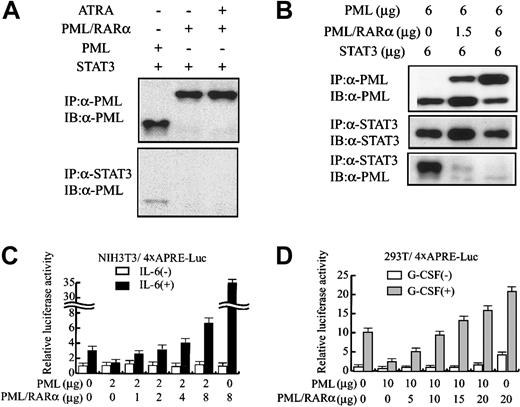
Next, we examined whether PML/RARα interacts with STAT3 in 293T cells. In contrast to PML (Figure 4A, lane 1), PML/RARα was not coimmunoprecipitated with STAT3 regardless of the treatment with all-trans retinoic acid (Figure 4A, lanes 2-3). However, cotransfected PML/RARα dose-dependently inhibited the in vivo binding between PML and STAT3 (Figure 4B). In addition, PML/RARα canceled the inhibitory effects of PML on STAT3 activity and augmented IL-6–induced 4 × APRE-Luc activity up to 33-fold in NIH3T3 cells (Figure 4C). Together, these results suggest that PML/RARα dissociates PML from STAT3 and cancels its inhibitory effects on STAT3 activity. To further provide insight into the effects of PML and PML/RARα on STAT3 activity, we performed a similar analysis with 293T cells, in which endogenous PML was hardly detectable by Western blotting (Figure 1). As was the case with NIH3T3 cells, PML/RARα restored gp130-mediated STAT3 activity suppressed by PML (Figure 4D). When PML/RARα was transfected alone, PML/RARα stimulated 4 × APRE-Luc activity up to 4-fold even without G-CSF stimulation. Given that PML/RARα did not stimulate the backbone reporter gene lacking the 4 × APRE element in 293T cells (data not shown), it was speculated that some proportion of STAT3 already was located in the nucleus and revealed its basal activity even without G-CSF stimulation and that PML/RARα enhanced this basal activity. Moreover, PML/RARα augmented G-CSF–induced STAT3 activity from 10-fold to 20-fold even in the absence of PML (Figure 4D). These results indicate that PML/RARα not only restores STAT3 activity repressed by PML but also augments STAT3 activity by itself.
PML/RARα restores STAT3 activity inhibited by PML.
(A-B) 293T cells were transfected with pEF-BOS-G-CSF-R/gp130 and the indicated amounts of expression vectors, and cultured with or without 1 μM all-trans retinoic acid (ATRA). Total cell lysates prepared after G-CSF treatment were subjected to coimmunoprecipitation analyses. (C) NIH3T3 cells were transfected with 4 × APRE-Luc and the effector genes indicated. After IL-6 stimulation, relative luciferase activities were quantitated. (D) 293T cells were transfected with pEF-BOS-G-CSF-R/gp130, 4 × APRE-Luc, and the indicated amounts of effector genes. After G-CSF stimulation, relative luciferase activities were quantitated. The results are shown as the means ± SDs of triplicate cultures. Similar results were obtained from at least 4 independent experiments.
PML/RARα restores STAT3 activity inhibited by PML.
(A-B) 293T cells were transfected with pEF-BOS-G-CSF-R/gp130 and the indicated amounts of expression vectors, and cultured with or without 1 μM all-trans retinoic acid (ATRA). Total cell lysates prepared after G-CSF treatment were subjected to coimmunoprecipitation analyses. (C) NIH3T3 cells were transfected with 4 × APRE-Luc and the effector genes indicated. After IL-6 stimulation, relative luciferase activities were quantitated. (D) 293T cells were transfected with pEF-BOS-G-CSF-R/gp130, 4 × APRE-Luc, and the indicated amounts of effector genes. After G-CSF stimulation, relative luciferase activities were quantitated. The results are shown as the means ± SDs of triplicate cultures. Similar results were obtained from at least 4 independent experiments.
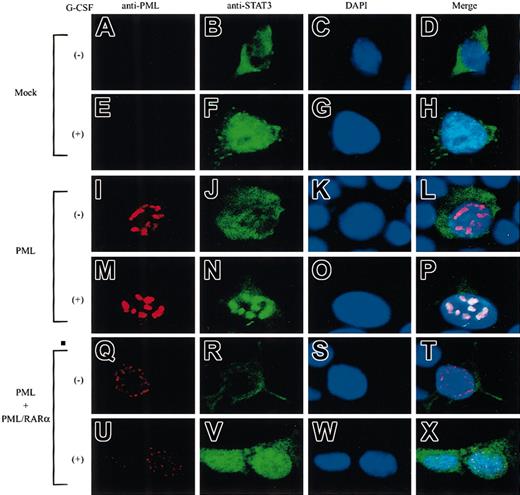
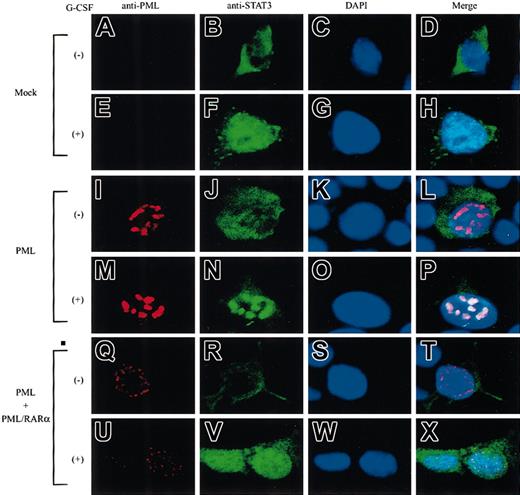
Next, we examined the localization of PML and STAT3 by immunocytochemical analyses in 293T cells. PML was hardly detectable in mock-transfected cells (Figure 5A,E), whereas it localized in the nucleus and formed NBs in PML-transfected cells (Figure 5I,M). When PML and PML/RARα were cotransfected, both molecules formed microspeckled distribution in the nucleus (Figure5Q,U). Before G-CSF stimulation, STAT3 was dispersed in the cytoplasm (Figure 5B). After G-CSF stimulation, STAT3 translocated into the nucleus and showed a diffuse distribution pattern in mock-transfected cells (Figure 5F), whereas it was concentrated into NBs in PML-transfected cells (Figure 5N). Superimposition of the staining for PML and STAT3 showed that PML-NBs were colocalized with STAT3-containing NBs (in 198 of 200 cells examined) (Figure 5P). Importantly, when PML and PML/RARα were cotransfected, activated STAT3 revealed a diffuse distribution pattern in the nucleus, as in mock-transfected cells (Figure 5V). In this condition, colocalization of PML and STAT3 was observed in only 19 of 200 cells examined. These results suggest that STAT3 colocalizes with PML in NBs and that this colocalization is perturbed by PML/RARα.
STAT3 colocalizes with PML but not with PML/RARα in NBs.
293T cells were transfected with STAT3 and G-CSF-R/gp130, together with a mock vector (A-H), PML alone (I-P), or PML + PML/RARα (Q-X). Cells were stained with an anti-PML Ab (A,E,I,M,Q,U), an anti-STAT3 Ab (B,F,J,N,R,V), and DAPI (C,G,K,O,S,W). Original magnifications, ×400.
STAT3 colocalizes with PML but not with PML/RARα in NBs.
293T cells were transfected with STAT3 and G-CSF-R/gp130, together with a mock vector (A-H), PML alone (I-P), or PML + PML/RARα (Q-X). Cells were stained with an anti-PML Ab (A,E,I,M,Q,U), an anti-STAT3 Ab (B,F,J,N,R,V), and DAPI (C,G,K,O,S,W). Original magnifications, ×400.
STAT3-dependent growth of Ba/F3 cells was inhibited by PML and enhanced by PML/RARα
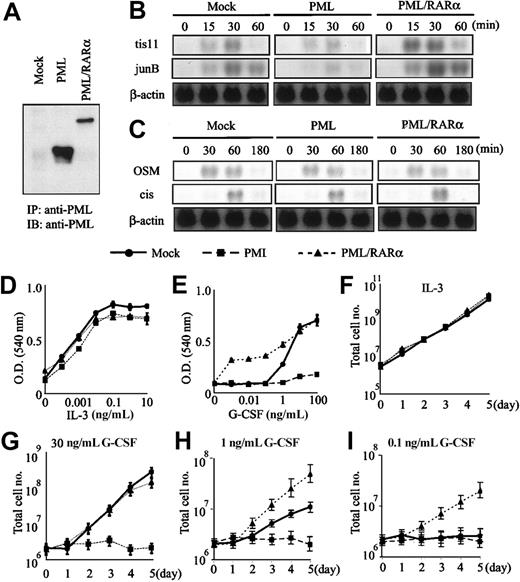
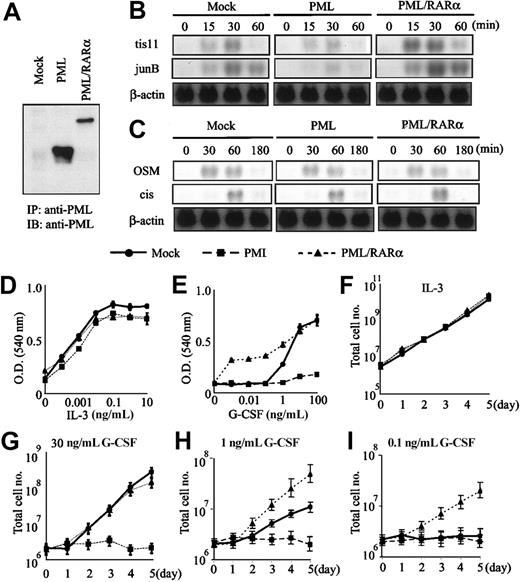
To assess the biologic significance of the interaction among PML, PML/RARα, and STAT3, we transfected PML, PML/RARα, and mock (an empty expression vector) into IL-3–dependent Ba/F3 cells expressing G-CSF-R/gp130, in which gp130-mediated growth is essentially dependent on STAT3 activity.21 At first, we screened the expression levels of each transgene and selected 5 single cell clones from the respective transfectants in which PML or PML/RARα was expressed efficiently (the expression levels of PML and PML/RARα in each representative clone is shown in Figure6A). When these clones were stimulated with G-CSF, the induction of STAT3-responsive genes, tis11 and junB, was reduced in PML-transfected clones as compared with that in PML/RARα-transfected clones (the data obtained from a representative clone is shown in Figure 6B). In contrast, PML and PML/RARα hardly affected IL-3–induced expression of STAT5-responsive genes, oncostatin M (OSM), and cis (Figure 6C). The similar pattern of gene responses was observed in the other 4 transfectants of PML or PML/RARα (data not shown). Next, we analyzed the growth characteristics of these transfectants under the culture with IL-3 or G-CSF. To avoid the biased results due to the analysis on the single clone, we mixed 5 clones with an equal ratio (each 20%) and prepared the mixed clones from the respective transfectants. Each of 5 single-cell clones and the mixed clone showed similar growth curves and dose responses under the culture with IL-3 or G-CSF (data not shown). We show the results obtained from the mixed clones in Figure 6D-I. As shown in Figure 6D, there was no significant difference in the dose responses to IL-3 among mock-transfected, PML-transfected, and PML/RARα-transfected mixed clones, and these clones showed similar growth curves under the culture with IL-3 for 5 days (Figure 6F). When these mixed clones were cultured with various concentrations of G-CSF, the PML-transfected mixed clone was hardly responsive to G-CSF even at a concentration of 100 ng/mL. In contrast, the PML/RARα-transfected mixed clone showed proliferative response to a low concentration of G-CSF, at which the mock clone hardly proliferated (Figure 6E). Furthermore, time course analysis showed that the PML-transfected mixed clone did not proliferate in response to G-CSF at the concentration of 0.1, 1, or 10 ng/mL for 5 days (Figure 6G-I). By contrast, the PML/RARα-transfected mixed clone was more responsive to 1 ng/mL G-CSF than the mock-transfected clone (Figure 6H). In addition, the PML/RARα-transfected clone was still able to proliferate in response to 0.1 ng/mL G-CSF, whereas the mock-transfected clone hardly proliferated (Figure 6I). These data indicate that STAT3-dependent growth of Ba/F3 cells is inhibited by PML, whereas it is augmented by PML/RARα. In spite of the marginal effect of PML on STAT3-dependent gene expression, PML completely abrogated gp130-mediated proliferation in Ba/F3 cells. As for this reason, we speculated that cyclin-dependent kinase activities regulated by STAT3 might be abrogated by a partial loss of its target genes such as c-myc and cyclin D1.16-21
STAT3-dependent growth of Ba/F3 cells is inhibited by PML and augmented by PML/RARα.
PML and PML/RARα were introduced into Ba/F3 cells expressing G-CSF-R/gp130. According to the expression levels of the transgene, 5 clones in which PML or PML/RARα was expressed efficiently were selected from the respective transfectants. These 5 clones were mixed with an equal ratio to prepare the mixed clones from the respective transfectants. (A) The protein expression level of PML and PML/RARα in a representative clone of each transfectant. (B-C) The responses to IL-3 and G-CSF in a representative clone of each transfectant. After IL-3 deprivation, the cells were stimulated with 30 ng/mL G-CSF (B) or 1 ng/mL IL-3 (C). Induction levels of STAT3-responsive (B) or STAT5-responsive (C) genes were examined by Northern blot analysis. (D-I) The growth characteristics of respective transfectants were examined in 5 single-cell clones and the mixed clone. All of 5 clones and the mixed clone showed similar growth responses to IL-3 or G-CSF. ● indicates mock; ▪ PML; ▴ PML/RARα. The data obtained from the mixed clones were shown as the means ± SDs of triplicate cultures. (D-E) Dose responses to IL-3 and G-CSF were examined by MTT assays. O.D. indicates optical density. (F-I) Total cell number was counted under the culture with 1 ng/mL IL-3 (F), 30 ng/mL (G), 1 ng/mL (H), or 0.1 ng/mL (I) G-CSF.
STAT3-dependent growth of Ba/F3 cells is inhibited by PML and augmented by PML/RARα.
PML and PML/RARα were introduced into Ba/F3 cells expressing G-CSF-R/gp130. According to the expression levels of the transgene, 5 clones in which PML or PML/RARα was expressed efficiently were selected from the respective transfectants. These 5 clones were mixed with an equal ratio to prepare the mixed clones from the respective transfectants. (A) The protein expression level of PML and PML/RARα in a representative clone of each transfectant. (B-C) The responses to IL-3 and G-CSF in a representative clone of each transfectant. After IL-3 deprivation, the cells were stimulated with 30 ng/mL G-CSF (B) or 1 ng/mL IL-3 (C). Induction levels of STAT3-responsive (B) or STAT5-responsive (C) genes were examined by Northern blot analysis. (D-I) The growth characteristics of respective transfectants were examined in 5 single-cell clones and the mixed clone. All of 5 clones and the mixed clone showed similar growth responses to IL-3 or G-CSF. ● indicates mock; ▪ PML; ▴ PML/RARα. The data obtained from the mixed clones were shown as the means ± SDs of triplicate cultures. (D-E) Dose responses to IL-3 and G-CSF were examined by MTT assays. O.D. indicates optical density. (F-I) Total cell number was counted under the culture with 1 ng/mL IL-3 (F), 30 ng/mL (G), 1 ng/mL (H), or 0.1 ng/mL (I) G-CSF.
Discussion
We demonstrated here that STAT3 activity is inhibited by PML and augmented by PML/RARα. As for this mechanism, we found that PML directly bound to STAT3 in NBs and thereby inhibited the DNA binding activity of STAT3. In contrast, PML/RARα did not associate with STAT3 but inhibited the binding between PML and STAT3, probably through the heterodimerization with PML. In the present study, GST pulldown assays revealed that STAT3 binds to the B-box and C-terminal domains of PML. Based on the previous report that PML/RARα and PML form a complex through the respective coiled-coil domains,12 it seems strange that PML/RARα inhibits the binding between PML and STAT3. However, considering that the PML oligomer is able to form NBs through the B-box domain26 while the PML-PML/RARα heterodimer lacks this ability, the structure of the B-box domain in the PML-PML/RARα heterodimer was supposed be somewhat different from that in the PML oligomer. Therefore, as a probable interpretation of this result, it was speculated that, if PML more preferentially binds to PML/RARα than to STAT3, STAT3 might not be capable of binding to the PML-PML/RARα heterodimer any more, due to the conformational change of the B-box domain. Taken together, our results suggest that PML/RARα would enhance STAT3 activity by reducing inhibitory effects of PML. However, we also found that PML/RARα augmented gp130-mediated STAT3 activity in 293T cells even in the absence of PML. Therefore, it was speculated that PML/RARα by itself could enhance STAT3 activity independently of PML through an as-yet-undetermined mechanism. Because STAT3 activity is modulated by various binding molecules such as Smad1, c-Jun, and p300,27-29 further studies focusing on the effects of PML/RARα on these interactions would undoubtedly provide more clear information as to this molecular mechanism.
STAT3 is activated by various growth factors such as G-CSF, IL-6, and OSM in hematopoietic cells. Activated STAT3 regulates cytokine-dependent growth of hematopoietic cells by promoting G1/S progression through the induction of c-myc and cyclin D1.16-21 In addition to its function in normal hematopoiesis, STAT3 was aberrantly activated in peripheral blood samples obtained from the patients with acute myeloid leukemia, lymphoblastic leukemia, and polycytemia vera.17Furthermore, STAT3 activity was shown to be enhanced by STAT5b-RARα, another type of fusion protein identified in APL.30 These lines of evidence suggest that dysregulated STAT3 activity would play some role in malignant transformation of normal cells. Supporting this hypothesis, dominant-negative STAT3 was reported to abrogate v-Src–dependent proliferation of hematopoietic cells.31Furthermore, Bromberg et al32 showed that constitutively active STAT3 enabled a fibroblast cell line to form colonies in soft agar assays and to develop tumors in nude mice.
In our study, PML/RARα by itself induced neither phosphorylation nor activation of STAT3 in Ba/F3 cells (data not shown). However, PML/RARα potentiated gp130-mediated STAT3 activity and consequent STAT3-dependent growth of Ba/F3 cells. Therefore, it was assumed that dysregulated STAT3 activity by PML/RARα would enhance cytokine-dependent growth of APL cells and affect some aspects of the pathophysiology of APL. This speculation is largely coincident with the previous findings that transgenic mice that express PML/RARα exclusively in myeloid/promyelocytic cells reveal abnormal hematopoiesis like myeloproliferative disorders before the onset of APL.33 Further studies to determine the role of dysregulated STAT3 in the growth and differentiation arrest of APL cells may be useful to understand the mechanism underlying leukemogenesis of APL.
We acknowledge Dr T. Kitamura and Dr M. Alcalay for providing the plasmids.
Prepublished online as Blood First Edition Paper, December 27, 2002; DOI 10.1182/blood-2002-08-2474.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Itaru Matsumura, Department of Hematology and Oncology, Osaka University Graduate School of Medicine, C9, 2-2 Yamada-oka, Suita, Osaka, 565-0871, Japan; e-mail:matumura@bldon.med.osaka-u.ac.jp.

![Fig. 1. PML binds to STAT3 and inhibits its activity. / NIH3T3 (A,F), 293T (B), HepG2 (C,E,G), and 32D (D) cells were transfected with an appropriate reporter gene (4 × APRE-Luc [A-D], 4 × ISRE-Luc [E], or 3 × β-Cas-Luc [F-G]) along with the indicated expression vectors. Also, 293T cells were transfected with pEF-BOS-G-CSF-R/gp130 and HepG2 cells with G-CSF-R. Relative luciferase activities induced by IL-6, G-CSF, EPO, IFNα, or 1*6-STAT5 were quantitated. The results are shown as the means ± standard deviations (SDs) of triplicate cultures. Similar results were obtained from at least 4 independent experiments. (H-I) 293T cells were transfected with pEF-BOS-G-CSF-R/gp130 and the indicated expression vectors. After G-CSF stimulation, total cell lysates were isolated and subjected to immunoprecipitation (IP) and immunoblotting (IB) with the indicated antibodies.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/9/10.1182_blood-2002-08-2474/4/m_h80934218001.jpeg?Expires=1767730608&Signature=hw~6iJ9rdz1F5JJOhom2u9HRqH5Ufwvihda~zkafeD7kk9k9dB-XwQwE3q8LDmvhRvps9gG5E9WMiLY0zvgpdwdr5pF-p2T1skC0bi~Gdb7jPx8T63Pda1le2mMqQDZQcTD-t4batxMhyxDr1TtM1ddywiyDssrcqn10tUSZV5NAdXfIxFLeLAsojKTurdRlqQ4fKUWTd~NJvq6lEXZ6-AfZtzOGSn-gTNyxSWuG4XakR3LxV~DjLBXUQhiSX7-hSuLAVtSHp-Q8kJaET06iLUxd4AejJNXOXP9~AklJYWuMmH4234Dvh51Iydm1PtXSKrKeu6EVx54LByouJVuuaw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 1. PML binds to STAT3 and inhibits its activity. / NIH3T3 (A,F), 293T (B), HepG2 (C,E,G), and 32D (D) cells were transfected with an appropriate reporter gene (4 × APRE-Luc [A-D], 4 × ISRE-Luc [E], or 3 × β-Cas-Luc [F-G]) along with the indicated expression vectors. Also, 293T cells were transfected with pEF-BOS-G-CSF-R/gp130 and HepG2 cells with G-CSF-R. Relative luciferase activities induced by IL-6, G-CSF, EPO, IFNα, or 1*6-STAT5 were quantitated. The results are shown as the means ± standard deviations (SDs) of triplicate cultures. Similar results were obtained from at least 4 independent experiments. (H-I) 293T cells were transfected with pEF-BOS-G-CSF-R/gp130 and the indicated expression vectors. After G-CSF stimulation, total cell lysates were isolated and subjected to immunoprecipitation (IP) and immunoblotting (IB) with the indicated antibodies.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/9/10.1182_blood-2002-08-2474/4/m_h80934218001.jpeg?Expires=1768329202&Signature=jur5qhvSDa7noiLwlB70tsO-ZBzCBFG8OvEQiNArEfdL-mT5SuviBaNO4rd1qDve-Nfoqo1L~ouv1urEipCVz0kNOZy-OnY47Ggw5gp6LnYceZVgMlOhOYpzRswEd4dWimFQt-rXUtiyuyFCvk7yC~u-dS8V-dLJScRPYM6MbZpmroOYO4V0K28Mi7s23~3k0NbXB7hxNtAfyl8-FK2Y2N1dTtCIzjGcVFGtMmVkswxNKu8xbegdwWY4NJtbezybEKD6aJTB-6n4kQSXIitnGeF9v8YHdOYc247cc8oPqsji5FJpatzwG4JLZMZa1myrL4bKoNPTDe~aQxT~vwOrLQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)