Abstract
To date, the diagnosis of polycythemia vera (PV) relies on clinical criteria. We have recently described the overexpression of a hematopoietic receptor, polycythemia rubra vera-1 (PRV-1), in patients with PV. Here, we report a quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) assay for the measurement of PRV-1 mRNA levels. We have determined PRV-1 expression in 71 patients with PV, 11 patients with secondary erythrocytosis (SE), as well as in 80 healthy controls. PV patients express significantly higher amounts of PRV-1 than healthy controls or patients with SE (P < .0001). Because there is no overlap between the PRV-1 expression in PV patients versus healthy controls or SE patients, the assay has a very high sensitivity and specificity for the diagnosis of PV in our population. In patients with erythrocytosis, the quantitative RT-PCR assay described here therefore provides a rapid, highly specific and sensitive tool for the diagnosis of PV. (Blood. 2003;102: 3569-3574)
Introduction
Primary polycythemia results from an intrinsic defect in a hematopoietic stem cell, which causes an increased production of erythrocytes. The most common form of primary polycythemia is polycythemia vera (PV), one of a group of 4 acquired disorders collectively termed the chronic myeloproliferative disorders (cMPDs). Besides PV, this category also includes essential thrombocythemia (ET), idiopathic myelofibrosis (IMF), and chronic myeloid leukemia (CML). Today, CML is regarded as a separate entity, defined by a 9;22(q34;q11) translocation, termed the “Philadelphia chromosome” (Ph-), which results in production of the Bcr/Abl fusion protein. The Philadelphia chromosome is not found in any of the 3 remaining subtypes of cMPD.1,2 Consequently, they have been termed the “Ph-negative cMPDs.”
Unlike CML, the 3 Ph-negative cMPDs lack a sensitive and specific molecular marker. Therefore, the differentiation between PV and secondary erythrocytosis (SE) and, hence, the diagnosis of PV relies on clinical criteria.3 Different sets of diagnostic criteria for PV have been proposed. The classical criteria were established by the Polycythemia Vera Study Group (PVSG)4,5 and for many years remained the gold standard for clinical trials. The PVSG criteria served as a basis for several more recent revisions.6-9
The revised criteria make use of methods that were not available when the PSVG criteria were defined. These include the formation of endogenous erythroid colonies (EECs) as one diagnostic tool. Unlike healthy progenitors, PV stem cells form erythroid colonies in semisolid medium in the absence of erythropoietin (Epo).10-12 In patients with erythrocytosis, EECs are specific for PV and can be used to distinguish PV from SE.13-16 However, the EEC assay is technically demanding and not available in most hematologic laboratories. It has therefore been included in the revised criteria only as a facultative major (“A”) criterion or as a minor (“B”) criterion. Thus, to date, there is no readily and widely available molecular marker for the diagnosis of PV.
We have recently shown that polycythemia rubra vera-1 (PRV-1), a novel member of the urokinase-type plasminogen activator receptor (uPAR) superfamily, is overexpressed in mature peripheral blood granulocytes from patients with PV, but not in a variety of controls including healthy individuals, patients with SE, and patients with CML.17 Here we describe the development of a quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) assay for the measurement of PRV-1 mRNA expression. Analysis of a large cohort of patients with PV and SE as well as healthy controls demonstrates that quantification of PRV-1 mRNA levels represents a highly sensitive and specific molecular assay for the diagnosis of PV in patients with erythrocytosis.
Patients, materials, and methods
Patients
Peripheral blood samples were obtained from 71 patients with PV, and 11 patients with SE treated at 7 hematologic centers. Venous blood was anticoagulated with EDTA (ethylenediaminetetraacetic acid) or heparin and shipped by courier to the central laboratory without cooling. Maximum time interval between venipuncture and arrival in the laboratory was 24 hours. Eighty blood donors served as healthy controls.
Of the 71 PV patients, 7 were diagnosed according to the PVSG criteria (hereafter called PV-PVSG), while 64 patients were classified according to the WHO criteria (PV-WHO), which are reproduced in Table 1. All clinical and laboratory procedures required for classification by either the WHO or PVSG criteria were the responsibility of the participating clinical institutions. Diagnostic criteria according to PVSG or WHO had to be met either at the time of diagnosis or at any time between diagnosis and blood sampling.
Patients with SE displayed a documented cause for secondary erythrocytosis; in all cases analyzed, this was chronic obstructive pulmonary disease (COPD).
Separation of cells
Anticoagulated blood from patients and healthy controls was used as a source of granulocytes and mononuclear cells. Granulocytes were purified by dextran sedimentation followed by Ficoll-Paque (Pharmacia, Freiburg, Germany) separation.18 Erythrocytes were eliminated by hypotonic lysis (0.2% NaCl for 30 seconds). This method consistently yielded granulocyte preparations of greater than 98% purity as judged by visual inspection of Wright-Giemsa-stained slide preparations. Peripheral blood mononuclear cells were isolated by Ficoll-Paque separation.
RNA isolation and Northern blots
Total cellular RNA was harvested using an acidic phenol extraction (Trizol; Gibco/BRL, Karsruhe, Germany or Trireagent; Sigma, Deisenhofen, Germany). RNA (10 μg) was analyzed in a Northern blot. The blots were hybridized in ExpressHyb Hybridization Solution (Clontech, Heidelberg, Germany) at 68°C. PRV-1 cDNA probes were labeled using the Prime-It-II labeling kit (Stratagene, Amsterdam, the Netherlands) and α-32 P deoxycytidine triphosphate (dCTP; Amersham, Freiburg, Germany). The blots were washed 3 times for 10 minutes in 2 × sodium sodium citrate (SSC), 0.05% sodium dodecyl sulfate (SDS) at room temperature and twice at 50°C for 20 minutes in 0.1 × SSC, 0.1% SDS. Hybridization was detected by autoradiography.
Taqman quantitative RT-PCR for PRV-1 and GAPDH
Triplicate measurements of PRV-1 and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA levels were made using 5 or 50 ng total RNA per reaction. Each PRV-1 reaction consisted of 25 μL 2 × MasterMix without uracil-DNA glycosylase (UNG), 1.25 μL 40 × Multiscribe and RNase Inhibitor Mix, 5 μL PRV-1 forward primer (9 μM; 5′-GCTGTCCACCAAAATGAGCAT-3′), 5 μL PRV-1 reverse primer (0.5 μM; 5′-TTCTCACGCGCAGAGAAGATC-3′), and 5 μL PRV-1 probe (2.5 μM; 5′-FAM-TTCTTGTTGAACCACACCAGACAAATCGG-TAMRA-3′) in a total volume of 50 μL. Each GAPDH reaction consisted of 25 μL 2 × MasterMix without UNG, 1.25 μL 40 × Multiscribe and RNase Inhibitor Mix, 1 μL GAPDH forward primer (10 μM), 1 μL GAPDH reverse primer (10 μM), and 1 μL GAPDH probe (5 μM) in a total volume of 50 μL. GAPDH primers and probe were taken from the Taqman GAPDH Control Reagents (no. 402869; Applied Biosystems, Darmstadt, Germany). Both 2 × MasterMix without UNG and 40 × Multiscribe and RNase Inhibitor Mix were components of the Taqman One Step RT-PCR Master Mix Reagent Kit (no. 4309169; Applied Biosystems). Reactions were carried out for 30 minutes at 48°C followed by 10 minutes at 95°C and a subsequent amplification using 40 cycles of 15 seconds at 95°C, 1 minute at 60°C. MicroAmp 96-well reaction plates (no. N801-0560; Applied Biosystems) sealed with MicroAmp optical caps (no. N801-0935; Applied Biosystems) were used in a Taqman ABI PRISM 7700 or 7000 Sequence Detection System. Data collected during cycling were analyzed with the FAM threshold set at 0.2 and the JOE threshold set at 0.04 on the Taqman ABI PRISM 7700, or with the FAM threshold set at 0.4 and the JOE threshold set at 0.15 on the Taqman ABI PRISM 7000. A mean cycle of threshold (CT) value for each triplicate measurement was calculated. Subsequently, a CT PRV-1/CT GAPDH ratio was determined for each sample.
For the report of quantitative PCR results, either the CT ratio or a ΔCT can be used. However, use of the ΔCT method is valid only if the 2 PCR reactions (PRV-1 and GAPDH) achieve the same efficiency of amplification. With the primer and probe combinations used here, the PRV-1 and GAPDH efficiencies are not equal, and therefore, use of the ΔCT method is not appropriate in our hands.
Statistics
The PRV-1/GAPDH ratios are not normally distributed. Therefore, the significance of the difference of the means of PRV-1/GAPDH ratios for PV patients, SE patients, and healthy controls was calculated using the Kruskal-Wallis analysis of variance on ranks and the Dunn method for pairwise multiple comparisons.19
Results
Quantification of PRV-1 mRNA by RT-PCR
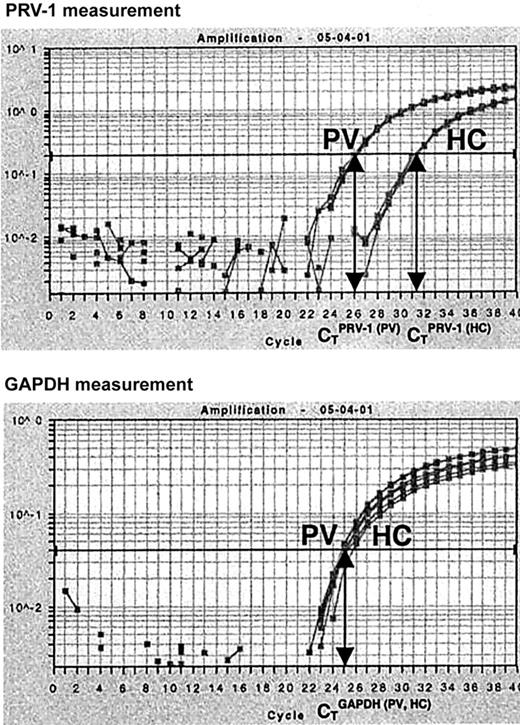
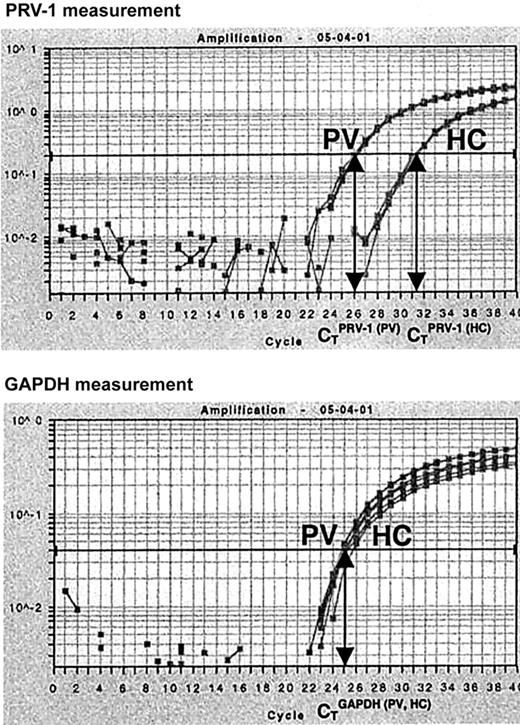
In order to quantify PRV-1 mRNA expression, we developed an RT-PCR assay based on TaqMan technology. Total RNA from purified peripheral granulocytes was subjected to reverse transcription and PCR amplification in a single reaction tube. For each RNA sample, both PRV-1 and GAPDH mRNA levels were determined. In a TaqMan PCR, the amount of cDNA generated is quantified by measuring the amount of fluorescence emitted from a gene-specific probe. Hence, for each reaction, the PCR cycle in which a preset level of fluorescence is reached can be recorded. This cycle is called the cycle of threshold (CT). We determined the CT for PRV-1 and GAPDH in each sample. Subsequently, a CTPRV-1/CTGAPDH ratio was calculated (Figure 1).
Quantification of PRV-1 mRNA by Taqman RT-PCR. Total RNA (5 ng) from a PV patient (PV) and from a healthy control (HC) was subjected to quantitative RT-PCR for PRV-1 and GAPDH in an ABI PRISM 7700 Sequence Detection System. Fluorescence emitted during the PCR amplification (y-axis) is plotted against the cycle number (x-axis). For each reaction, the cycles of threshold (CT) are determined when a preset level of fluorescence is reached: one CT is obtained for PRV-1, another for GAPDH. The PRV-1/GAPDH ratio of a sample is determined by dividing the CTPRV-1by the CTGAPDH.
Quantification of PRV-1 mRNA by Taqman RT-PCR. Total RNA (5 ng) from a PV patient (PV) and from a healthy control (HC) was subjected to quantitative RT-PCR for PRV-1 and GAPDH in an ABI PRISM 7700 Sequence Detection System. Fluorescence emitted during the PCR amplification (y-axis) is plotted against the cycle number (x-axis). For each reaction, the cycles of threshold (CT) are determined when a preset level of fluorescence is reached: one CT is obtained for PRV-1, another for GAPDH. The PRV-1/GAPDH ratio of a sample is determined by dividing the CTPRV-1by the CTGAPDH.
Linearity, reproducibility, and accuracy of the PRV-1 RT-PCR assay
In each sample, the PRV-1 and GAPDH CT values were measured in triplicate. The average of the 3 values was used to calculate the PRV-1/GAPDH ratio. In the 136 individuals assayed, all CT triplicates measured fell within 0.3 of the individual average. Thus, the intra-assay variation is minimal. Nonetheless, triplicate measurements were performed throughout the study.
In order to determine the interassay variability, RNA samples from 48 individuals (13 PV patients, 4 SE patients, and 31 healthy controls) were analyzed in 2 separate sets of reactions on different days. The average difference in PRV-1/GAPDH ratios between the 2 measurements was 0.03 ± 0.02 (mean of the differences ± standard deviation of the mean). Furthermore, the RNA from one PV patient (UPN 533) was measured on 14 separate occasions. The mean ± standard deviation of these measurements was 1.11 ± 0.03. Finally, 2 separate blood samples, taken between 2 and 95 weeks apart, were analyzed from 15 individuals (10 PV patients, 1 SE patient, and 4 healthy controls). The mean difference in PRV-1/GAPDH ratios between the 2 samples was 0.02 ± 0.017. These data demonstrate that both inter- and intra-assay variation is minimal under the conditions used.
Using serial dilutions of RNA samples, we determined that the assay is linear for an input between 0.5 and 50 ng total RNA. In this range, the PRV-1/GAPDH ratio remains constant. Thus, if 5 ng RNA is routinely assayed, even an under- or overestimation of the RNA concentration by one order of magnitude does not alter the result.
PRV-1 expression in PV patients, SE patients, and healthy controls
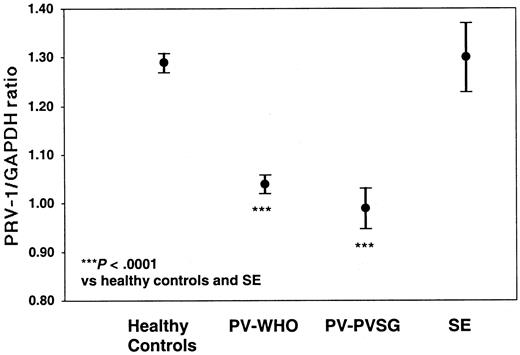
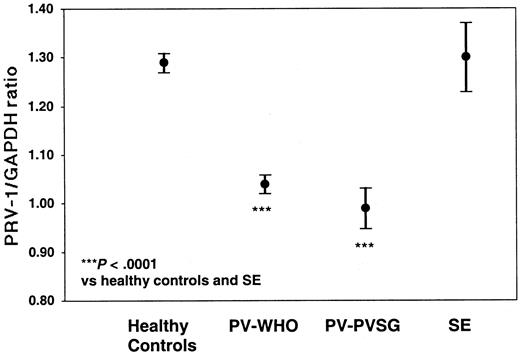
Of the 71 PV patients, 68 individuals (61 diagnosed by WHO and 7 diagnosed by PVSG) as well as 7 patients with SE and 61 healthy controls were analyzed for PRV-1 expression by quantitative RT-PCR (Table 4; Figure 2). PV-WHO patients display a mean PRV-1/GAPDH ratio of 1.04 ± 0.07 (mean ± SD; 95% confidence interval [CI] of the mean: 1.02-1.06) with values ranging from 0.85 to 1.17. Likewise, PV-PVSG patients display a mean PRV-1/GAPDH ratio of 0.99 ± 0.05 (mean ± SD; 95% confidence interval [CI] of the mean: 0.95-1.03) with values ranging from 0.94 to 1.10. In contrast, the PRV-1/GAPDH ratio in healthy controls averages 1.31 ± 0.07 (mean ± SD; 95% CI of the mean: 1.29-1.33) with a range from 1.20 to 1.52. The PRV-1/GAPDH ratios of SE patients fall within the range of healthy controls (1.22-1.49) with a mean of 1.31 ± 0.09 (mean ± SD; 95% CI of the mean: 1.24-1.38). Because there is no overlap between the PRV-1/GAPDH ratios of PV patients and those of SE patients or healthy controls (P < .0001 for the significance of the differences of the means, Figure 1), the quantification of PRV-1 mRNA levels provides a highly sensitive and specific assay for the differentiation of PV from SE or healthy hematopoiesis.
PRV-1/GAPDH ratios of patients with PV and SE, and of healthy controls. The mean PRV-1/GAPDH ratio of 61 patients with PV-WHO, 7 patients with PV-PVSG, 7 patients with SE, and 61 healthy controls is shown together with the 95% confidence interval of the mean. The significance of the difference of the means was assessed by the Kruskal-Wallis analysis of variance on ranks and the Dunn method for pairwise multiple comparisons.
PRV-1/GAPDH ratios of patients with PV and SE, and of healthy controls. The mean PRV-1/GAPDH ratio of 61 patients with PV-WHO, 7 patients with PV-PVSG, 7 patients with SE, and 61 healthy controls is shown together with the 95% confidence interval of the mean. The significance of the difference of the means was assessed by the Kruskal-Wallis analysis of variance on ranks and the Dunn method for pairwise multiple comparisons.
PRV-1 expression in patients with erythrocytosis who do not meet the WHO criteria for a diagnosis of PV
In addition to the 71 confirmed PV patients, 4 patients given a clinical diagnosis of PV by the referring physician but not fulfilling either the WHO or the PVSG criteria for the diagnosis of PV were analyzed. Patient 319 was found to carry a bcr/abl translocation; all 20 metaphases analyzed cytogenetically displayed a Philadelphia chromosome, establishing the diagnosis of CML. This patient had a PRV-1/GAPDH ratio of 1.25, well within the normal range.
The PRV-1/GAPDH values of the remaining 3 patients (324, 335, and 358) were within the range of the 71 patients who fulfill the WHO or the PVSG criteria. One patient (358), who fulfills WHO criteria A2, A3, A5, B1, and B3, which, however, do not suffice for a positive diagnosis of PV (Table 1), remains on treatment with phlebotomy. However, 2 of these patients (324 and 335), upon follow-up after PRV-1 measurement, subsequently fulfilled the WHO criteria for the diagnosis of PV. Hence, PRV-1 mRNA quantification may allow a molecular diagnosis of PV in patients whose clinical status is equivocal.
Comparison of RT-PCR and Northern blot analysis
In a subgroup of patients (52 PV-WHO patients, 7 SE patients, and 34 healthy controls) we analyzed PRV-1 expression by Northern blot. PRV-1 was overexpressed in all PV patients tested, but not in any of the SE patients or healthy controls (data not shown, summarized in Table 4). Consequently, in the 49 PV patients, 2 SE patients, and 14 healthy controls for which both assays were conducted there was a 100% concurrence between RT-PCR and Northern blot results.
Discussion
The molecular etiology of Ph--cMPDs remains elusive. Consequently, molecular markers for the diagnosis of these disorders have been lacking. Here we report a highly sensitive and specific assay for the differential diagnosis of PV from SE and healthy hematopoiesis. In our cohort, PRV-1 mRNA quantification has a very high specificity and sensitivity for the diagnosis of PV in patients with erythrocytosis or elevated hematocrits.
Our data are supported by several recent independent investigations in smaller cohorts of PV patients.20-24
Of these groups, 3 find no overlap between the quantity of PRV-1 mRNA expressed in PV patients and that in healthy controls.20,21,23 Of these, Brohee et al20 as well as Bux et al23 measured PRV-1 expression in purified granulocytes, similar to the data presented here. Spinelli et al21 used buffy coat cells for the quantification of PRV-1.
In 2 studies, PRV-1 quantification was slightly less discriminatory. Ricksten et al,22 who analyzed buffy coat cells, reported that in a cohort of 26 PV patients analyzed, only 23 (88%) overexpress PRV-1. The difference in sensitivity may be explained by the presence of lymphocytes in the buffy coat fraction, which contribute GAPDH mRNA but not PRV-1 mRNA to the measurement. Larger amounts of GAPDH mRNA but constant PRV-1 mRNA alters the PRV-1/GAPDH ratio, causing patients to score in the normal range, even though they overexpress PRV-1 mRNA in the granulocyte fraction. Since Spinelli et al21 also use total leukocyte preparations for PRV-1 measurement, upon analyzing larger numbers of patients, these investigators will most likely experience the problem already encountered by Ricksten et al.22 Kralovics et al demonstrate elevated PRV-1 mRNA levels in purified granulocytes of 91% (21/23) of PV patients analyzed.24 In addition, these authors investigated a family with hereditary thrombocythemia (HT), who carry a mutation in the TPO gene. Interestingly, several affected patients in this pedigree demonstrate a moderate elevation of PRV-1.24 Since granulocyte colony-stimulating factor (G-CSF) can stimulate PRV-1 expression in normal granulocytes,17 Kralovics et al24 discuss whether the elevated TPO levels in HT patients may likewise increase PRV-1 expression.
Liu et al have investigated PRV-1 mRNA expression in purified granulocytes in a cohort of 13 PV patients.25 In this group, only 9 of 13 patients overexpress PRV-1. However, the range of PRV-1 expression in the healthy controls used for comparison is much larger than that observed by us (Table 4), which may cause some patients to score within the normal range. In contrast, while there are methodologic differences in the calculation of the PRV-1/GAPDH ratio between the 2 studies (“Patients, materials, and methods”), these do not explain the differences in results between the study by Liu et al25 and the data presented here.
Rather, it is likely that technical aspects account for the discrepancy. Storage or shipment of the blood samples for more than 24 hours can lead to the release of inhibitory substances that disturb the PCR reaction.
In addition to PRV-1, Liu et al25 have also investigated clonality and EEC formation in MPD patients. In this cohort, there was a close correlation between clonality and EEC formation, but less correlation between EEC formation and PRV-1 expression.
Because of the differences between the data presented here and the results obtained by Liu et al,25 further studies, in the form of prospective trials, will be required to resolve these discrepancies and to determine the specificity and sensitivity of the PRV-1 assay.
PRV-1 quantification is not the only molecular marker available for the diagnosis of PV. Moliterno et al have previously described a decreased expression of the thrombopoietin receptor, c-Mpl, in patients with PV.26,27 Using an internally controlled Western blot assay, these authors demonstrate a sensitivity of 96% and a specificity of 95% for the discrimination of PV from SE.28 Other groups have reported less favorable data for either the diagnosis of PV or the discrimination of ET from reactive thrombocytosis (RT).29,30 However, these discrepancies can be explained by technical differences, especially the use of different antibodies. Nonetheless, these differences emphasize the logistic difficulties involved in using Western blotting as a diagnostic tool.
Prior to inclusion in the analysis, we confirmed that German patients assigned a clinical PV diagnosis by the referring physician met the WHO criteria for the positive diagnosis of PV. We used the WHO criteria rather than the PVSG criteria for the following reason. Besides the inclusion of the EEC assay, the 2 sets differ in that the former no longer requires the use of the red cell mass for the diagnosis of erythrocytosis. Since the red cell mass is almost never determined in Germany and we chose a retrospective analysis of previously diagnosed patients, red cell masses at the time of diagnosis were not available to us.
However, there is concern that without determination of the red cell mass, absolute erythrocytosis cannot be unequivocally diagnosed in patients with elevated hematocrits.31,32 Therefore, we analyzed an additional cohort of 7 PV patients in whom the red cell mass had been determined at diagnosis and was found to be elevated more than 2 standard deviations (25%) of the mean or of the expected value for this patient. In these patients, PRV-1 mRNA expression was also markedly elevated, even slightly higher than in the PV-WHO cohort (Table 4), demonstrating that PRV-1 is overexpressed in patients with stringently diagnosed PV.
We demonstrate that PRV-1 mRNA quantification identified 3 patients who had received a diagnosis of PV from their hematologist, but who did not, at the time of diagnosis, meet either the PVSG or the WHO criteria for a positive diagnosis of PV (patients 324, 335, and 358). Patient 324 was analyzed at the time of presentation to the hematology service and thus exemplifies a frequent clinical situation: a patient presenting with erythrocytosis but not qualifying for a positive diagnosis of PV because the necessarily strict criteria of the PVSG or the WHO are not met. Upon follow-up after PRV-1 measurement, this patient subsequently fulfilled the diagnostic criteria of the WHO. For such patients, the availability of a molecular diagnostic would represent a substantial advance in ascertaining a firm diagnosis.
A 70-year-old female (patient 319), given the diagnosis of PV, was referred to one of us (H.H.) for a second opinion. The initial diagnosis was supported by a moderate increase in the hematocrit (.485 [48.5%]) and the hemoglobin (Hb) concentration (164 g/L [16.4 g/dL]) as well as by increased numbers of neutrophils and platelets. At the time of re-evaluation, PRV-1 expression was determined and found to be normal (PRV-1/GAPDH ratio: 1.25). However, this result was not made available to the clinician until the clinical diagnosis of the patient had been repeated. Upon clinical work-up, the patient was found to have a bcr/abl translocation, establishing the diagnosis of CML. Concomitantly, the patient displayed a unilateral paralysis of the diaphragm of unknown origin, possibly explaining the moderate increase in the hematocrit and the Hb concentration. PRV-1 quantification correctly identified the misdiagnosis of PV in this patient.
In PV patients, PRV-1 expression is not dependent on the stage of disease, as it is elevated in patients at diagnosis as well as in patients with a known history of disease exceeding 30 years (Tables 2,4). We detected no correlation between the extent of PRV-1 mRNA overexpression and the duration of disease.
Our data do not allow us to determine whether cytoreductive treatment will alter PRV-1 expression. Of the 71 patients, 36 were treated by cytoreductive therapy before or at the time of sampling. This includes 5 patients treated with interferon-α. Preliminary data suggest that prolonged interferon treatment may normalize PRV-1 expression in selected patients,33 but longer observation periods are required to confirm these data. None of our cohort of 71 PV patients displayed normal PRV-1 mRNA levels. Likewise, no correlation between prior cytoreductive therapy and PRV-1 expression was found.
Control subjects and patients without erythrocytosis but with a reactive leukocytosis can also overexpress PRV-1. Such is the case in healthy individuals functioning as peripheral stem cell donors and treated with G-CSF for several days.17 Likewise, patients with leukocytosis due to polytrauma, following a prolonged operation, or during sepsis express elevated levels of PRV-1 (H.L.P. et al, unpublished observation). Thus, it is mandatory that a patient present with erythrocytosis and that an infectious diathesis is ruled out before PRV-1 expression is used to render a diagnosis of PV.
Among patients with Ph--cMPDS, PRV-1 overexpression is not unique to PV. We have demonstrated that 50% of ET patients also overexpress PRV-1 (Klippel et al34 ; and M.G. et al, manuscript submitted). All PRV-1-overexpressing ET patients also grow EECs, while PRV-1-negative ET patients do not show EEC growth. Shih and Lee first demonstrated that EEC formation in patients with idiopathic thrombocytosis but normal hemoglobin levels predicts the onset of overt PV within several years.35 The differences in clinical course between PRV-1-negative and PRV-1-positive ET patients are currently being evaluated.
PRV-1 is also overexpressed in 50% of patients with IMF (H. Gisslinger et al, manuscript in preparation). Again, PRV-1-positive patients also display EEC growth, while PRV-1-negative patients do not. Therefore, PRV-1 quantification cannot be used to distinguish the 3 clinically defined subtypes of Ph-cMPDs. Rather, it is to be used in patients with erythrocytosis, elevated hematocrit, and/or increased hemoglobin for the differential diagnosis between secondary erythrocytosis and polycythemia vera.
Based on preliminary presentation of this data,34 the latest edition of the diagnostic guidelines of the German Society for Haematology has recommended the use of PRV-1 mRNA quantification for the diagnosis of PV.36 An international initiative, the MPD Research Consortium, has been formed, which will conduct large prospective trials to determine the role of PRV-1 quantification in clinical diagnosis and therapeutic decisions.
Prepublished online as Blood First Edition Paper, July 31, 2003; DOI 10.1182/blood-2003-03-0919.
Supported by the Alfried Krupp Förderpreis für Junge Hochschullehrer, by the Sonderforschungsbereich (SFB) 364, Project A12, both granted to H.L.P., and by the Kompetenznetzwerk Leukämien (Project 25).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors sincerely thank Prof Dr K. Geiger for his continuing support. We are indebted to H. Iwan for excellent technical assistance and to Dr T. Loop for critical review of the data and help with statistical analysis. We extend a very special thank you to the helpful staff of the hematology clinic and the therapy ward of the University Hospital Freiburg, to Prof Dr R. Mertelsmann, PD Dr M. Lübbert, PD Dr Ch. v. Kalle, and to the referring physicians from other institutions: Prof Dr A. Ganser and Dr R. Müller, Hannover; PD Dr E. Lengfelder, Mannheim; Prof Dr A. Wehmeier, Remscheid; Prof Dr H. Döhner, Ulm; Dr I. Hagner, Berlin; Dr D. Frommherz, Herbolzheim; Dr E. Schäfer and Dr M. Just, Bielefeld; Prof Dr H. Gisslinger, Wien; Dr H.-R. Bruch, Bonn; PD Dr S. Fruehauf, Dr J. Villalobos, and Prof Dr R. Skoda, Heidelberg.