Abstract
We examined the hypothesis that filamin A binding to the cytoplasmic tail of platelet glycoprotein Ibα (GpIbα) is regulated by pathologic shear stress and modulates von Willebrand factor (VWF)–induced platelet activation. To begin, we examined filamin binding to GpIbα in Chinese hamster ovary cells coexpressing mutant human GpIb-IX and wild-type human filamin A. We observed that many different deletions and truncations N-terminal to GpIbα's cytoplasmic domain residue 594 disrupted filamin A binding, but that binding was unaffected by 14 different point mutations in hydrophilic residues between amino acids 557 and 593. To try to narrow GpIbα's filamin A–binding domain, we next measured the effect of several cytoplasmic domain peptides on human filamin A binding to a GST-GpIbα cytoplasmic domain fusion protein. One peptide (residues 557-575; designated “A4 peptide”) inhibited filamin A binding to the GST-GpIbα cytoplasmic domain fusion protein and competed with GpIbα for binding to filamin A. When the A4 peptide was delivered to intact human platelets using a carrier peptide, we observed the dose-dependent inhibition of VWF-induced platelet aggregation in response to both ristocetin and shear stress. The effect of the A4 peptide on shear-induced platelet aggregation was accompanied by the attenuation of shear-induced filamin A binding to GpIbα and diminished shear-dependent protein tyrosine phosphorylation. These results suggest that shear-dependent VWF-induced platelet activation affects filamin A binding to GpIb-IX-V, and that filamin A binding to the cytoplasmic tail of GpIbα regulates proaggregatory tyrosine kinase signaling.
Introduction
Pathologic wall shear stress develops in atherosclerotic arteries affected by progressive stenosis or plaque rupture. Pathologic shear stress exerts frictional forces that enhance blood cell-vessel wall collisions and directly modulate platelet adherence to freshly exposed subendothelial collagen admixed with insoluble and soluble von Willebrand factor (VWF). Shear-induced VWF binding to the glycoprotein (Gp) Ib-IX-V complex initiates stop-and-go platelet adherence.1,2 Platelet arrest and accrual resulting in pathologic arterial thrombosis require the activation of the αIIbβ3 integrin receptor complex.3,4
Mechanisms of platelet αIIbβ3 activation are multiple and complex.5 The current model proposes that activation results from signal pathways converging on a final common pathway mediated by talin binding to the β3 subunit.6 When the cytoplasmic domain of β3 binds the head domain of the cytoskeletal protein talin, hydrophobic and electrostatic interactions between the cytoplasmic tails of αIIb and β3 are relaxed. When these intracellular interactions are relaxed, a ligand-receptive conformational change develops in the extracellular domains of the complex.7
The mechanism by which talin binding to αIIbβ3 is regulated is uncertain. Data show that calpain cleavage exposes a β3-binding site on the head domain of talin.8 Other data indicate that phosphatidylinositol 4-phosphate (PIP) and phosphatidylinositol 4,5-bisphosphate (PIP2) bridge the β1 cytoplasmic domain to full-length talin,9 suggesting the possibility that PIP or PIP2 or both may regulate αIIbβ3 activation. Assuming that one or both of these mechanisms operate to effect αIIbβ3 activation following shear-induced VWF binding to GpIb-IX-V, perhaps the most important question that needs to be addressed is “how are αIIbβ3-activating signals relayed between GpIb-IX-V and talin?” We hypothesize that signals are relayed through cytoskeletal proteins that directly link the cytoplasmic domain of GpIbα with the β3 tail: GpIbα binds filamin A, which binds to filamentous actin cross-linked by α-actinin that binds to vinculin that binds to talin that interacts with the cytoplasmic domain of the β3 tail. This hypothetical model is supported by data demonstrating that shear-induced VWF binding to GpIb-IX-V results in such sequential associations.10-12 The model is further buttressed by data demonstrating that platelets generate VWF-dependent signals that could regulate these associations, including VWF-stimulated changes in cytosolic ionized calcium13-16 and VWF-triggered activation of tyrosine kinases17-19 and phosphatidylinositol 3-kinases. 12,20,21
To examine the hypothesis that the cytoskeleton transduces signals from GpIb-IX-V to αIIbβ3, we first focused on the proximal connection between GpIbα and filamin A. We sought to determine if the GpIbα/filamin A interaction is regulated by shear-induced VWF binding to GpIb-IX-V and if the GpIbα/filamin A interaction affects signaling leading to αIIbβ3 activation.
Materials and methods
Preparation of CHO cells expressing recombinant GpIb-IX and filamin A
Recombinant wild-type and mutant GpIbα, coexpressed with recombinant GpIbα and GpIX in Chinese hamster ovary (CHO) cells, were prepared and used as described previously.22 Human filamin A cDNA (in PREP4 vector, a gift from Dr John Hartwig, Brigham and Women's Hospital, Boston, MA) was stably expressed in CHO-GpIbα/β-IX and CHO-GpIbβ-IX cells. Levels of expression in various cell lines cloned by limiting dilution were measured by Western blotting whole cell lysates with an antihuman filamin A antibody (Transduction Laboratories, Lexington, KY).
Site-directed mutagenesis of the cytoplasmic tail of GpIbα targeted the following residues: RGS 557-559 to AGA 557-559, RSS 564-566 to AAA 564-566; RPNGR 572-576 to APAGA 572-576; RR 584-585 to AA 584-585; SALS 587-590 to AALA 587-590; SQGR 590-593 to AAGA 590-593; and LPTFRSSLFLW 560-570 to deletion 560-570. The primers used in these constructions are as follows: M557 and 559-1: 5′CTCTTCCTTGCAGGTGCGCTTCCC3′; M557 and 559-2: 5′GGGAAGCGCACCTGCAAGGAAGAG3′; M564-66-1: 5′CCCACTTTCGCCGCCGCCCTCTTCCTG3′; M564-66-2: 5′CAGGAAGAGGGCGGCGGCGAAAGTGGG3′; M572-1: 5′CTGTGGGTAGCGCCTAATGGC3′; M572-2: 5′GCCATTAGGCGCTACCCACAG3′; M572 and 574-1: 5′GTAGCGCCTGCAGGCCGTGTG3′; M572 and 574-2: 5′CACACGGCCTGCAGGCGCTAC3′; M572 and 574 and 576-1: 5′CCTGCAGGCGCAGTGGGGCCT3′; M572 and 574 and 576-2: 5′AGGCCCCACTGCGCCTGCAGG3′; M584-85-1: 5′GTGGCAGGAGCGGCGCCCTCAGCT3′; M584-85-2: 5′AGCTGAGGGCGCCGCTCCTGCCAC3′; M587 and 590-1: 5′AGGCCCGCAGCTCTGGCTCAGGGT3′; M587 and 590-2: 5′ACCCTGAGCCAGAGCTGCGGGCCT3′; M590 and 591 and 593-1: 5′GCTCTGGCAGCGGGTGCAGGTCAGGAC3′; M590 and 591 and 593-1: 5′GTCCTGACCTGCACCCGCTGCCAGAGC3′; M560-570-1: 5′CTCTTCCTGGTACCCGTACGGCCT3′; M560-570-2: 5′GAAAGTGGGTACCGAACCTCG3′; PGM6: 5′ATGCAGCATCTCGAGCTTTGTCTTGTC3′ (3′ primer nucleotides 2131-2158); Gpseq: 5′CATGTGAAACCACAGGCCCTGGAC3′ (5′ primer nucleotides 1633-1659).
Polymerase chain reaction (PCR) products were cloned into pBluescript SK (Stratagene, La Jolla, CA), sequenced by an ABI Prism 3100 Genetic Analyzer, and ligated to form full-length mutated cDNA sequences. The desired inserts were then transferred to a pcDNA3.1/Zeo vector (Invitrogen, San Diego, CA) for expression in CHO-GpIbβ-IX-filamin A cells as described.22
Wild-type and mutant CHO-GpIbα/β-IX-human filamin A cells were lysed in phosphate-buffered saline (PBS) containing 1% triton X-100, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM EGTA (ethylene glycol tetraacetic acid), 1 mM NaF, 1 mM Na3VO4,1 μg/mL pepstatin A, 1 μg/mL aprotinin, 1 μg/mL leupeptin, and 10 μg/mL 4-(2-aminoethyl)-benzenesulfonyl fluoride. Cell lysates were then centrifuged at 14 000 rpm for 10 minutes. GpIbα was immunoprecipitated using the mouse monoclonal antibody AN51 (Dako, Carpinteria, CA). Mouse IgG was used as a control. Lysates were incubated with the immunoprecipitating antibody at 4°C overnight followed by 1 hour incubation with 40 μL protein G at 4°C. Samples were washed 4 times with ice cold PBS, resuspended in 50 μL2 × sample buffer and boiled for 4 minutes. Proteins were separated in 7.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. Membranes were blocked with 5% nonfat milk for 1 hour and incubated with the primary antifilamin A antibody at 4°C overnight. Immunoreactive bands were reported by enhanced chemiluminescence (ECL; Amersham Pharmacia, Piscataway, NJ).
Preparation of the cytoplasmic domain-GST fusion protein
The entire GpIbα cytoplasmic tail (H 515-L 610) was synthesized by PCR and fused to glutathione-S-transferase (GST) in pGEX-2T at BamHI and EcoRI sites (Amersham Pharmacia). The primers used were 5′GGATCCCATGTGAAACCACAG3′ and 5′GAATTCAGAGGCTCTTCTCTC3′. Synthetic peptides corresponding to regions in the cytoplasmic domain of GpIbα were either provided to us as gifts from Dr Michael C. Berndt23 (Monash University, Victoria, Australia) or synthesized by Sigma Genosys (The Woodlands, TX). All peptides were purified by reverse-phase high-performance liquid chromatography (HPLC) and characterized by mass spectroscopy. The GST-GpIbα-C-terminal fusion protein was expressed in Escherichia coli and purified by glutathione Sepharose 4B beads (Amersham Pharmacia Biotech, Uppsala, Sweden). Recombinant human filamin A from CHO cells was used for measurements of filamin A binding to the GST-GpIbα-C-terminal fusion protein. CHO cells expressing human GpIbα, GpIX, and filamin A were lysed as described in “Preparation of CHO cells expressing recombinant GpIb-IX and filamin A.” Cell lysates were centrifuged at 14 000 rpm for 10 minutes and their supernatants incubated with the GST-GpIbα-C-terminal fusion protein beads in the presence of the synthetic peptides, always used at a final concentration of 25 μg/mL. The beads were then washed 3 times with PBS containing 0.5% Triton X-100. Samples were separated by SDS-PAGE, transferred to nitrocellulose, and blotted with a rabbit anti–GpIbα-C-terminal antibody (a gift from Dr Xiaoping Du, University of Illinois, Chicago, IL, used as described previously22 ) and the antifilamin A antibody.
Measurement of A4 peptide binding to filamin A
Protein binding to A4 peptide was measured following covalent linkage of the target A4 peptide (RGSLPTFRSSLFLWVRPNG corresponding to residues 557-575 of GpIbα) or the scrambled A4 peptide (LWRTSVNPGLSFTRGLPSRF) to Affi-Gel beads (activated immunoaffinity support Affi-Gel-10 and Affi-Gel-15 manufactured by Bio-Rad, Hercules, CA). 0.5 mL Affi-Gel-10 and 0.5 mL Affi-Gel-15 were mixed, washed in 8 mL dimethyl sulfoxide (DMSO), added to 8 mL 100 mM MOPS (3-[N-morpholino]propanesulphonic acid) buffer (pH 7.5, 4°C), and then divided into 2 aliquots of 4 mL. Then, 50 μM A4 peptide or scrambled A4 peptide (S-A4) was coupled to Affi-Gel by shaking for 1 hour at room temperature. The unreacted peptides were blocked by the addition of ethanolamine. The peptides coupled to Affi-Gel were washed in MOPS buffer. CHO cells transfected with filamin A plus GpIbβIX, GpIbαβIX, or mutant GpIbαβIX (GpIbα truncated at residues 594 or 583) were lysed and lysate supernatants were incubated with A4-or S-A4–coupled Affi-Gel overnight, washed in PBS, separated by SDS-PAGE, and blotted with antifilamin A or GpIbα antibodies.
Peptide delivery to intact washed human platelets
The peptide carrier designated “Pep-1” was used as described by Morris et al.24 In brief, the peptide KETWWETWWTEWSQPKKKRKV, acetylated at its N-terminus with a cysteamine residue at its C-terminus, was synthesized and purified by Sigma Genosys. The A4 peptide or scrambled A4 peptide was mixed with Pep-1 at a molar ratio of 1:20 in buffer, as described in “Preparation of platelets for shear and aggregometry analyses,” for 30 minutes at 37°C. Platelet-rich plasma (PRP) or washed platelets (2.5 × 108/mL) were then incubated with the A4/pep-1 complex for an additional 30 minutes at 37°C. For determining the efficiency of A4 peptide delivery, the A4 peptide was conjugated to fluorescein isothiocyanate (FITC) and uptake of fluorescence measured by flow cytometry. In addition, some of the fixed platelets were transferred to a glass slide and fluorescence uptake examined by a Nikon fluorescent microscopy. To analyze the effect of the A4 peptide on filamin A binding to GpIbα in intact human platelets stimulated by shear stress, sheared washed platelet suspensions were removed from the viscometer and microcentrifuged at 15 000g for 5 seconds. The supernatant was discarded and the pellet immediately lysed in ice-cold lysis buffer. These specimens were then subjected to immunoprecipitation using the mouse monoclonal antibody AN51, and coimmunoprecipitating filamin A was measured by immunoblotting.
Preparation of platelets for shear and aggregometry analyses
Venous blood was obtained from healthy adult volunteer donors with a 19-gauge needle and collected in 15% acid-citrate-dextrose (ACD). Approval for these studies was obtained from the Baylor College of Medicine institutional review board. Informed consent was provided according to the Declaration of Helsinki.
Blood was centrifuged at 270g at 24°C for 15 minutes to separate PRP. PRP was collected, the pH was adjusted to 6.5 with ACD, and it was treated with phosphocreatine (CP, 5 mM) and creatine phosphokinase (CPK, 25 U/mL). Platelets were then separated from the PRP by a second centrifugation at 1600g at 24°C for 15 minutes. The pellet was suspended in Tyrode buffer (138 mM NaCl, 2.9 mM KCl, 12 mM NaHCO3, 0.36 mM sodium phosphate, 5.5 mM glucose, pH 6.5) containing CP and CPK, and then centrifuged at 1200g at 24°C for 10 minutes. Washed platelets were resuspended in JNL buffer (6 mM glucose, 130 mM NaCl, 9 mM NaHCO3, 10 mM sodium citrate, 10 mM Tris [tris(hydroxymethyl)aminomethane] base, 3 mM KCl, 2 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid] and 0.9 mM MgCl2, pH 7.35 to which 1 mM of CaCl2 and 5 μg/mL purified VWF [Calbiochem, San Diego, CA] were added).
Specimens were subjected to fluid shear stress (120 dyne/cm2) in a cone-plate viscometer at 24°C as previously described.10-13 Then, 5 μL of the sheared platelet suspension was fixed in 500 μL 1% paraformaldehyde and aggregation measured by flow cytometry as previously described.11,21 For experiments using peptide delivery to washed platelets, an extra washing step was used to eliminate the Pep-1/A4 mixture prior to stimulation. This was done to eliminate nonspecific inhibitory effects of the delivery peptide, which were seen at concentrations of Pep-1 more than 150 μM and more than 250 μM in washed platelets and PRP, respectively. For this washing step, CP and CPK were added to the reaction mixture and platelets were washed free of Pep-1/peptide by centrifugation at 1200g at 24°C for 10 minutes. The washed platelet pellet was resuspended in JNL buffer and shear-induced aggregation measured.
For measurement of 0.9 mg/mL ristocetin-induced platelet aggregation, stirring citrate-anticoagulated PRP (containing the Pep-1/peptide mixture) was stimulated in a ChronoLog aggregometer at 37°C. For measurement of aggregation in response to 20 μM adenosine diphosphate (ADP), 1 mg/mL purified human fibrinogen was added to washed platelets prior to the addition of agonist.
Resting and sheared platelets were lysed in an equal volume of ice-cold PBS buffer containing 2% Triton X-100, 2 mM Na3VO4, 2 mM NaF, 2 mM EDTA (ethylenediaminetetraacetic acid), 2 mM PMSF, 2 mg/mL deoxycholic acid, and 2 μg/mL aprotinin, leupeptin, and pepstatin A, and briefly sonicated. Platelet lysates were centrifuged at 15 000g at 4°C for 15 minutes and the supernatant collected. GpIbα was immunoprecipitated using the mouse monoclonal antibody AN51 and coimmunoprecipitating filamin A measured by immunoblotting.
To analyze the effect of the A4 peptide on shear-induced tyrosine phosphorylation, sheared platelet lysates were subjected to Western blotting with the phosphotyrosine-specific monoclonal antibody 4G10 (Upstate, Charlottesville, VA). To identify the tyrosine phosphorylated substrates, blots were stripped and reprobed with the following specific antibodies: FAK (clone 4.47; Upstate), α-actinin (BM-75.2; Sigma), Syk (4D10; Santa Cruz Biotechnology, Santa Cruz, CA), Fyn (15; Santa Cruz) and Src (GD11; Upstate).
Data analysis
Quantitation of Western blot signals was made using a Hewlett-Packard Scan Jet 7400C scanner connected to a PC and processed using the Bio-Rad Quantity One image analysis software. Quantitation of fluorescence was done using a Becton Dickinson FACStar flow cytometer (Mountain View, CA). Group comparisons were made using the Student t test calculated by SigmaPlot (Sigma, St Louis, MO).
Results
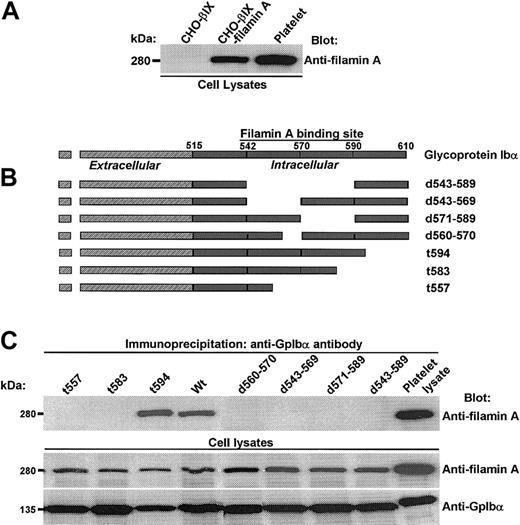
To begin our exploration of the regulation and function of filamin A binding to GpIbα, we first established several genetically engineered cell lines coexpressing human GpIbα, GpIbβ, GpIX, and filamin A. Figure 1A shows the coexpression of filamin A protein in CHO cells transduced with the cDNAs of GpIbβ, GpIX, and filamin A. We chose to express recombinant human filamin A for 2 reasons. The first reason is that we wanted to be able to control for all of the primary structural determinants being examined and, although the sequences of both GpIbα and filamin A (its major platelet filamin partner) are well-known, the protein sequences of filamin isoforms present in the hamster have not yet been elucidated. Secondly, we wanted to ensure that filamin A binding to GpIbα was maximally saturated, because it is estimated that more than 90% of all GpIbα molecules are bound to filamin A in human platelets.25 And although we are not certain if CHO cell filamin saturates recombinant GpIbα expressed in CHO cells, we are sure that recombinant human filamin A shows saturable binding to GpIbα in our CHO cells coexpressing filamin A, GpIbα, GpIbβ, and GpIX (data not shown). Of note, we observed that the coexpression of human filamin A increased the expression of GpIbα in several CHO cell lines, as has been previously reported by others (data not shown).26
Filamin A binds to a discrete domain in the cytoplasmic tail of GpIbα. (A) Expression of human filamin A in CHO cells transduced with the cDNAs of GpIbβ, GpIX, and filamin A. (B) Constructs used to identify GpIbα's primary structural requirements for filamin A binding. (C) Several truncations and deletions N-terminal to residue 594 in GpIbα's cytoplasmic domain eliminate filamin A binding.
Filamin A binds to a discrete domain in the cytoplasmic tail of GpIbα. (A) Expression of human filamin A in CHO cells transduced with the cDNAs of GpIbβ, GpIX, and filamin A. (B) Constructs used to identify GpIbα's primary structural requirements for filamin A binding. (C) Several truncations and deletions N-terminal to residue 594 in GpIbα's cytoplasmic domain eliminate filamin A binding.
To identify GpIbα's primary structural requirements for filamin A binding, we examined the capacity of wild-type and mutant GpIbα to coimmunoprecipitate recombinant human filamin A from CHO cells coexpressing it and GpIb-IX. These experiments assay the endogenous interaction between GpIbα and filamin A, and the constructs used are schematized in Figure 1B. Figure 1C shows that several truncations and deletions N-terminal to residue 594 in GpIbα's cytoplasmic domain eliminate filamin A binding.
These results, which are consistent with results previously published by others using alternative27 and similar28 methods, suggest that filamin A contacts several amino acids widely spaced throughout the cytoplasmic domain of GpIbα and/or that filamin A binds to GpIbα through complex higher-order structural interactions disrupted by changes in the primary structure of GpIbα shown in Figure 1.
To try to perturb contact points without major disruption in the secondary structure of the tail of GpIbα, several point mutations in GpIbα were generated in regions ascribed by us and others (residues 557-568, 569-579, and 580-590)27,28 as essential for filamin A binding. These point mutations were made to try to identify hydrophilic residues presented on the outer face of the cytoplasmic domain of GpIbα that interact with filamin A. Figure 2A shows these mutations, and Figure 2B shows that such mutations have no effect on the endogenous interaction between recombinant GpIbα and filamin A in CHO cells.
Filamin A binding is unaffected by mutating several hydrophilic residues in GpIbα's large filamin A–binding domain. (A) Mutations used to try to disrupt filamin A contact points without major disruption of the secondary structure of the tail of GpIbα. (B) Such mutations have no affect on the endogenous interaction between recombinant human GpIbα and recombinant human filamin A in CHO cells.
Filamin A binding is unaffected by mutating several hydrophilic residues in GpIbα's large filamin A–binding domain. (A) Mutations used to try to disrupt filamin A contact points without major disruption of the secondary structure of the tail of GpIbα. (B) Such mutations have no affect on the endogenous interaction between recombinant human GpIbα and recombinant human filamin A in CHO cells.
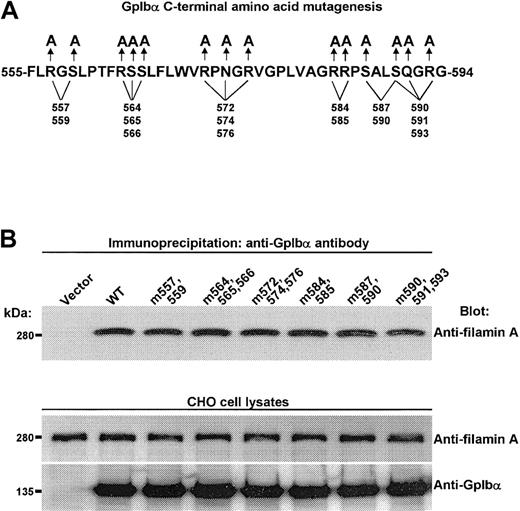
To develop an alternative approach that might provide more detailed information about GpIbα's primary structural requirements for filamin A binding, a GST-GpIbα-C-terminal construct was made (Figure 3A), and the effects of several cytoplasmic domain peptides (Figure 3B) on the binding of human filamin A to the GST-GpIbα-C-terminal peptide was measured. The first 2 and penultimate lanes in Figure 3C show that filamin A binds specifically to the GST-GpIbα-C-terminal peptide, confirming that GpIbα alone is sufficient for filamin A binding to the GpIb-IX-V complex. The remaining lanes in Figure 3C show the effects of the cytoplasmic domain peptides. Only one peptide (designated A4) corresponding to amino acids 557 to 575 blocked filamin A binding to the GST-GpIbα-C-terminal peptide. Figure 3 also shows that several related peptides have no effect on filamin A binding to the GST-GpIbα-C-terminal peptide, including a peptide with overlapping N-terminal sequence (the A3 peptide), 2 peptides with overlapping C-terminal sequence (the A5 and X1 peptides), and one peptide with a fully overlapping sequence (the X0 peptide).
Filamin A binding to the cytoplasmic tail of GpIbα is affected only by a peptide spanning residues 557 to 575. (A) The GST-GpIbα-C-terminal construct. (B) Several cytoplasmic domain peptides examined for affects on the binding of recombinant human filamin A to the GST-GpIbα-C-terminal peptide. (C) The first 2 and last 2 lanes show that filamin A binds specifically to the GST-GpIbα-C-terminal peptide. The remaining lanes show the effects of the cytoplasmic domain peptides. (D) Effect of each peptide on recombinant filamin A binding to the GST-GpIbα-C-terminal peptide (n = 3; means + SDs; **P < .01 in comparison to no peptide by Student t test).
Filamin A binding to the cytoplasmic tail of GpIbα is affected only by a peptide spanning residues 557 to 575. (A) The GST-GpIbα-C-terminal construct. (B) Several cytoplasmic domain peptides examined for affects on the binding of recombinant human filamin A to the GST-GpIbα-C-terminal peptide. (C) The first 2 and last 2 lanes show that filamin A binds specifically to the GST-GpIbα-C-terminal peptide. The remaining lanes show the effects of the cytoplasmic domain peptides. (D) Effect of each peptide on recombinant filamin A binding to the GST-GpIbα-C-terminal peptide (n = 3; means + SDs; **P < .01 in comparison to no peptide by Student t test).
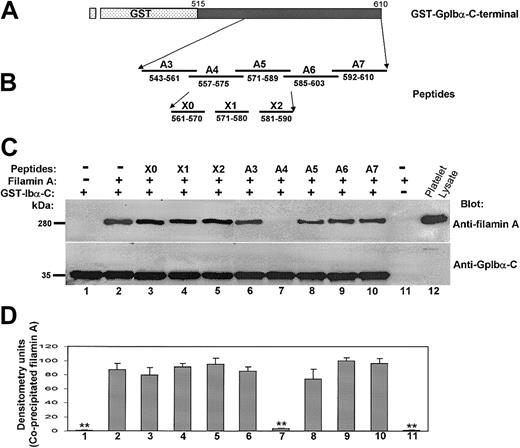
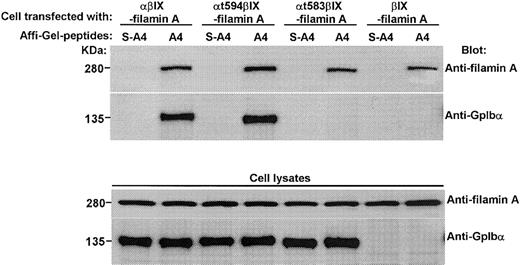
To prove that the A4 peptide binds specifically to filamin A, it—or a scrambled A4 peptide—was covalently bound to Affi-Gel beads. Figure 4 shows that only the A4 peptide binds to filamin A and that it coprecipitates wild-type GpIbα bound to filamin A. When GpIbα is mutated such that it cannot bind to filamin A (Figure 1) or when lysates from CHO cells expressing only GpIbβ, GpIX, and filamin are incubated with the Affi-Gel-bound A4 peptide, only filamin A is bound. These results suggest that the stoichiometry of GpIbα binding to filamin A is more than 1:1 and that A4 binds to filamin A with high affinity.
The A4 peptide binds specifically to filamin A. Affi-Gel-bound A4 peptide or Affi-Gel-bound scrambled A4 peptide was incubated overnight with lysates from CHO cells expressing filamin A plus GpIbα/βIX, filamin A plus GpIbαt594βIX, filamin A plus GpIbαt583βIX, or filamin A plus GpIbβIX.
The A4 peptide binds specifically to filamin A. Affi-Gel-bound A4 peptide or Affi-Gel-bound scrambled A4 peptide was incubated overnight with lysates from CHO cells expressing filamin A plus GpIbα/βIX, filamin A plus GpIbαt594βIX, filamin A plus GpIbαt583βIX, or filamin A plus GpIbβIX.
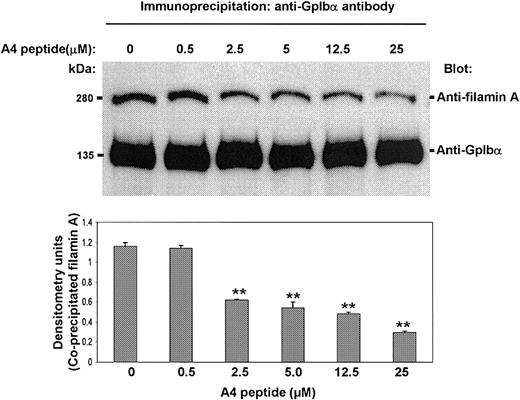
To determine if the A4 peptide competes for platelet filamin A binding with platelet GpIbα, we examined its effect on the coimmunoprecipitation of filamin A with GpIbα. Washed resting human platelets were immunoprecipitated with the GpIbα-specific monoclonal antibody AN51 overnight, and the amount of coimmunoprecipitating filamin A was measured by immunoblotting. Figure 5 shows that the A4 peptide, when it is added to the platelet lysates simultaneously with AN51, causes a dose-dependent diminution in the amount of immunodetectable filamin A present in the GpIbα immunoprecipitates. These results suggest the possibility that the A4 peptide, by competing with GpIbα for filamin A binding, could affect the endogenous interaction between filamin A and GpIbα in intact human platelets.
The A4 peptide decreases the amount of filamin A coimmunoprecipitating with GpIbα. The A4 peptide, when it is added to platelet lysates simultaneously with the GpIbα-specific antibody AN51, decreases the amount of immunodetectable filamin A that coimmunoprecipitates with GpIbα (bars present means + SDs; ** P < .01 when compared to 0 A4 peptide by Student t test).
The A4 peptide decreases the amount of filamin A coimmunoprecipitating with GpIbα. The A4 peptide, when it is added to platelet lysates simultaneously with the GpIbα-specific antibody AN51, decreases the amount of immunodetectable filamin A that coimmunoprecipitates with GpIbα (bars present means + SDs; ** P < .01 when compared to 0 A4 peptide by Student t test).
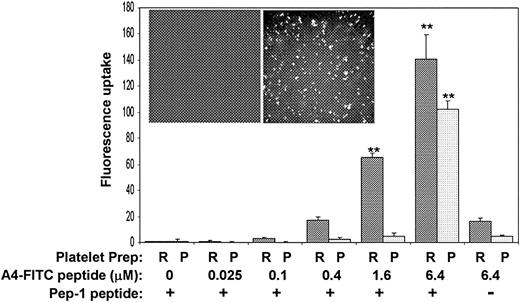
To investigate the interaction between filamin A and GpIbα in intact human platelets, we attempted to develop an A4 peptide delivery system using a peptide carrier, KETWWETWWTEWSQPKKKRKV, designated Pep-1. It has been reported that this peptide carrier can deliver both peptides and proteins into living cells and thereby target specific endogenous protein-protein interactions.24 To target the platelet filamin A/GpIbα interaction with the A4 peptide, it was essential to first prove that Pep-1 is capable of delivering the A4 peptide into platelets. Figure 6 shows that Pep-1 delivers FITC-conjugated A4 into platelets. The bar graph shows fluorescent uptake into platelets when the FITC-A4 peptide plus Pep-1 are mixed with platelets for 30 minutes at 37°C. Data in this figure showing greater uptake by platelets in PRP corroborate other's data that Pep-1–mediated delivery is enhanced in the presence of serum.24 The molar ratio of FITC-A4 peptide to Pep-1 is maintained at 1:20, and Figure 6 shows that optimal uptake as measured by flow cytometry occurs with 6.4 μM FITC-A4 peptide plus 128 μM Pep-1. The inset shows fluorescent micrographs after washed platelets are incubated with 6.4 μM FITC-A4 peptide plus 0 μM Pep-1 (left) or 6.4 μM FITC-A4 peptide plus 128 μM Pep-1 (right) and allowed to adhere to glass slides. Nonspecific attachment of FITC-A4 to platelets or glass is not observed.
Pep-1 delivers FITC-conjugated A4 peptide into platelets. The bar graph shows fluorescent uptake into platelets when the A4 peptide plus Pep-1 are mixed with PRP (dark bars) or washed platelet suspensions (light bars) for 30 minutes at 37°C. The molar ratio of A4 peptide to Pep-1 is maintained at 1:20 and optimal uptake as measured by flow cytometry occurs with 6.4 μM A4 peptide plus 128 μM Pep-1. The inset shows fluorescent micrographs after washed platelets are incubated with 6.4 μM A4 peptide plus 0 μM Pep-1 (left) or 6.4 μM A4 peptide plus 128 μM Pep-1 (right) and allowed to adhere to glass slides (n = 3; error bars present means + SDs; **P < .01 when compared to 0 A4-FITC by Student t test).
Pep-1 delivers FITC-conjugated A4 peptide into platelets. The bar graph shows fluorescent uptake into platelets when the A4 peptide plus Pep-1 are mixed with PRP (dark bars) or washed platelet suspensions (light bars) for 30 minutes at 37°C. The molar ratio of A4 peptide to Pep-1 is maintained at 1:20 and optimal uptake as measured by flow cytometry occurs with 6.4 μM A4 peptide plus 128 μM Pep-1. The inset shows fluorescent micrographs after washed platelets are incubated with 6.4 μM A4 peptide plus 0 μM Pep-1 (left) or 6.4 μM A4 peptide plus 128 μM Pep-1 (right) and allowed to adhere to glass slides (n = 3; error bars present means + SDs; **P < .01 when compared to 0 A4-FITC by Student t test).
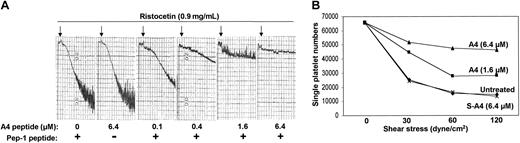
Having convinced ourselves that we can deliver the A4 peptide into intact human platelets, we next undertook a set of experiments to test the hypothesis that the A4 peptide, by disrupting filamin A binding to GpIbα, will affect platelet aggregation in response to VWF binding to GpIb-IX-V. Figure 7A shows that the A4 peptide inhibits ristocetin-induced aggregation of PRP. The anti-aggregatory effect shown here correlates with flow cytometry measurements of FITC-A4 uptake into platelets in PRP (Figure 6). Because ristocetin causes platelet aggregation by inducing VWF binding to GpIbα, these results suggest that the filamin A/GpIbα interaction positively regulates VWF binding to GpIb-IX-V. This interpretation is consistent with data from CHO cells demonstrating that mutations in the tail of GpIbα that disrupt filamin binding cause decreased shear-induced CHO cell adherence to VWF.28
The A4 peptide inhibits ristocetin- and shear-induced platelet aggregation. (A) A4 peptide inhibits ristocetin-induced aggregation of PRP. (B) Shows that 1.6 μM A4 peptide plus 32 μM Pep-1 or 6.4 μM A4 peptide plus 128 μM Pep-1 inhibits shear-induced aggregation of washed platelets after 2 minutes. The use of 6.4 μM scrambled A4 peptide plus 128 μM Pep-1 (S-A4) does not inhibit shear-induced platelet aggregation (in the far left panel of A, the amount of Pep-1 added was 128 μM; representative of 2 separate experiments).
The A4 peptide inhibits ristocetin- and shear-induced platelet aggregation. (A) A4 peptide inhibits ristocetin-induced aggregation of PRP. (B) Shows that 1.6 μM A4 peptide plus 32 μM Pep-1 or 6.4 μM A4 peptide plus 128 μM Pep-1 inhibits shear-induced aggregation of washed platelets after 2 minutes. The use of 6.4 μM scrambled A4 peptide plus 128 μM Pep-1 (S-A4) does not inhibit shear-induced platelet aggregation (in the far left panel of A, the amount of Pep-1 added was 128 μM; representative of 2 separate experiments).
To examine platelet aggregation in response to shear-induced VWF binding to platelet GpIb-IX-V, washed platelets were subjected to different shear stresses in a cone-plate viscometer. Figure 7B shows that 1.6 μM A4 peptide plus 32 μM Pep-1 or 6.4 μMA4 peptide plus 128 μM Pep-1 inhibits shear-induced aggregation after 2 minutes. The scrambled A4 peptide (S-A4; 6.4 μM plus 128 μM Pep-1) has no effect on shear-induced platelet aggregation. Aggregation is represented by a loss in the number of single platelets from the washed platelet suspension, and the inhibition of aggregation is observed at all shear stresses examined. We speculate that the apparent discrepancy between platelet uptake data (Figure 6 shows that 1.6 μM FITC-conjugated A4 peptide is not delivered to washed platelets) and aggregation data (Figure 7B shows that 1.6 μM A4 peptide inhibits shear-induced aggregation) is explained by enhanced delivery of the unconjugated A4 peptide compared with the FITC-conjugated peptide. Of note is that 6.4 μM A4 peptide plus 128 μM Pep-1 does not inhibit either thrombin-induced (0.5 U/mL) or ADP-induced (20 μM) aggregation of washed platelets, and that concentrations of Pep-1 more than or equal to 150 μM alone nonspecifically inhibit ADP and thrombin-induced aggregation of washed platelets (data not shown).
The mechanism of the effect of the A4 peptide should theoretically involve the interaction between filamin A and GpIbα. There are no published data indicating that this interaction is dynamically regulated in activated platelets or that it is modulated specifically by VWF binding to GpIb-IX-V. To examine for these possibilities, washed platelets activated by 120 dyne/cm2 shear stress were subjected to immunoprecipitation with the GpIbα-specific antibody AN51, and the presence of coprecipitating filamin A was measured by immunoblotting. Figure 8A shows that 6.4 μM A4 peptide plus 128 μM Pep-1 causes decreased filamin A binding to GpIbα. Please note that the immunoprecipitations were carried out in lysates from pellets following microcentrifugation of the sheared platelet suspensions, so that the concentration of extracellular A4 peptide during the immunoprecipitation procedure was negligible. Please also note that delivery of 6.4 μM A4 to resting platelets (the third lane from the left) does not cause decreased filamin A to coimmunoprecipitate with GpIbα, suggesting that the intraplatelet concentrations of A4 are insufficient to compete with endogenous GpIbα for filamin A binding during the overnight incubation. It is therefore reasonable to conclude that data in Figure 8A suggest that the A4 peptide competes effectively with GpIbα for a pool of filamin A made accessible as a consequence of shearing at 120 dyne/cm2 for 2 minutes.
The effect of the A4 on intact platelets. (A) The A4 peptide decreases filamin A binding to GpIbα. (B) Lysates from the same samples after they were subjected to immunoblotting for tyrosine phosphorylated proteins. (C) Quantitative evidence that delivery of the A4 peptide decreases the tyrosine phosphorylation of FAK, α-actinin, Syk, Fyn, and Src when platelets are sheared at 120 dyne/cm2 (n = 3; bars present means + SDs; *P ≤ .05; **P ≤ .01 when compared to sheared untreated platelets by the Student t test).
The effect of the A4 on intact platelets. (A) The A4 peptide decreases filamin A binding to GpIbα. (B) Lysates from the same samples after they were subjected to immunoblotting for tyrosine phosphorylated proteins. (C) Quantitative evidence that delivery of the A4 peptide decreases the tyrosine phosphorylation of FAK, α-actinin, Syk, Fyn, and Src when platelets are sheared at 120 dyne/cm2 (n = 3; bars present means + SDs; *P ≤ .05; **P ≤ .01 when compared to sheared untreated platelets by the Student t test).
Figure 8B shows lysates from the same samples after they were subjected to immunoblotting for tyrosine phosphorylated proteins. Each tyrosine phosphorylated band was identified by stripping the phosphotyrosine blot and then reprobing it for FAK, α-actinin, Syk, Fyn, and Src (data not shown). Figure 8C shows densitometric measurements of tyrosine phosphorylated bands indicating that the A4 peptide decreases the tyrosine phosphorylation of FAK, α-actinin, Syk, Fyn, and Src. These phosphorylations have been previously shown to be initiated by VWF binding to GpIb-IX-V.17-21 Such results indicate that filamin A binding to GpIbα regulates proaggregatory tyrosine kinase signaling.
Discussion
There is evidence that filamin A binding to the cytoplasmic domain of GpIbα enhances the ligand-binding adhesive function of GpIb-IX-V.28 This effect is almost certainly due to the capacity of filamin A to tether the GpIb-IX-V complex to the submembranous cytoskeleton, but the molecular mechanism of cytoskeletal-mediated “inside-out” signaling to GpIb-IX-V is unknown.10,29,30 By contrast, numerous data implicate the cytoskeleton in “inside-out” signaling to αIIbβ3, including a growing repertoire of specific molecules that modulate the activation of αIIbβ3's ligand binding function.5-8,31 Some of these molecules are affected by shear-induced VWF binding to GpIb-IX-V and therefore have the potential to provide both structural and functional coupling between GpIb-IX-V and αIIbβ3. We and others have shown that the cross-linked filamentous actin/α-actinin network is required for GpIb-IX-V–induced α β 10,30,31 IIb 3 activation. Such a network is not inert or metabolically inactive, and data indicate that GpIb-IX-V stimulates dynamic changes in individual cytoskeletal proteins associated with both protein and lipid kinases. In particular, α-actinin serves as scaffold protein that responds to shear-induced VWF binding to GpIb-IX-V by dynamically binding Syk, phosphatidylinositol 3-kinase IA, and phosphatidylinositol 4,5-bisphosphate, and regulating shear-induced tyrosine phosphorylation and phosphatidylinositol 3,4,5-trisphosphate production, both of which are critically important in the initiation and maintenance of αIIbβ3 activation.11,12,20,21 Others have shown that the cytoskeleton directs the final step in the activation of αIIbβ3, probably by polyphosphoinositide- or calcium-activated protease-mediated changes in the structure of talin altering its association with the integrin-β tail.6-9 3 And although neither of these mechanisms has yet been directly shown to operate following shear-induced VWF binding to GpIb-IX-V, either appears feasible based on evidence that both polyphosphoinositide turnover21 and calcium responses13-15 are induced when GpIbα engages VWF under shearing conditions.
If one accepts the premise that the cytoskeleton transduces activation signals emanating from GpIb-IX-V and impinging on αIIbβ3, it is essential to explore the role of filamin A, which is the first link in the chain of structural proteins that connect GpIb-IX-V with αIIbβ3. To begin this exploration, we tried to identify the filamin A binding domain in GpIbα. Using recombinant human filamin A, we corroborated the many published data on this subject that GpIbα has a large folded domain in its cytoplasmic tail (residues 543-590) comprised of several critical subdomains (residues 557-568, 560-570, 569-579, 580-590) that regulate filamin A binding to the cytoplasmic domain of GpIbα.27-30 In addition, we have also presented new data that introduce a derivative hypothesis about GpIbα's cytoplasmic domain tertiary structure. This hypothesis, which is based on results in Figure 3, suggests that in the middle of the broad filamin A–binding domain there is a smaller region organized into a primary filamin A–binding site (residues 557-575). This region could extend a surface that is covered by and grips the 3 compact β-sandwich repeats (repeats 17-19) used by filamin A for binding to the cytoplasmic domain of GpIbα.32 Although this model is consistent with mutagenesis data,28 it will require structural analyses for its confirmation.
The peptide inhibition data may also provide more than inferential structural information. The observation that the A4 peptide, encompassing cytoplasmic tail amino acids 557 to 575, uniquely inhibits filamin A binding to GpIbα, suggests that the A4 peptide could be a tool useful for examining the hypothesis that filamin A binding to platelet GpIb-IX-V is regulated, and that such regulation affects platelet responses to shear-induced VWF binding to GpIb-IX-V. In other words, if the A4 peptide competes with GpIbα's primary filamin A binding site, could it be used to modify filamin A binding to GpIbα in intact platelets activated by pathologic shear stress? And if it affects filamin A binding to GpIbα, does this influence platelet responses to pathologic shear stress?
Results in Figure 8A provide support for the hypothesis that shear-induced VWF binding to GpIb-IX-V affects filamin A binding. This figure shows that sheared platelets preincubated for 30 minutes with 6.4 μM A4 peptide plus 128 μM Pep-1 have decreased filamin A binding to GpIbα, suggesting that there may be changes in affinity of filamin A for GpIbα resulting from shear-induced VWF binding to GpIb-IX-V, and that such changes permit the A4 peptide to compete with GpIbα for binding to filamin A within the pathophysiologically relevant time-frame of 2 minutes. The idea that filamin A binding to GpIbα may be regulated by GpIbα-engaging ligand expands the repertoire of regulatory mechanisms operating to control filamin's role in cellular function. Currently, evidence indicates that filamin A is regulated through transcriptional means33 and by targeting to specific cytoskeletal compartments directed by the small GTPase RalA,34 protein kinase-dependent tyrosine35 and serine36,37 phosphorylation, and by establishing associations with adaptor proteins38 or protein kinases.39 In only one case has a regulatory event been considered to develop directly downstream of ligand binding to a transmembranous receptor (a growth factor receptor),36 and in no cases have these regulatory mechanisms been shown to affect filamin A binding to a target protein, particularly a target that also appears to initiate the signal directing the filamin A response.
Results in Figure 7 demonstrate that delivery of the A4 peptide to intact platelets blunts the aggregation response to VWF stimulated by either ristocetin or pathologic shear stress. Because aggregation of PRP in response to 0.9 mg/mL ristocetin is predominantly due to VWF binding GpIb-IX-V, its inhibition by the A4 peptide suggests that filamin A binding to GpIbα in intact platelets positively regulates VWF binding to GpIb-IX-V. This finding corroborates data from heterologous cells showing that filamin A binding to GpIb-IX-V regulates “inside-out” signaling. Specifically, it confirms data from CHO cells demonstrating that the disruption of the filamin A/GpIbα interaction by mutagenesis attenuates GpIb-IX-V-dependent anchorage to VWF under high shear conditions.28
Because shear-dependent platelet aggregation is critically dependent on the activation of integrin αIIbβ3, the inhibition of shear-induced aggregation by the A4 peptide implies that filamin A binding to GpIbα in intact platelets regulates αIIbβ3 activation. Results in Figure 8 suggest that filamin A binding to GpIbα regulates VWF-dependent shear-induced αIIbβ3 activation through its effect on tyrosine kinase signaling. This idea may be very important because there are many published data focusing on tyrosine kinases as signals for αIIbβ3 activation that are activated immediately downstream of VWF binding to GpIb-IX-V, but the mechanism by which they are switched “on” is unknown.17-19,40,41 Results in Figure 8 could help to narrow the focus of investigations to molecules closely associated with filamin A.
In summary, we have confirmed that there is a large filamin A–binding domain in the cytoplasmic tail of GpIbα, and we have shown data that this domain includes a smaller region that may be a primary filamin A–binding site regulated by VWF binding. We have also presented data that introduce the hypothesis that shear-induced modulation of filamin A/GpIbα interactions may affect proaggregatory tyrosine kinase signaling. Finally, we suggest that the A4 peptide, which disrupts filamin A binding to GpIbα, could serve as a prototype for a new class of antiplatelet agents targeted to treat diseases due to arterial thrombosis, such as myocardial infarction and stroke. Further investigations of filamin A/GpIbα interactions could provide insight into mechanisms of platelet activation that specifically operate to direct arterial thrombosis.
Prepublished online as Blood First Edition Paper, June 5, 2003; DOI 10.1182/blood-2002-12-3805.
Supported through grants by the Research Service of the Department of Veterans Affairs and the National Institutes of Health (HL65967).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Michael Berndt, Xiaoping Du, John Hartwig, and José López for providing us with essential reagents.