Abstract
After apoptosis or necrosis, macrophages clear dead cells by phagocytosis. Although this process is efficient, circulating nucleosomes can occur in certain diseases, presumably reflecting either increased production or impaired clearance. To investigate the generation of blood nucleosomes, graded numbers of apoptotic and necrotic cells were administered to healthy mice, and levels of blood nucleosomes and DNA were determined. Using Jurkat cells as a model, nucleosomes and DNA were detected in the blood after the administration of 108 apoptotic or necrotic cells per mouse by the intraperitoneal route. The kinetics of the response were similar for both types of cells. The role of macrophages was assessed by eliminating these cells with clodronate liposomes or silica. Although clodronate treatment alone produced a peak level of blood DNA, the subsequent administration of dead cells caused no change in DNA levels. In contrast, silica treatment alone did not elicit a blood DNA response, though this treatment limited the rise in DNA from administered cells. Molecular studies showed that the blood DNA following the administration of apoptotic or necrotic cells arose from the mouse and the Jurkat cells, and its size distribution was consistent with apoptosis. Together, these findings suggest that the generation of blood nucleosomes depends on macrophages, with apoptosis a concomitant of a high burden of dead and dying cells.
Introduction
Cell death is integral to the biology of the mammalian organism and varies in form and mechanism. Two types of cell death have been broadly defined. Apoptotic or programmed cell death is a regulated and orderly process mediated by a cascade of enzymes that rearrange and degrade cell constituents. Apoptosis occurs in physiologic and pathologic settings and can be triggered by a variety of stimuli that initiate a stereotyped death program. In contrast, necrotic or accidental cell death results from cell injury and is frequently a concomitant of inflammation, whether induced by physical or chemical means.1-4 Death by necrosis does not involve the regulated action of enzymes and leads to cellular changes that differ in phenotype and structure from apoptotic death. Although this dichotomy may be an oversimplification,5,6 it is nevertheless useful in assessing the impact of dead and dying cells on the organism.
In addition to their structure and phenotype, apoptotic cells and necrotic cells may differ in their interaction with the immune system. As shown in in vitro studies, apoptotic cells are rapidly engulfed by mononuclear phagocytes that bind an array of cell surface molecules, including phosphatidylserine. Phosphatidylserine appears on the surface of the apoptotic cell with the loss of membrane symmetry.7-12 In addition to signaling uptake by phagocytosis, phosphatidylserine binding may induce an anti-inflammatory state because of the production of mediators such as transforming growth factor-β (TGF-β).13-15 Like apoptotic cells, necrotic cells are engulfed by phagocytosis, though the recognition structures may differ.16 Unlike apoptotic cells, necrotic cells may induce inflammation.17-19 In the case of apoptotic cells, the high efficiency of uptake may explain the paucity of these cells by tissue histology.
Although macrophages can engulf and degrade apoptotic and necrotic cells, the subsequent disposition of the contents of these cells has not been well investigated. Furthermore, the disposition of dead cells that escape phagocytosis in vivo is unknown. These issues may be important, however, because of the evidence that the remains of apoptotic or necrotic cells may enter the circulation and exert immunologic effects. Thus, a number of clinical conditions are characterized by high blood levels of nucleosomes or DNA that presumably arise from dead cells.20-26 Among these conditions, systemic lupus erythematosus (SLE) is a prototypic autoimmune disease characterized by the production of antinuclear antibodies.27,28 In SLE, nucleosomes may serve as a source of self-antigen to drive pathogenic antibody responses and to perturb cytokine production when in the form of immune complexes.29,30 In animal models, circulating DNA and nucleosomes occur after exposure to lipopolysaccharide (LPS), which can induce apoptosis among other effects.31
In the current experiments, we wanted to explore the mechanisms for the generation of nucleosomes in the circulation from dead or dying cells. As a model, we used the administration of necrotic or apoptotic Jurkat cells into healthy mice by the intraperitoneal route, assaying nucleosomal material by either immunoassay of nucleosomes or fluorometric assay of DNA. We chose this model to allow the use of graded numbers of genetically identifiable cells and to avoid confounding effects resulting from agents like LPS that induce multiple cellular changes. Furthermore, we can manipulate the recipient to assess the role of macrophages in the handling of both cell types. In results presented herein, we show that the administration of apoptotic or necrotic cells produces circulating nucleosomal material, with DNA derived from the recipient mouse and administered cells. We also show that the expression of blood DNA requires the presence of macrophages. Together, these findings support the importance of macrophages in the disposal of dead and dying cells and indicate that the generation of blood DNA depends on more than one cell type.
Materials and methods
Administration of cell preparations
Female BALB/c mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and were used for experiments at ages 6 to 12 weeks. For the induction of apoptosis, Jurkat or J774 cells were treated with either camptothecin at 10 μg/mL or etoposide at 30 μg /mL (both products of Sigma Chemical, St Louis, MO) for 24 hours. For the induction of necrosis, cells were either heated at 56°C for 30 minutes or were treated with 70% ethanol for 10 minutes. After treatment, cells were washed 2 times with phosphate-buffered saline (PBS) and suspended in PBS. Cells were administered by the intraperitoneal (IP) route in doses and schedules as indicated in “Results” and in the figure legends. Mice were bled at regular times thereafter. Blood samples were collected into tubes with 3 to 5 μL 0.5 M EDTA (ethylenediaminetetraacetic acid), followed immediately by centrifugation. The plasma was removed and stored at –20°C until use.
Detection of nucleosomes in plasma
Nucleosome levels were determined by a capture enzyme-linked immunosorbent assay (ELISA) method using a pair of monoclonal antinucleosome antibodies, described previously.32,33 Briefly, Immulon II 96-well plates (Dynatech, Chantilly, VA) were coated overnight in PBS at 4°C with 5 μg/mL immunoglobulin G2a (IgG2a) monoclonal antinucleosome antibody denoted PA-1. The plates were blocked at 37°C for 1 hour with PBS containing 1% bovine serum albumin (BSA; Sigma) and 0.05% Tween 20. Plasma concentrations were diluted 1:200 in PBS, pH 7.4, containing 0.5% BSA and 0.05% Tween 20 and were incubated at room temperature for 1 hour. After washing, an IgG2b antinucleosome antibody, PL2-6, was added. Horseradish peroxidase (HRP)–conjugated goat antimouse IgG2b (Southern Biotechnology Associates, Birmingham, AL) was used as the detecting antibody. Color was developed using TMB substrate (0.015% 3,3′, 5,5′-tetramethylbenzidine HCl [0.01% H2O2]; Sigma) in 0.1 M citrate buffer, pH 4.0. Plates were read at OD380 on an automatic plate reader (UV MAX; Molecular Devices, Sunnyvale, CA).
For these experiments, the values were determined as relative units (RUs) according to the value of a rat nucleosome preparation purified from rat liver using a sucrose gradient sedimentation method.34 Nucleosome concentrations were determined by comparing the mean value of OD380 with a standard curve of the titrated rat nucleosomes to calculate the RU.
Plasma DNA quantitation
In some experiments, plasma DNA was assessed by a fluorometric assay. For this purpose, plasmas in various dilutions were mixed at a 1:1 ratio with the dye PicoGreen (Molecular Probes, Eugene, OR) diluted 1:200 in TE buffer (10 mM Tris, 1 mM EDTA, pH 8) in a black 96-well microtiter plate (Costar; Corning, Corning, NY). Fluorescence was measured using a TECAN GENios microplate fluorescence reader (Salzburg, Austria) with an excitation wavelength at 485 nm and an emission wavelength at 535 nm. Values were obtained as relative fluorescence units (RFUs). DNA concentrations in plasma were calculated according to a standard curve made from double-stranded calf thymus DNA (Sigma).
Flow cytometry analysis
Jurkat or J774 cells were pelleted and washed in PBS/0.5% BSA. Cell concentrations were adjusted to 1 × 107/mL in PBS. One hundred microliters (1 × 106 cells) was used for staining. After blocking the Fcγ receptor using an antimouse CD16/CD32 antibody (BD PharMingen, San Diego, CA), cells were resuspended in 100 μL binding buffer (10 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], 150 mM NaCl, 1 mM MgCl2, 5 mM KCl, 1.8 mM CaCl2, pH 7.4) containing 10 μL Annexin V–fluorescein isothiocyanate (FITC) (BD PharMingen) and 0.5 μg propidium iodide (PI) (Sigma) and then were incubated at room temperature for 1 hour. Cells were analyzed by FACScan flow cytometer (Becton Dickinson, Mansfield, MA). Data analysis was performed using CellQuest software (Becton Dickinson Immunocytometry System, San Jose, CA).
Plasma DNA purification
Blood was collected in 50 μL 0.5 M EDTA and was centrifuged for 2 minutes. Plasma DNAs were purified with a MasterPure DNA Purification kit (Epicentre, Madison, WI) according to the manufacturer's instructions.
PCR analysis
Primers specific to the Y chromosome were Y1.5 (5′-CTA GAC CGC AGA GGC GCC AT-3′) and Y1.6 (5′-TAG TAC CCA CGC CTG CTC CGG-3′). Primers for GAPDH were GAPDHf1 (5′-AGT ATG ATG ACA TCA AGA AGG-3′) and GAPDHr (5′-ATG GTA TTC AAG AGA GTA GGG-3′). Polymerase chain reaction (PCR) was performed for 2 minutes at 94°C, followed by 30 cycles of 94°C for 30 seconds; 60°C for 60 seconds; 72°C for 60 seconds; and a final extension at 72°C for 10 minutes.
Inhibition of macrophage function
Clodronate liposomes were prepared according to the method by Van Rooijen and Sanders.35 Briefly, 86 mg phosphatidylcholine and 8 mg cholesterol (Sigma) were mixed to form a phospholipid film by gentle rotation in a round-bottomed flask. Then 2.5 g clodronate (Sigma) in 10 mL PBS was added to the lipids. Nonencapsulated clodronate was removed by centrifugation. Encapsulated clodronate was washed twice with PBS and resuspended in 4 mL PBS. Clodronate-liposomes were injected intraperitoneally in a dose of either 25 μL or 100 μL. Silica (Sigma) was resuspended in PBS and injected intraperitoneally at 1 mg, 10 mg, or 50 mg per mouse at day 0 and day 3.
To assess the effects of clodronate on macrophage numbers, flow cytometry was performed on preparations of peritoneal washout cells. Briefly, each mouse was administered 100 μL clodronate/liposomes or 100 μL liposomes alone or was left untreated. At 90 hours, mice were killed by cervical dislocation, and peritoneal cells were harvested by lavage with 10 mL cold RPMI 1640 with 10% fetal bovine serum (HyClone, Logan, UT). After washing with PBS with 0.5% BSA, cells were treated with antimouse CD16/CD32 antibody (PharMingen) and then stained with phycoerythrin-F4/80 (PE-F4/80; Serotec, Raleigh, NC) or PE-Mac1 (PharMingen). Cell cytometry was performed as described.
In vitro treatment of macrophages with clodronate liposomes
To investigate macrophage responses in an in vitro system, the effects of clodronate liposomes were assessed using RAW264.7 cells as a model. Briefly, RAW264.7 cells were plated in 12-well plates (Costar; Corning) at 5 × 105 cells/well in 1 mL RPMI 1640 (Gibco, Carlsbad, CA) with 10% fetal bovine serum (HyClone) with 20 μg/mL gentamicin (Sigma) and were allowed to adhere overnight. Clodronate or control liposomes were added at 100, 10, or 1 μL/well, and cells were incubated for 24 hours. Supernatants were then removed, and the concentration of dsDNA was determined using PicoGreen. Cells were then washed 3 times with PBS; liposomes not phagocytosed were removed. Jurkat cells, made necrotic by heating at 56°C for 30 minutes, were added at 5 × 106 cells/well, and cells were incubated together in complete media. After 24 hours, supernatants were collected for the determination of dsDNA using PicoGreen.
Results
Plasma nucleosome and DNA levels after administration of apoptotic or necrotic cells
To determine the conditions for the release of nucleosomal material into the blood, apoptotic and necrotic Jurkat cells were administered to healthy mice, and plasma levels of nucleosomes or DNA were determined. For these experiments, apoptosis of Jurkat cells was induced by treatment with camptothecin or etoposide, and necrosis was induced by heating for 56°C for 30 minutes or by treatment with 70% ethanol for 10 minutes. Cells were administered at various doses, and plasma was sampled thereafter for analysis of nucleosomes by immunoassay. Figure 1 presents results of these experiments. As these data indicate, the administration of apoptotic and necrotic Jurkat cells led to the appearance of nucleosomes with similar kinetics. In these experiments, the highest levels of blood nucleosomes occurred with cells made apoptotic by treatment with etoposide.
Plasma nucleosome levels from apoptotic or necrotic Jurkat cells. After the administration of apoptotic (camptothecin, A) (etoposide, C) or necrotic (heat, B) (70% ethanol, D) Jurkat cells in varying doses, plasma samples were collected, and nucleosome levels were measured by immunoassay. Results are presented as mean (± SEM) of 3 mice for each dose as indicated. Nucleosome values are expressed as RUs.
Plasma nucleosome levels from apoptotic or necrotic Jurkat cells. After the administration of apoptotic (camptothecin, A) (etoposide, C) or necrotic (heat, B) (70% ethanol, D) Jurkat cells in varying doses, plasma samples were collected, and nucleosome levels were measured by immunoassay. Results are presented as mean (± SEM) of 3 mice for each dose as indicated. Nucleosome values are expressed as RUs.
To assess whether cell types other than Jurkat would produce similar findings, the experiments were repeated with J774 cells that were made apoptotic with etoposide or necrotic with ethanol. As shown in Figure 2, the administration of ethanol-treated necrotic J774 cells led to levels comparable to those of similarly treated Jurkat cells. In contrast, nucleosome levels after the administration of etoposide-treated J774 cells were low. To explore the basis of these differences, cell cytometry was performed to determine the cell populations found in the treated cells (Table 1). For this purpose, cells were analyzed in terms of staining with Annexin V and PI.
Plasma nucleosome and DNA levels from Jurkat and J774 cells. Plasma nucleosome levels (A) were assessed after the administration of 108 Jurkat or J774 cells, treated with varying methods as indicated. In comparison, plasma DNA levels (B) were determined using a fluorometric assay. Data are the mean (± SEM) of 3 mice.
Plasma nucleosome and DNA levels from Jurkat and J774 cells. Plasma nucleosome levels (A) were assessed after the administration of 108 Jurkat or J774 cells, treated with varying methods as indicated. In comparison, plasma DNA levels (B) were determined using a fluorometric assay. Data are the mean (± SEM) of 3 mice.
The data in Table 1 indicate 2 notable findings. The first concerns the differences between J774 and Jurkat cells in the induction of apoptosis by these agents. Thus, etoposide induced fewer J774 cells than Jurkat cells to undergo apoptosis. The second concerns the phenotype of ethanol-treated cells. Although these cells were positive for staining with PI, they differed from heat-treated cells in terms of Annexin V binding. The inability to detect Annexin V staining could result from chemical changes caused by ethanol. Together, these data suggest that levels of nucleosomes in the blood relate to the number of dead or dying cells in the population and that the induction of necrosis by heat or ethanol leads to different membrane changes. Despite these differences, heat-treated and ethanol-treated cells led to the appearance of blood DNA for either cell type.
Role of macrophages in the induction of plasma DNA
To explore the mechanisms for the generation of blood nucleosomes, the role of the macrophage was investigated. Among scavenger-type cells in the body, macrophages have been implicated in the uptake of apoptotic and necrotic cells, with phagocytosis mediated by a variety of cell surface molecules.7,8,12 The capacity for uptake is large, and, though engulfment and intracellular digestion of dead cells occur readily, the role of this process in the generation of nucleosomes into the extracellular milieu has not been previously investigated.
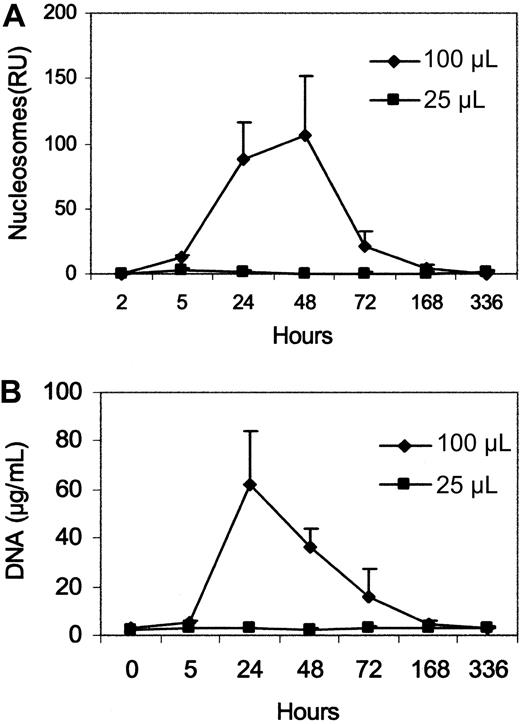
To determine whether macrophages play a role in the blood nucleosome response, we first investigated the consequences of eliminating macrophages by treatment with clodronate liposomes. Clodronate is a bis-phosphonate that can induce apoptosis after internalization.36,37 This technique has been commonly used to explore the role of macrophages in a variety of settings and leads to prolonged macrophage depletion. This depletion results from the uptake of liposomes by phagocytosis, with macrophages dying by apoptosis. Figure 3 presents results of experiments in which mice were treated with 2 doses of clodronate liposomes or empty liposomes and plasma levels of nucleosomes or DNA were measured.
Effect of clodronate liposomes on plasma nucleosome and DNA levels. Plasma nucleosome (A) and DNA (B) levels were measured after the administration of clodronate liposomes at 2 doses. Clodronate liposome doses are given in microliters. Data represent the mean (± SEM) of 3 mice. Empty liposomes (not shown) did not produce changes in levels of nucleosomes and DNA.
Effect of clodronate liposomes on plasma nucleosome and DNA levels. Plasma nucleosome (A) and DNA (B) levels were measured after the administration of clodronate liposomes at 2 doses. Clodronate liposome doses are given in microliters. Data represent the mean (± SEM) of 3 mice. Empty liposomes (not shown) did not produce changes in levels of nucleosomes and DNA.
As the data in Figure 3 indicate, treatment of healthy mice with the high dose clodronate liposomes caused a prompt and sustained nucleosome response that peaked at 24 to 48 hours and gradually declined. Under those conditions, empty liposomes had no effect on plasma DNA levels (data not shown). Because clodronate induces apoptosis, blood nucleosome and DNA responses most likely reflected the release of DNA from dead and dying macrophages.
To determine whether macrophage death in fact occurs under the conditions used in these experiments, we performed fluorescence-activated cell sorter (FACS) analysis of peritoneal washout cells using 2 different monoclonal antibodies to macrophages. As shown in Figure 4, at 90 hours after treatment of mice with clodronate, there was a marked diminution in the number of macrophages measured. Although treatment of mice with empty liposomes produced a change in the profile of staining, cell numbers were not reduced. These findings indicate that macrophage loss occurred after clodronate treatment and could be the source of DNA in the blood. We cannot exclude the possibility, however, that nucleosome release reflects the failure of macrophages to take up and process apoptotic cells that arise ordinarily. In the absence of a major clearance mechanism, these cells may break down and release nuclear contents.
Effect of clodronate on peritoneal macrophage cell populations. Mice were treated with either 100 μL control liposomes or clodronate liposomes. Ninety hours later, peritoneal cells were harvested by lavage, and macrophage levels were assessed by staining with F4/80 or Mac-1. Results are presented as FACS profiles for unstained cells, normal washout cells, washout cells from a mouse treated with clodronate alone, or a mouse treated with clodronate liposomes.
Effect of clodronate on peritoneal macrophage cell populations. Mice were treated with either 100 μL control liposomes or clodronate liposomes. Ninety hours later, peritoneal cells were harvested by lavage, and macrophage levels were assessed by staining with F4/80 or Mac-1. Results are presented as FACS profiles for unstained cells, normal washout cells, washout cells from a mouse treated with clodronate alone, or a mouse treated with clodronate liposomes.
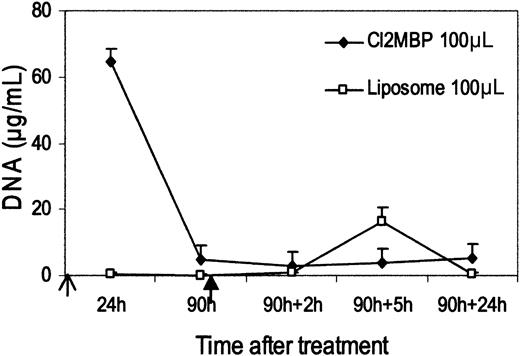
Figure 5 presents results of an experiment in which necrotic Jurkat cells were administered to mice that were treated with clodronate liposomes or empty liposomes. Cells were administered after the blood DNA level returned to baseline. As these data indicate, mice treated with clodronate failed to display a significant increase in blood DNA level, whereas control mice exposed to liposomes alone produced blood DNA levels similar to those of untreated mice. These results indicate that macrophages are required for the generation of blood DNA after the administration of dead cells.
Effect of clodronate treatment on plasma DNA release induced by the administration of dead cells. Open arrow: time of injection of clodronate (Cl2 MBP) liposomes. Solid arrow: injection of 108 Jurkat cells treated with ethanol. Mean (± SEM) of 4 mice in clodronate group and 3 control mice in liposome group are shown.
Effect of clodronate treatment on plasma DNA release induced by the administration of dead cells. Open arrow: time of injection of clodronate (Cl2 MBP) liposomes. Solid arrow: injection of 108 Jurkat cells treated with ethanol. Mean (± SEM) of 4 mice in clodronate group and 3 control mice in liposome group are shown.
To confirm these findings, the effects of silica were tested. Silica can impair macrophage function after uptake, with the occurrence of apoptosis dependent on the crystalline nature of the preparation.38-40 In these experiments, a preparation denoted as amorphous was used. This preparation consists of finely ground crystals and has previously been shown to cause prolonged functional impairment of macrophages. As shown in Figure 6, though the administration of silica to mice did not cause a blood DNA response, this treatment, in a dose-dependent way, impaired the generation of a blood DNA response after the administration of cells. These results support the idea that macrophages are required for the generation of blood DNA after exposure to dead cells.
Effect of silica on plasma DNA levels. Silica in varying doses was injected intraperitoneally (open arrow). Plasma samples were collected and assayed for DNA (A). In a separate experiment, 108 necrotic Jurkat cells were administered (solid arrow) following silica treatments (open arrows), and plasma DNA levels were measured (B). Each point represents the mean (± SEM) for 2 mice.
Effect of silica on plasma DNA levels. Silica in varying doses was injected intraperitoneally (open arrow). Plasma samples were collected and assayed for DNA (A). In a separate experiment, 108 necrotic Jurkat cells were administered (solid arrow) following silica treatments (open arrows), and plasma DNA levels were measured (B). Each point represents the mean (± SEM) for 2 mice.
Molecular properties of blood DNA
Although the administration of dead cells to healthy mice leads to the generation of blood DNA, the source of this DNA and the mechanism of its release cannot be determined from quantitation of DNA levels alone. To define further the properties of blood DNA, 2 analyses were performed. The first concerns molecular analysis of the circulating DNA by PCR to determine whether the DNA was derived from the Jurkat cells or the mouse. Because Jurkat cells are human in origin and are derived from a male, their DNA can be readily detected by amplifying human Y-chromosome sequences.41 In contrast, mouse DNA can be detected by amplifying mouse sequences. The second analysis of the DNA concerns the size of the DNA. For this purpose, plasma DNA was purified and analyzed by gel electrophoresis with staining by ethidium bromide. Whereas DNA from necrotic cells has high molecular weight, circulating DNA derived from apoptotic cells has lower molecular weight, with DNA in fragments from nuclease digestion.2,3
Figure 7 shows the results of these analyses. As the data indicate, healthy mice did not display DNA that could be visualized by gel electrophoresis, nor could it be amplified. In contrast, plasma from mice treated with clodronate showed DNA with a laddering pattern and a band corresponding to the mouse housekeeping gene GAPDH. As expected, the Y-chromosome sequence was not amplified. Results for the administration of dead cells showed similarity, whether the administered cells were apoptotic or necrotic. Thus, the circulating DNA showed laddering associated with an apoptotic source and with amplification of 2 DNA bands. One band corresponded to the Y-chromosome–specific TSPY/CYS14 gene and presumably originated from the administered Jurkat cells. The other band corresponded to the mouse GAPDH gene. Thus, even if the administered cells were necrotic, the resultant size distribution resembled that of apoptotic DNA. These results are thus consistent with the induction of cell death in recipient mice and the release of DNA that has been cleaved in a manner associated with apoptosis.
Properties of plasma DNA. Peak plasma DNA levels from various preparations, as indicated, were extracted and analyzed on a 0.8% agarose gel (A). These DNAs were also used as templates in PCR to amplify the Y chromosome (B) and the GAPDH gene (C). Numbers indicate DNA size according to a DNA standard. Results are representative of several experiments.
Properties of plasma DNA. Peak plasma DNA levels from various preparations, as indicated, were extracted and analyzed on a 0.8% agarose gel (A). These DNAs were also used as templates in PCR to amplify the Y chromosome (B) and the GAPDH gene (C). Numbers indicate DNA size according to a DNA standard. Results are representative of several experiments.
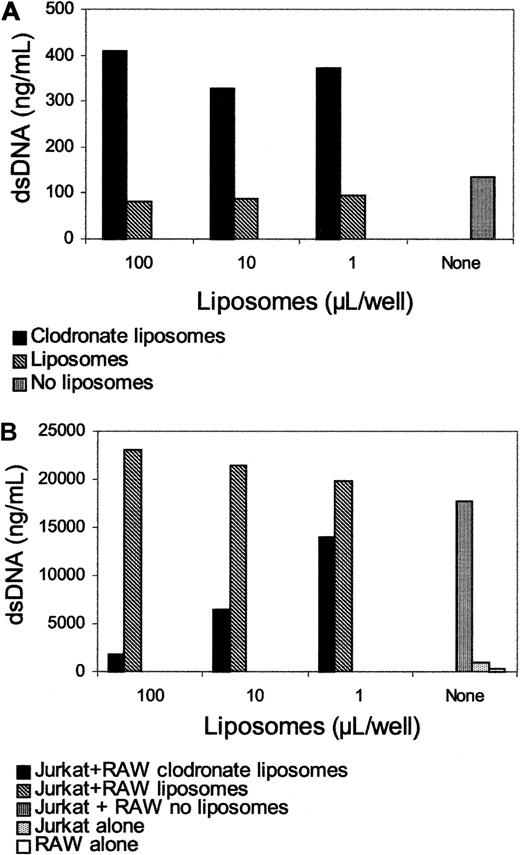
To define further the role of macrophages in the extracellular release of DNA, an in vitro model was established to assess salient features of the in vivo model. The first aspect tested concerned the effect of clodronate on DNA release by macrophages. Figure 8 shows the effects on the DNA levels in the media of RAW264.7 cells incubated with either clodronate liposomes, liposomes alone, or no other addition. As these data show, macrophages incubated with clodronate liposomes released DNA into the media. Because this agent induces apoptosis, these findings are consistent with in vivo experiments in suggesting that clodronate-induced apoptosis leads to DNA release. Furthermore, coculture of necrotic Jurkat cells with RAW264.7 cells leads to DNA release (Figure 8B), whereas RAW264.7 cells treated with clodronate liposomes, but not with liposomes alone, failed to release DNA into the medium. Similar to the in vivo situation, clodronate treatment of macrophages in vitro prevented DNA release, providing further evidence for the role of functional macrophages in the generation of extracellular DNA from dead or dying cells.
In vitro effects of clodronate on macrophages. RAW264.7 cells were incubated with various doses of clodronate, and DNA levels in the media were determined by PicoGreen after 24 hours (A). At that point, heated Jurkat cells were added to the macrophages, and DNA levels in the media were determined after 24 hours (B). Results are presented as nanograms per milliliter DNA as measured by fluorometry.
In vitro effects of clodronate on macrophages. RAW264.7 cells were incubated with various doses of clodronate, and DNA levels in the media were determined by PicoGreen after 24 hours (A). At that point, heated Jurkat cells were added to the macrophages, and DNA levels in the media were determined after 24 hours (B). Results are presented as nanograms per milliliter DNA as measured by fluorometry.
Discussion
Results presented here provide new insight into the clearance of dead and dying cells and into the origin of plasma nucleosomes and DNA. Thus, we have shown that the administration of apoptotic and necrotic cells to healthy mice is associated with the release of nucleosomes and DNA into plasma. This release is dependent on the presence of macrophages and appears to result from host cells and administered cells. Furthermore, because the administration of necrotic cells and apoptotic cells produced plasma DNA with a laddering profile, these results are also consistent with the induction of in vivo apoptosis or the degradation of necrotic cell DNA into nucleosomal fragments. Together, these findings indicate that the generation of plasma DNA may involve more than one cell type, with macrophages playing a central role.
As shown in a variety of in vivo and in vitro studies, the uptake and clearance of dead cells is a highly efficient process mediated by macrophages that bind dead cells, either apoptotic or necrotic, by cell surface receptors.7,8,12 This recognition event involves a variety of surface molecules, with phosphatidylserine a major target of recognition. The binding of phosphatidylserine to macrophages may also deliver a signal to these cells, causing an anti-inflammatory state.9,10,15 Although the uptake of necrotic cells may involve other surface molecules, this process is also highly efficient, though it may lead to a proinflammatory rather than an anti-inflammatory state.16-19
Although macrophages engulf and digest apoptotic and necrotic cells, the mechanisms for the disposal of the degraded contents have not been well characterized. DNA levels in the blood are elevated, however, in patients with systemic lupus erythematosus, malignancy, pulmonary embolus, and trauma and in women who are pregnant.20-26 In healthy mice, treatment with agents such as LPS elevates blood nucleosome levels, presumably the result of apoptosis.31 Although these situations are associated with increased blood DNA levels, the mechanisms by which DNA is released into the circulation and the cellular origin of this material have been unclear.
As an approach to these issues, we have developed a model based on the IP administration of apoptotic cells and necrotic cells to healthy mice, measuring nucleosomes or DNA in the blood as a marker of the degradation and release process. With this system, we have shown that the appearance of DNA in the blood occurs rapidly after cell administration, with apoptotic and necrotic cells producing a plasma nucleosome response of similar magnitude. Furthermore, the kinetics of this response were similar with dead cells of either type. Although we tested only 2 cell lines, it is not unlikely that this reaction is generalized and does not depend on cell lineage. In this experiment, we used the IP route for administering the apoptotic and necrotic cells. Preliminary experiments indicated that intravenous administration of such cell populations also led to the appearance of DNA, though we noted increased mortality rates among those mice (N.J., preliminary observation). At present, it is unclear whether DNA release varies depending on the site in which the administration of dead cells occurs. These experiments are in progress.
To determine the cellular basis for the release of nucleosomes and DNA, we initially focused on the role of the macrophage because this cell has been considered the major scavenger of apoptotic and necrotic cells, capable of phagocytosing and efficiently removing these cells. We used 2 approaches to eliminate macrophages, clodronate liposomes and silica.35-40 With clodronate liposome treatment, there were initial DNA and nucleosome responses in the blood. Clodronate liposome induces macrophage apoptosis and has been used in a variety of model systems to assess the role of macrophages in such diverse responses in mice as those to endotoxin release, allograft rejection, and hemolytic anemia.42-46 Clodronate liposome treatment affects of a wide variety of macrophage functions in vivo and leads to the loss of macrophages in liver, spleen, and other sites (eg, lung, peritoneum), depending on the route of administration. Indeed, FACS results presented in this study point to substantial macrophage elimination in at least the peritoneum. Given the effects of clodronate, treatment with this agent could elicit blood DNA from either the dead or dying macrophages or from the failure to clear other cell populations undergoing apoptosis.
Silica treatment also functionally eliminates macrophages and, depending on preparation, can induce apoptosis.38-40 In our experiments, we did not observe a blood DNA response, suggesting that apoptosis was not induced and that blood nucleosome and DNA levels after clodronate did not reflect a failure to clear apoptotic debris from ordinary processes. Nevertheless, in mice treated with silica at the high dose, the amount of DNA released into the plasma after the administration of necrotic cells was reduced in comparison with the controls. These findings indicate that macrophages are needed for the release of DNA and that the presence of dead and dying cells, even in large numbers, may be insufficient for release.
Because macrophages efficiently take up apoptotic and necrotic cells, at least 2 possibilities exist for their roles in the release of DNA from dead cells. In the first mechanism, dead cells are engulfed and digested by macrophages, and nucleosomal material is extruded. According to this scenario, the macrophages function efficiently, with large numbers of administered cells needed to produce enough debris for DNA levels in blood to increase. In the absence of macrophages, the contents of dead cells may be degraded by autolytic processes and gradually enter the circulation at levels below detection.
According to a second mechanism, the release of DNA into the blood represents a failure of the clearance systems, which ordinarily degrade dead cells without release of this molecule. When the clearance capacity of macrophages is exceeded—for example, with the administration of large numbers of dead cells—a number of events may ensue. First, dead cells may be degraded without uptake by macrophages or other cell types, releasing DNA. Second, with excessive amounts of engulfed cells, degradation processes may be incomplete, and nucleosomes may extrude or “overflow” from macrophages unable to keep up with the burden. Third, when their uptake and degradation capacities are exceeded, macrophages themselves undergo apoptosis.
Our results provide some credence for the role of dead and dying cells in the induction of apoptosis in macrophages (or other scavenger cell types). Thus, in the absence of macrophages, we did not observe the release of DNA. It is therefore unlikely that blood nucleosome and DNA responses reflect a failure of the clearance mechanisms. Furthermore, the studies on the properties of DNA in the circulation indicate that the administration of Jurkat cells leads to the appearance of mouse and human DNA and of DNA fragments. These findings suggest that high levels of apoptotic and necrotic cells can induce in vivo apoptosis, with the macrophage the most likely cell to die because of its scavenger activity. In vitro experiments support these possibilities.
In view of these results, including the molecular properties of circulating DNA, we propose that cell death, when extensive, can induce macrophage apoptosis, which leads to the release of nuclear contents (both the engulfed dead cell's and its own) in a cleaved form into the blood. In our studies, we administered dead cells to healthy mice as a model, but large-scale cell death could occur in patients who experience trauma, shock, and toxin exposure.47-50 Macrophage loss could contribute to the immune dysfunction that characterizes these conditions and could lead to complications such as infection. Modulating the clearance of dead and dying cells may represent an important avenue of therapy in a number of medical conditions.
Prepublished online as Blood First Edition Paper, May 29, 2003; DOI 10.1182/blood-2002-10-3312.
Supported by a VA Merit Review grant, an Alliance for Lupus Research grant, and National Institutes of Health grant AI44808.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Marc Monestier for providing the antinucleosome antibodies.