Abstract
The mechanisms by which agonists activate glycoprotein (GP) IIb-IIIa function remain unclear. We have reported data on a patient with thrombocytopenia and impaired receptor-mediated aggregation, phosphorylation of pleckstrin (a protein kinase C [PKC] substrate), and activation of the GPIIb-IIIa complex. Abnormalities in hematopoietic transcription factors have been associated with thrombocytopenia and platelet dysfunction. To define the molecular mechanisms, we amplified from patient platelet RNA exons 3 to 6 of core-binding factor A2 (CBFA2) cDNA, which encompasses the DNA-binding Runt domain; a 13-nucleotide (nt) deletion was found (796-808 nt). The gDNA revealed a heterozygous mutation (G>T) in intron 3 at the splice acceptor site for exon 4, leading to a frameshift with premature termination in the Runt domain. On immunoblotting, platelet CBFA2, PKC-θ, albumin, and IgG were decreased, but pleckstrin, PKC-α, -βI, -βII, -η, -ϵ, -δ, and -ζ, and fibrinogen were normal. Our conclusions are that (1) CBFA2 mutation is associated with not only thrombocytopenia, but also impaired platelet protein phosphorylation and GPIIb-IIIa activation; (2) proteins regulated by CBFA2 are required for inside-out signal transduction-dependent activation of GPIIb-IIIa; and (3) we have documented the first deficiency of a human PKC isozyme (PKC-θ), suggesting a major role of this isozyme in platelet production and function. (Blood. 2004;103:948-954)
Introduction
Signal transduction-dependent activation of integrin glycoprotein (GP) IIb-IIIa complex and subsequent fibrinogen binding is a prerequisite for platelet aggregation on exposure to receptor-mediated agonists. However, the molecular mechanisms by which GPIIb-IIIa function is regulated on cell activation are not fully understood. We have previously reported1 detailed studies in a patient with lifelong mucocutaneous bleeding manifestations, mild thrombocytopenia, and markedly abnormal platelet aggregation (including in primary wave) and dense granule secretion in response to multiple agonists. The patient's father and grandfather also had a history of easy bruising, and the father (who had thrombocytopenia) had died of acute leukemia. Phosphorylation of pleckstrin (a protein kinase C [PKC] substrate) and myosin light chain (MLC) was diminished in the patient's platelets on activation with platelet-activating factor (PAF) and thrombin. PKC activation has been linked to inside-out signal transduction-dependent activation of GPIIb-IIIa on platelets.2,3 In detailed studies in this patient, we demonstrated by flow cytometry1 that the platelets had a full complement of GPIIb-IIIa complexes but receptor-mediated signal transduction-dependent activation of GPIIb-IIIa was impaired on stimulation with PAF, adenosine diphosphate (ADP), and protease-activated receptor 1 (PAR-1) agonist SFLLRN. The expression of ligand-induced binding sites (LIBSs) induced by a number of agonists was normal. We concluded1 that the patient's platelets had a defect in GPIIb-IIIa activation due to an upstream defect in the signaling mechanisms rather than in the GPIIb-IIIa complex itself. The genetic defect in this patient has remained unknown.
Recent studies have shown that abnormalities in hematopoietic transcription factors may be associated with thrombocytopenia and platelet dysfunction, including in the activation (fibrinogen binding) of GPIIb-IIIa. For example, Shiraga et al4 demonstrated that transcription factor p45 NF-E2 knockout mice have thrombocytopenia and the megakaryocytes have impaired activation of GPIIb-IIIa despite the GPIIb-IIIa complexes being present in normal numbers. Deficiency of another transcription factor, GATA-1, is also associated with thrombocytopenia.5 In humans, deficiency of core-binding factor A2 (CBFA2; also called RUNX1, AML1), which regulates expression of several genes involved in hematopoiesis,6 is associated with familial thrombocytopenia, predisposition to acute leukemias, and poorly characterized platelet dysfunction.7 We, therefore, postulated that our patient may have a defect in one of these transcription factors. In the present study we demonstrate that the propositus has a heterozygous mutation in CBFA2, indicating that proteins regulated by this transcription factor play a role in the activation of GPIIb-IIIa. In addition, we show that the platelets are deficient in one of the PKC isozymes, PKC-θ. To our knowledge, this is the first report of a human platelet deficiency in a PKC isozyme.
Patient, materials, and methods
Patient information
Details regarding the clinical presentation and the studies on platelet aggregation, secretion, activation of GPIIb-IIIa, protein phosphorylation, and Ca2+ mobilization on activation have been described previously.1 All studies in human subjects were performed after approval by the Institutional Review Board of the Temple University School of Medicine.
Materials
TRIzol RNA extraction reagent and Platinum PCR Supermix were purchased from Invitrogen (Carlsbad, CA). The DNA extraction kit was from Gentra Systems (Minneapolis, MN). All polymerase chain reaction (PCR) primers were synthesized by Integrated DNA Technologies (Caralville, IN). Antibodies against CBFA2 and CBFA2+ control (Jurkat nuclear extract) were purchased from Active Motif (Carlsbad, CA). Antibodies against PKC isoforms, PKC-βI (mouse monoclonal), -βII (rabbit), -ϵ (rabbit), -δ (rabbit), -η (rabbit), and -θ (goat), anti-PKC-ϵ antibody-blocking peptide, and horseradish peroxidase (HRP)-conjugated donkey antigoat IgG were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-PKC-α (mouse monoclonal) and antipleckstrin (mouse monoclonal) antibodies were from Transduction Laboratories (San Diego, CA). Antialbumin (rabbit), antifibrinogen (mouse), and anti-IgG (rabbit) antibodies were from Sigma-Aldrich (St Louis, MO). HRP-conjugated antirabbit IgG and antimouse IgG were purchased from Promega (Madison, WI). Antibody CD41a (GPIIb-IIIa) conjugated with fluorescein isothiocyanate (FITC) and isotype control were from PharMingen (San Diego, CA). PAC-1 antibody was obtained from Becton Dickinson (San Diego, CA). Ten percent resolving and 10% to 20% gradient density sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and Western blotting buffers were purchased from Bio-Rad (Hercules, CA). Gelcode blue stain reagent and Restore Western blot stripping buffer were from Pierce Chemicals (Rockford, IL). Other chemicals and reagents were from Sigma Chemical (St Louis, MO).
Preparation of platelet RNA and proteins
Four tubes of 42.5 mL blood were collected from the patient and healthy volunteer donors into 7.5 mL each of acid citrate-dextrose (ACD) solution (71.4 mM citric acid, 85 mM sodium citrate dihydrate, 11.1 mM dextrose). A stable prostacyclin analog, carbacyclin (30 nM), was added to the blood, and incubated for 20 minutes at room temperature. Platelet-rich plasma (PRP) was obtained by centrifugation at 200g for 20 minutes at room temperature. Only the upper two thirds of the PRP was collected and carbacyclin (30 nM) and ACD (one-tenth volume) were added to the PRP. The platelets were pelleted by centrifugation at 650g for 15 minutes at room temperature. The platelet pellet was resuspended in HEPES buffer (pH 6.5, 20 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], 5.5 mM dextrose, 0.376 mM NaH2PO4, 1 mM MgCl2, 2.7 mM KCl, 137 mM NaCl, and 1 mg/mL bovine serum albumin), and washed twice using the same buffer by centrifugation at 650g for 10 minutes at room temperature. Platelet pellets were resuspended in 6 mL TRIzol reagent and total RNA was isolated as recommended by the manufacturer. The final RNA preparations were stored in diethylpyrocarbonate-treated water at -80°C.
For preparing platelet protein, platelet pellet from 45 mL blood was resuspended in 0.6 mL M-Per Mammalian Protein Extraction Reagent (Pierce Chemical) with proteinase inhibitors (1 mM EGTA [ethylene glycol tetraacetic acid], 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 μg/mL leupeptin, 10 μg/mL aprotinin) by repeated pipetting and incubated on ice for 10 minutes. Platelet protein in supernatant was collected after centrifugation at 14 000 rpm for 20 minutes at 4°C and stored at -80°C.
Preparation of neutrophil RNA
Blood was drawn into one-tenth volume of 3.8% sodium citrate and overlaid on neutrophil isolation medium (NIM) essentially as described by the manufacturer (Cardinal Associates, Santa Fe, NM). Red blood cells (RBCs) were sedimented by centrifugation at 400g for 40 minutes at room temperature. The neutrophil layer was collected and washed twice by centrifugation with Hanks balanced salt solution (HBSS). The RBCs were lysed by incubation with 2.5 volumes of erythrocyte-lysing buffer (E-lyse; Cardinal Associates) for 10 minutes at room temperature. The neutrophils were recovered from the lysis solution by centrifugation at 250g for 5 minutes at room temperature. The preparation was assessed for viability by trypan blue dye exclusion. The RNA was extracted as described for platelets.
Preparation of leukocyte gDNA
Whole blood (300 μL) anticoagulated with one-tenth volume of either ACD or 3.8% sodium citrate was transferred to a 1.5-mL Eppendorf tube containing 900 μL RBC lysis solution. Leukocyte gDNA was extracted according to manufacturer's guidelines using DNA extraction kit (Purgen, Gentra Systems).
PCR amplification of cDNA
First-strand cDNA was synthesized from total RNA using oligo(dT)12-18 primers with Superscript II reverse transcription kit (Invitrogen). PCR was performed using the Platinum PCR SuperMIX reagent, which contains 22 U/mL complexed recombinant Taq DNA polymerase with PLATINUM Taq antibody, 22 mM Tris (tris(hydroxymethyl)aminomethane-HCl (pH 8.4), 55 mM KCl, 1.65 mM MgCl2, 220 μM each of deoxyguanosine triphosphate (dGTP), deoxyadenosine triphosphate (dATP), deoxythymidine triphosphate (dTTP), and deoxycytidine triphosphate (dCTP), and stabilizers. PCR reagent (45 μL) was mixed with 4 μL cDNA and 200 nM primers. The sample mixture for each PCR was heated for 2 minutes at 94°C and subjected to 32 cycles with heating at 94°C for 30 seconds, at specific annealing temperature as indicated below for 30 seconds, and at 72°C for 30 seconds, followed by final elongation at 72°C for 5 minutes. PCR amplification of cDNA was performed in a Perkin Elmer DNA thermal Cycler (Perkin Elmer Cetus, Norwalk, CT). p45 NF-E2 cDNA was amplified with primers: sense 5′-GTCCAGCAGTGTCAGCTCAG-3′ (119-138), antisense 5′-AATCCCATCAGCAGTTCCAC-3′ (1419-1400 at an annealing temperature of 56°C, and sequenced with an additional primer 5′-CATACTCCTATGGCAACATG-3′ (550-569) (numbering as per GenBank accession no. BC0005044). GATA-1 cDNA was amplified with primers, sense 5′-AACCACCAGCCCAGGTTAAT-3′ (80-100) and antisense AGAGGAGAAGGACACCA CCC (1401-1382) at an annealing temperature of 58°C, and sequenced using primer 5′-CTTGTAGTAGAGGCCGCAGG-3′ (954-937) (accession number NM-002049). The Runt domain of CBFA2 cDNA spanning exons 3 to 6 was amplified with 2 sets of primers S21/AS21 (591-613/1231-1206) and S22/AS22 (592-611/1136-1118) as described by Osato et al.8 The numbering in parentheses indicates nucleotide positions in CBFA2 cDNA according to GenBank entry D43969. PCR was first performed with primers S21/AS21 (annealing temperature 57°C) and then one tenth of the first PCR solution was used as template to run a second PCR with primers S22/AS22 (annealing temperature 57°C). In addition, a short segment of platelet and neutrophil CBFA2 cDNA (179 bp) spanning exons 3 and 4 (769-947) was amplified with primers and conditions as described.7 Finally, exon 8 of CBFA2 was amplified using primers: sense 5′-CAATACCTGGGATCCATTGC-3′ (1300-1320) and antisense 5′-CCTCAGTAGGGCCTCCA CAC-3′ (1889-1870) for 40 cycles with annealing for 50 seconds at 54°C. The PCR products were visualized by 0.05% ethidium bromide-stained agarose gel electrophoresis and excised. The amplified product was purified using the Qiagen purification kit (Chatsworth, CA). PCR products were sequenced by automatic sequencing performed using the PRISM Ready Reaction DyeDeoxy Terminator Cycle Sequencing Kit on the Applied Biosystems model 377 DNA Sequencing System (Foster City, CA).
PCR amplification of gDNA
A 343-bp segment of CBFA2 genome encompassing the splicing site between intron 3 and exon 4 was amplified in the propositus and the family members using leukocyte gDNA and primers: sense 5′-CATTGCTATTCCTCTGCAACC-3′ (82553-82533) and antisense CCATGAAACGTGTTTCAAGC (82220-82239) (GenBank no. AP000172.11). DNA (200 ng) was added to the PCR mixture, and the reaction was carried out at 94°C for 5 minutes to denature DNA, and then for 34 cycles of 1 minute each with heating at 94°C, at 54°C, and at 72°C. The patient's CBFA2 exons 1, 2, 3, 6, 7a, and 7b were amplified with primers and conditions as described7 (GenBank accession no. D43969). PCR products were purified and sequenced as described in “PCR amplification of cDNA.”
Flow cytometry analysis
This was carried out essentially as described previously.1 Blood was collected into one-tenth volume of 3.8% sodium citrate. PRP was collected by centrifuging the citrated blood at 200g for 20 minutes at room temperature, and platelets were diluted to 2 × 108/mL platelets with Tyrode buffer (pH 7.4, 5 mM HEPES, 2 mM MgCl2, 0.3 mM NaH2PO4, 3 mM KCl, 134 mM NaCl, 12 mM Na2HPO4, 0.1% glucose, and 0.35% bovine serum albumin). To assess activation of the GPIIb-IIIa complex, 10 μL (1 × 106 platelets) of platelet suspension was incubated with PAC-1 antibody (3.25 ng/mL) and agonist for 15 minutes at room temperature. To assess PAC-1 binding to resting platelets, PRP was incubated with 0.5 μM carbacyclin for 20 minutes at room temperature, and the platelet suspension was incubated with PAC-1 antibody without agonists. To assess the total GPIIb-IIIa complexes on platelets, antibody CD41a was incubated with 1 × 106 platelets for 15 minutes at room temperature. The antigen-antibody reaction was stopped by adding ice-cold Tyrode buffer to a final volume of 0.5 mL, and the samples were analyzed on a FACScan Flow Cytometer (Becton Dickinson). A total of 10 000 platelets were analyzed per sample. Results were expressed as histograms of log platelet fluorescence intensity in arbitrary units on the abscissa and platelet number on the ordinate.
Platelet aggregation and secretion of 14C serotonin
These studies were carried out using PRP and ADP, collagen, PAF, and SFLLRN as agonists, as described previously.1
Immunoblotting for CBFA2, pleckstrin, PKC isoforms, and α-granule proteins
The protein content in platelet homogenates was determined by the bicinchoninic acid (BCA) method using a commercial assay (Pierce Chemical). Equal amounts of total protein from different subjects were loaded and separated by SDS-PAGE (10% resolving gel, or 10%-20% gradient gel) and electrophoretically transferred to polyvinylidene fluoride (PVDF) nitrocellulose membranes (Millipore, Bedford, MA). The membranes were washed in Tris-buffered saline (TBS) with 0.1% Tween-20 and incubated for 1 hour at room temperature with the specific antibody in TBS with 1% bovine serum albumin and 0.1% Tween-20. All primary antibodies were used at a dilution of 1:2000 (1:4000 for antifibrinogen antibody). In experiments with the blocking peptide for anti-PKC-ϵ, the primary antibody was first incubated with 5 × blocking peptide concentration at 4°C overnight, and then incubated with PVDF membrane (1:2000 dilution). Antibodies bound to nitrocellulose were detected using a peroxidase-conjugated second antibody (1:5000) for 1 hour at room temperature, developed by Western Blot Chemiluminecence Reagent Plus (NEN, Boston, MA) and analyzed on a Kodak X-OMAT Blue imaging film (Perkin Elmer Life Sciences, Boston, MA). In some experiments, the PVDF membranes were stripped with stripping buffer and then probed with a different antibody. In experiments assessing platelet levels of IgG and fibrinogen, the platelet proteins were separated by PAGE under nonreducing conditions. Platelet levels of β-thromboglobulin were measured by radioimmunoassay.9
Quantitation of platelet PKC-θ mRNA by real-time PCR
First-strand cDNA synthesis was performed using 1.0 μg total RNA from platelets and SuperScript II Reverse Transcriptase (Gibco BRL, Carlsbad, CA) according to the manufacturer's instructions. Fluorescent PCR analysis was performed using the LightCycler (Roche Molecular Biochemicals, Indianapolis, IN) and FastStart DNA Master SYBR Green I Kit (Roche Diagnostics, Indianapolis, IN). Each reaction contained 1 μL cDNA, 2 μL 10 × PCR mixture, and 0.5 μM primer in a 20-μL volume. The amplification of PKC-θ (NM_006257) and the housekeeping gene hydroxymethyl-bilane synthase (HMBS; accession no. NM_000190)10 were performed in separate capillaries. The PCR primers for PKC-θ were sense 5′-AGGCCGAATGCTAATGAATG-3′ (357-376) and antisense 5′-GGAACATGGTTTCTCGGCTA-3′ (685-666) and those for HMBS were sense 5′-CCTGAGGATGACCCACAGTTGG-3′ (1076-1097) and antisense 5′-GGGGTAA TCACTC CCCAGAT-3′ (1337-1318). Neutrophil cDNA from normal controls was applied as a calibrator, which was used to normalize all samples within one run and for providing a constant calibration point between several PCR runs.
In the LightCycler instrument, each reaction capillary underwent a 10-minute incubation at 95°C followed by 43 cycles of 95°C for 12 seconds, 58°C for 12 seconds, and 72°C for 15 seconds, and this was combined with melting curve analysis. Fluorescence was monitored at the end of each elongation. The fluorescence detected in channel F1 was analyzed by LightCycler Analysis Software (Roche Molecular Biochemicals) at the end of the run. The crossing points (beginning of the PCR exponential phase) for each reaction were determined by the second derivative maximum algorithm and arithmetic baseline adjustment. For size estimation, the amplified product was confirmed on 2% agarose gels with 0.05% ethidium bromide. Analysis of results was performed with Relative Quantification software (Roche Molecular Biochemicals) without efficiency correction. The final results are shown as the normalized ratio of PKC-θ mRNA concentration to HMBS mRNA concentration (Light Cycler Technical Notes L, 13/2001).
Results
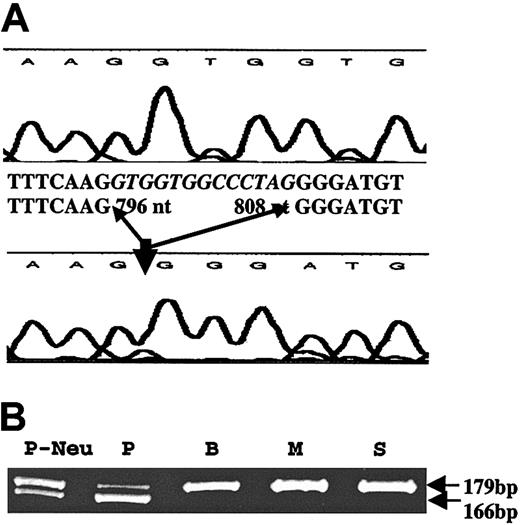
We first amplified platelet RNA from the patient by RT-PCR using primers specific for p45 NF-E2; no mutations were detected in the cDNA (1663 bp). Likewise, GATA-1 cDNA (1498 bp) revealed no mutations. We amplified and sequenced exons 3 to 6 CBFA2 (cDNA), which encompasses the conserved DNA-binding Runt domain.7 These studies revealed a 13-nt deletion in the patient cDNA sequence corresponding to 796 to 808 nt in exon 4 (accession no. D43969; Figure 1). Amplification of platelet RNA by RT-PCR (forward primer in exon 3 and reverse primer in exon 4) showed the presence of 2 transcripts of 179 bp (expected size) and 169 bp (Figure 1B), consistent with a heterozygous abnormality in the patient. The same results were obtained using neutrophil RNA.
Amplification of platelet RNA by RT-PCR. (A) Platelet RNA from a control subject (top tracing) and the patient (lower tracing) were subjected to RT-PCR using primers flanking exons 3 to 6 of CBFA2. The sequence shows a loss of 13 nt (796-808) in the patient in the Runt domain. (B) Amplification of platelet RNA from the patient (P), and the patient's brother (B), mother (M), and sister (S), and of patient's neutrophil RNA (P-Neu) by RT-PCR using a forward primer in exon 3 and reverse primer in exon 4. Two transcripts of 179 bp (expected size) and 169 bp are noted in the patient with only a single 179-bp transcript in the family members.
Amplification of platelet RNA by RT-PCR. (A) Platelet RNA from a control subject (top tracing) and the patient (lower tracing) were subjected to RT-PCR using primers flanking exons 3 to 6 of CBFA2. The sequence shows a loss of 13 nt (796-808) in the patient in the Runt domain. (B) Amplification of platelet RNA from the patient (P), and the patient's brother (B), mother (M), and sister (S), and of patient's neutrophil RNA (P-Neu) by RT-PCR using a forward primer in exon 3 and reverse primer in exon 4. Two transcripts of 179 bp (expected size) and 169 bp are noted in the patient with only a single 179-bp transcript in the family members.
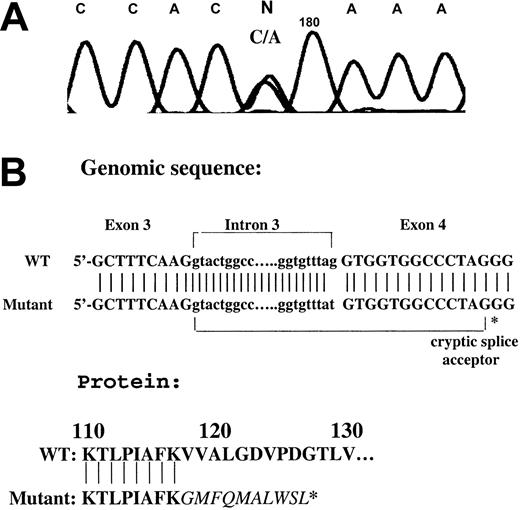
Amplification of gDNA using a forward primer in intron 3 and reverse primer in exon 4 revealed a heterozygous mutation in intron 3 (G>T; Figure 2A) at the splice acceptor site for exon 4, leading to the use of an enforced cryptic splice acceptor site in exon 4 and a frameshift with premature termination in the Runt domain (Figure 2). This results in a loss of 87 amino acids from the carboxy terminus of the Runt homology region; the abnormality is identical to the mutation previously described in an unrelated family with familial thrombocytopenia with predisposition to acute leukemia.7 No other mutations were detected in CBFA2 exons 1, 2, 3, 6, 7a, 7b, and 8 of the patient.
Amplification of a gDNA by PCR using a forward primer in intron 3 and reverse primer in exon 4. (A) A heterozygous point mutation (G>T) was noted in the patient. The tracing shows the reverse strand. (B) Mutational analysis showing a mutation in the splice acceptor site of exon 4 leading to the use of an enforced cryptic splice acceptor site in exon 4 and a frameshift with premature termination in the Runt domain. WT indicates wild type.
Amplification of a gDNA by PCR using a forward primer in intron 3 and reverse primer in exon 4. (A) A heterozygous point mutation (G>T) was noted in the patient. The tracing shows the reverse strand. (B) Mutational analysis showing a mutation in the splice acceptor site of exon 4 leading to the use of an enforced cryptic splice acceptor site in exon 4 and a frameshift with premature termination in the Runt domain. WT indicates wild type.
Studies in family members
The platelet counts were normal in the patient's mother, sister, and brother (all asymptomatic). The binding of monoclonal antibody CD41a that binds to both resting and activated GPIIb-IIIa complex was normal in all subjects, including the propositus (Figure 3), indicating that the platelets had a full complement of GPIIb-IIIa on the surface. The binding of antibody PAC-1 by platelets activated with PAF and U46619 was diminished in the patient but normal in the family members (Figure 3). The platelet aggregation and 14C secretion responses on activation of PRP with ADP, PAF, collagen, U46619, and PAR-1 agonist SFLLRN were normal in the mother and sister.
Activation of GPIIb-IIIa using flow cytometry. Binding of antibody CD41a (which binds to the GPIIb-IIIa complex) and PAC-1 (which binds to the activated GPIIb-IIIa complex) in the patient, brother, mother, and sister. CD41a binding was normal in all subjects, indicating that the GPIIb-IIIa complexes were present in normal numbers. PAC-1 binding in response to PAF and U46619 was diminished in the patient but normal in the family members. For PAC-1 binding, the open peak represents the resting state; the filled peak is after activation.
Activation of GPIIb-IIIa using flow cytometry. Binding of antibody CD41a (which binds to the GPIIb-IIIa complex) and PAC-1 (which binds to the activated GPIIb-IIIa complex) in the patient, brother, mother, and sister. CD41a binding was normal in all subjects, indicating that the GPIIb-IIIa complexes were present in normal numbers. PAC-1 binding in response to PAF and U46619 was diminished in the patient but normal in the family members. For PAC-1 binding, the open peak represents the resting state; the filled peak is after activation.
The gDNA CBFA2 sequence did not reveal the mutation at the splice junction between intron 3 and exon 4 in any of the other family members. We amplified platelet mRNA using a forward primer in exon 3 and reverse primer in exon 4; only one band (179 bp) was seen in the family members with 2 bands in the propositus (Figure 2). We believe that the patient has an autosomal dominant defect because the father also had thrombocytopenia and developed acute leukemia, recently recognized features of CBFA2 deficiency.7
Platelet levels of CBFA2, pleckstrin, PKC isozymes, and α-granule proteins
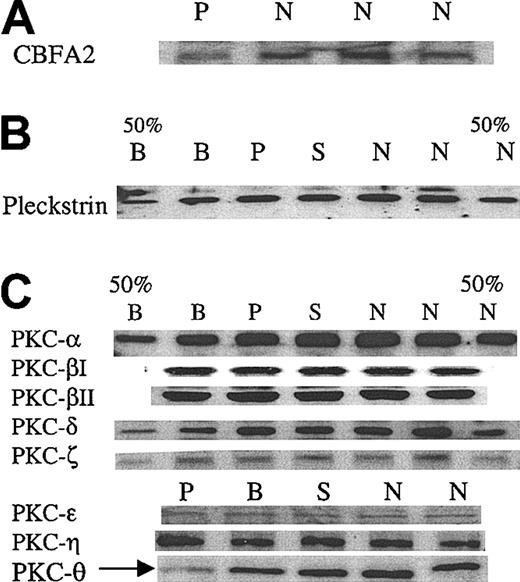
We assessed platelet CBFA2 protein level in the patient by immunoblotting and found it to be decreased (Figure 4A), although not very strikingly. A major signaling abnormality reported by us1 in the propositus was decreased phosphorylation of the PKC substrate pleckstrin in response to platelet activation with thrombin and PAF. To obtain insights into this abnormality, we assessed pleckstrin content in platelets by immunoblots and it was normal (Figure 4) in the patient. We then examined levels of several PKC isozymes including PKC-α, -βI, -βII, -δ, -ϵ, -η, -ζ, and -θ, previously shown to be present in platelets. PKC-θ levels were decreased in the patient, whereas the other PKC isozymes were intact (Figure 4C). These findings were consistently noted in at least 3 separate experiments. In some experiments, the immunoblot for PKC-θ was stripped and probed again with antibodies against PKC-δ and PKC-ζ. The results showed that only PKC-θ levels were decreased with normal levels of the other 2 PKCs, confirming that the protein amount loaded for each sample was consistent (data not shown).
Immunoblot analyses of CBFA2, pleckstrin, and PKC isozymes in platelets. (A) Immunoblot analysis of CBFA2 in platelets. Platelet lysates were subjected to SDS-PAGE and immunoblotting using an anti-CBFA2 antibody. Shown are the results in the patient (P) and 3 control subjects (N). (B) Immunoblot analysis of pleckstrin in platelets. Platelet lysates were subjected to SDS-PAGE and immunoblotting using an antipleckstrin antibody. Shown are the results in the patient (P), brother (B), sister (S), and 2 control subjects (N). 50% B and 50% N represent lanes with 50% protein applied. Pleckstrin levels are normal in the propositus. (C) Immunoblot analysis of PKC isozymes. The details are as described for panel B. PKC-θ levels were decreased in the patient (arrow). The results shown are representative of at least 3 separate experiments. To confirm that protein loading was comparable between the patient and the control subjects, the immunoblots used for PKC-θ were stripped and reblotted using antibodies against PKC-δ and PKC-ζ, which showed that the levels of these PKC in the patient were comparable to those of the other subjects (not shown).
Immunoblot analyses of CBFA2, pleckstrin, and PKC isozymes in platelets. (A) Immunoblot analysis of CBFA2 in platelets. Platelet lysates were subjected to SDS-PAGE and immunoblotting using an anti-CBFA2 antibody. Shown are the results in the patient (P) and 3 control subjects (N). (B) Immunoblot analysis of pleckstrin in platelets. Platelet lysates were subjected to SDS-PAGE and immunoblotting using an antipleckstrin antibody. Shown are the results in the patient (P), brother (B), sister (S), and 2 control subjects (N). 50% B and 50% N represent lanes with 50% protein applied. Pleckstrin levels are normal in the propositus. (C) Immunoblot analysis of PKC isozymes. The details are as described for panel B. PKC-θ levels were decreased in the patient (arrow). The results shown are representative of at least 3 separate experiments. To confirm that protein loading was comparable between the patient and the control subjects, the immunoblots used for PKC-θ were stripped and reblotted using antibodies against PKC-δ and PKC-ζ, which showed that the levels of these PKC in the patient were comparable to those of the other subjects (not shown).
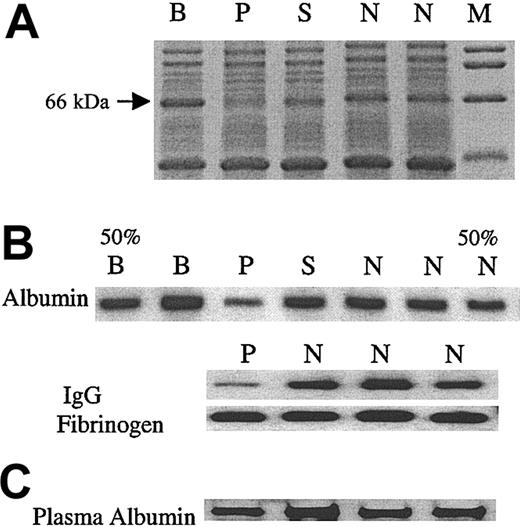
An interesting observation in the initial protein-stained SDS gels was a consistent decrease in a 66-kDa band in the patient (Figure 5). We, therefore, assessed the levels of albumin (68 kDa) in the platelet lysates by immunoblotting and found it to be decreased (Figure 5B) in the patient. The plasma albumin level was normal in the patient (Figure 5C). We assessed 2 other platelet proteins that are incorporated into the α granules from the plasma; platelet levels of IgG, but not fibrinogen, were decreased (Figure 5B). Moreover, platelet β-thromboglobulin, an α-granule protein synthesized by megakaryocytes, was normal in the patient (patient, 50.3 μg/109 platelets; control subjects, 42.5 ± 9.9 μg/109 platelets, mean ± SD, n = 10).
Analysis of platelet proteins, albumin, IgG, and fibrinogen. (A) Platelet lysates were subjected to SDS-PAGE. Gelcode blue-stained gel showing a decrease in an approximate 66-kDa band in the patient (P) but not the family members (B, S) or control subjects (N). M represents molecular weight markers. (B) Immunoblot analyses of albumin, IgG, and fibrinogen in platelet lysates. In the first (50% B) and the last (50% N) lanes, half of the corresponding sample (B, N) was applied, to assess the sensitivity of the analysis. (C) Immunoblot analysis of albumin in plasma. The albumin in patient plasma sample was comparable to that of 3 control subjects (N).
Analysis of platelet proteins, albumin, IgG, and fibrinogen. (A) Platelet lysates were subjected to SDS-PAGE. Gelcode blue-stained gel showing a decrease in an approximate 66-kDa band in the patient (P) but not the family members (B, S) or control subjects (N). M represents molecular weight markers. (B) Immunoblot analyses of albumin, IgG, and fibrinogen in platelet lysates. In the first (50% B) and the last (50% N) lanes, half of the corresponding sample (B, N) was applied, to assess the sensitivity of the analysis. (C) Immunoblot analysis of albumin in plasma. The albumin in patient plasma sample was comparable to that of 3 control subjects (N).
Assessment of platelet PKC-θ mRNA levels by real-time PCR
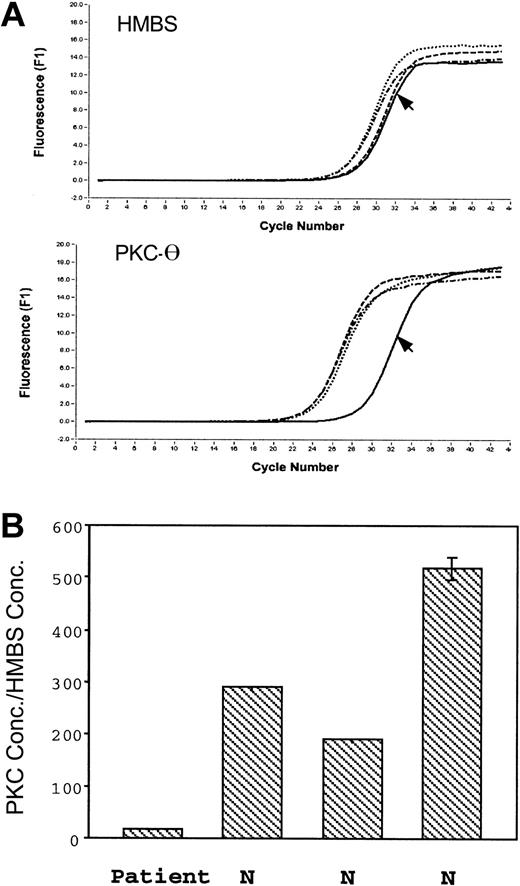
Platelet PKC-θ and HMBS mRNA levels were assessed in the patient and 3 control subjects (Figure 6). The PKC-θ crossing point, which indicates the beginning of the PCR exponential phase, was shifted to the right in the patient (Figure 6A); the ratio of PKC-θ to HMBS mRNA was strikingly decreased in the patient relative to the control subjects (Figure 6B), indicating the patient platelet PKC-θ mRNA levels are decreased.
Real-time PCR amplification of the HMBS and PKC-θ from platelet RNA. (A) The top tracings show the fluorescence recordings using RNA from the patient (arrow) and 3 control subjects. (B) The bars show the ratio of expression of PKC-θ to that of HMBS in the patient and 3 healthy subjects (N). Shown are results (mean ± SE) of 3 separate experiments.
Real-time PCR amplification of the HMBS and PKC-θ from platelet RNA. (A) The top tracings show the fluorescence recordings using RNA from the patient (arrow) and 3 control subjects. (B) The bars show the ratio of expression of PKC-θ to that of HMBS in the patient and 3 healthy subjects (N). Shown are results (mean ± SE) of 3 separate experiments.
Discussion
CBFA2 is a transcription factor that transactivates the expression of multiple genes required for normal hematopoiesis.6 Our studies document a heterozygous mutation in the CBFA2 gene in a patient previously described by us1 with abnormal platelet function, including diminished protein phosphorylation on activation. The observed mutation results in a premature termination with a loss of 87 amino acids in the conserved DNA-binding Runt domain of CBFA2. Mutations in the Runt domain with a loss of as few as 10 amino acids result in disrupted DNA-binding activity,11 predicting a nonfunctional protein in our patient. CBFA2 deficiency has been shown to be associated with familial thrombocytopenia and propensity to develop acute leukemia7 ; to our knowledge, our findings constitute the first demonstration that it is also associated with platelet dysfunction characterized by diminished agonist-induced phosphorylation of the PKC substrate pleckstrin (and myosin light chain), and receptor-mediated activation of the GPIIb-IIIa complex. Our studies indicate that one or more proteins regulated by the CBFA2 are required for protein phosphorylation and the inside-out signal transduction-dependent activation of GPIIb-IIIa in human platelets. We document a deficiency in one of the PKC isozymes, PKC-θ, which provides a plausible explanation for several of the observed platelet abnormalities. The link between CBFA2 deficiency and impaired activation of GPIIb-IIIa complex is analogous to the studies of Shiraga et al4 documenting a role of transcription factor NF-E2 in regulating GPIIb-IIIa activation in murine platelets. Two studies have described mice deficient in CBFA212,13 and this is associated with embryonic lethality and a lack of platelets; to our knowledge platelet function responses have not been reported in the heterozygotes.
A major signaling abnormality in our patient is the impaired phosphorylation of pleckstrin,1 a 47-kDa substrate of PKC. We, therefore, focused on PKC isozymes, a family of serine- and threonine-specific protein kinases comprising at least 12 related isozymes that phosphorylate a wide array of proteins involved in signal transduction.3 Human platelets possess several PKC isozymes including PKC-α, -βI, -βII, -δ, -ϵ, -ζ, -η, and -θ3,14 ; most of them are distinct gene products and their specific role in platelet function remains to be elucidated. Our studies show that patient platelets are deficient in PKC-θ but have normal levels of PKC-α, -βI, -βII, -δ, -ϵ, -η, and -ζ. This is supported by decreased platelet PKC-θ mRNA levels (Figure 6). PKC-θ is classified as one of the novel PKC isozymes and is expressed predominantly in hematopoietic cells.15 To our knowledge, this is the first demonstration of a human platelet PKC isozyme deficiency and of its association with a CBFA2 mutation. It provides a cogent explanation for the diminished pleckstrin phosphorylation and some of the other abnormalities.
Multiple lines of evidence link PKC to critical aspects of megakaryocyte differentiation, platelet formation and function, and GPIIb-IIIa activation.3 Several of these are highly relevant to the pathophysiologic findings in our patient and provide insights into the potential mechanisms. (1) PKC signaling has a critical role in megakaryocyte differentiation; phorbol esters activate PKC and induce progenitor cells to differentiate along megakaryocyte line and express GPIIb-IIIa.16-19 (2) Recent studies implicate PKC-ϵ and PKC-θ in megakaryocyte lineage commitment.16-21 In human progenitors, PKC-θ exhibits a lineage-restricted expression being expressed in megakaryocytes and erythroblasts but not granulocytes/monocytes.20 (3) PKC activation has been implicated in receptor-mediated GPIIb-IIIa activation in platelets.2,3 (4) GATA-1 plays a critical role in megakaryopoiesis and in activation of the GPIIb promoter. PKC-ϵ cooperates with GATA-1 in the activation of the GPIIb promoter.16 In CBFA2 deficiency, the patients have thrombocytopenia and megakaryopoiesis is impaired.7 Our studies extend these observations to demonstrate a deficiency of PKC-θ with CBFA2 mutation, suggesting that this isozyme is transcriptionally regulated by CBFA2. Moreover, this provides a potential mechanism for the thrombocytopenia and impaired megakaryopoiesis, as well as for the abnormal platelet aggregation responses noted in some patients with CBFA2 mutation.7,22-24 Elagib et al25 have recently shown that CBFA2 is up-regulated during megakaryocytic induction, that it is strongly expressed in megakaryoblasts but not in erythroblasts, and that it cooperates with GATA-1 in the activation of megakaryocytic promoters, including of GPIIb.
The extent of phosphorylation of pleckstrin on platelet activation is an integration of a number of opposing processes involving kinases and phosphatases. In our patient the pleckstrin phosphorylation defect may be explicable by the PKC-θ deficiency; however, additional mechanisms warrant consideration. Abnormal expression of other involved phosphatases or their inhibitors (which may also be regulated by CBFA2) may contribute to the decreased pleckstrin phosphorylation. Of note, MLC phosphorylation was also impaired in this patient.1 PKC activation plays a regulatory role in phosphorylation of MLC. For example, activation of PKC enhances MLC phosphorylation by inhibiting myosin phosphatase through phosphorylation of a protein that inhibits the phosphatase.26 The impact of a potential down-regulation in our patient of other relevant kinases, including MLC kinase, cannot presently be excluded. Despite the evidence linking PKC and activation of GPIIb-IIIa2,3 and the demonstrated PKC-θ deficiency, the possibility that the impaired activation of GPIIb-IIIa in our patient arises due to an associated deficiency of one or more of the large number of proteins known to bind to the cytoplasmic tails of the integrin complex2 needs to be considered. Expression of some of these proteins may also be regulated by CBFA2.
An interesting finding in this patient is the decrease in platelet albumin and IgG (Figure 5), which are present in α granules.27,28 In the absence of convincing evidence that megakaryocytes synthesize albumin or IgG, the platelet albumin and IgG reflect their uptake from plasma by megakaryocytes and platelets,27,28 a mechanism recognized also for other proteins, such as fibrinogen.29 The decreased platelet albumin and IgG levels in our patient suggest a defect in the uptake mechanisms; the normal platelet fibrinogen levels indicate a selectivity in this abnormality and that the incorporation of these plasma proteins into α granules is regulated by different mechanisms. In endothelial cells, activation of PKC by phorbol myristate acetate (PMA) or through phospholipase C plays an important role in the uptake of albumin30 ; this provides a relevant mechanism for the observed defect in our patient.
Overall, our studies demonstrate that a mutation in the hematopoietic transcription factor CBFA2 is associated with not only familial thrombocytopenia, but also impaired platelet function, pleckstrin phosphorylation, and receptor-mediated activation of GPIIb-IIIa. The present studies provide evidence that one or more proteins regulated by CBFA2 are required for full inside-out signal transduction-dependent activation of GPIIb-IIIa in human platelets. Moreover, they document for the first time a deficiency in one specific PKC isozyme (PKC-θ), indicating a major role of this isozyme in platelet production and function. Further studies in this family will provide important information on PKC isozymes in platelets and on the mechanisms of platelet GPIIb-IIIa activation.
Prepublished online as Blood First Edition Paper, October 2, 2003; DOI 10.1182/blood-2003-07-2299.
Supported by grant FY00-128 from the March of Dimes Birth Defects Foundation; grant HL 56724 from the National Heart, Lung, and Blood Institute; and grant RR-349 (General Clinical Research Center).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank the family members for participation in these studies; Dr Thomas Rogers for his guidance and assistance with the real-time PCR studies; Drs Danny Dhanasekaran and Satya P. Kunapuli for helpful discussions; and Mary Merrick and JoAnn Hamilton for administrative and secretarial assistance.