Abstract
Endomitosis (EnM) in megakaryocytes (MKs) is characterized by abortion of mitosis in late anaphase and failure of cytokinesis; subsequent reinitiation of DNA synthesis results in polyploidy. Ablation of chromosomal passenger proteins including Aurora-B kinase causes defects in late anaphase and cytokinesis in diploid cells; thus one hypothesis is that the expression or function of these proteins in polyploid MKs is abnormal. It has been reported that Aurora-B kinase mRNA is decreased in polyploid megakaryocytic cells, suggesting that deficiency of Aurora-B kinase is responsible for EnM. We examined the localization of Aurora-B kinase and additional members of the chromosomal passenger protein and aurora kinase families in MKs. We found that in EnM MKs (1) Aurora-B kinase is present and appropriately localized to centromeres in early EnM; (2) in low-ploidy human MKs, centromeric localization of survivin and inner centromere protein (INCENP) can also be demonstrated; (3) the function of Aurora-B kinase, as measured by Ser10 phosphorylation of histone H3, is intact; and (4) aurora-A kinase localizes appropriately to centrosomes in EnM. These results suggest that EnM MKs appropriately express functional Aurora-B kinase and related proteins in early anaphase, making a simple deficiency of this protein an unlikely explanation for polyploidy in this cell type.
Introduction
Megakaryocytes (MKs) are large cells that reside in the bone marrow and produce platelets, the cellular component of the coagulation system. As they mature, MKs undergo a distinctive mode of cell cycling known as endomitosis (EnM). EnM is currently thought to represent an abortive mitotic process in which the MKs do not progress beyond late anaphase and instead reenter G1 and a new round of DNA synthesis without nuclear or cytoplasmic division, becoming polyploid.1 Other mammalian cell types including endothelial cells and hepatocytes are known to undergo limited polyploidization, especially under conditions of stress, but none to the extent achieved by MKs (up to 128N in murine cells). The purpose for EnM is unknown, but it is theorized to enable MKs to attain the large cytoplasmic volume necessary for efficient platelet production.
From a biologic viewpoint, MKs raise several fundamental questions regarding the regulation of the cell cycle. During diploid mitosis, multiple checkpoints exist to ensure equal chromosomal segregation into 2 daughter cells and to couple DNA synthesis with cell division so that euploidy is maintained. EnM represents a marked departure from these conservative mechanisms, yet the cells continue to live and to cycle. Studies using either primary cells, cell lines, or transgenic mouse models have investigated mitotic cyclins (including cyclins B, D, and E) and mitotic checkpoint proteins (such as BubR1) for their potential contribution to the mechanism of polyploidy in MKs.2-11 Recently, several reports have suggested that the failure of normal cytokinesis in EnM MKs involves the deficiency of a mitotic kinase, Aurora-B kinase.12-14
The family of aurora kinases now includes many described members in several species, and for simplicity they can be grouped according to their primary localization and function into type A, type B, and type C (for reviews, see Adams et al15 and Giet and Prigent16 ). Aurora-A kinase (also known as IAK1) localizes to the centrosome at the mitotic spindle pole and is proposed to function in late anaphase, promoting spindle elongation and centrosome separation.17 Abnormal regulation of aurora-A kinase is thought to be oncogenic and result in the production of multiple centrosomes and aneuploidy.18,19 The kinetochore is a specialized complex of proteins assembled at the centromere of the mitotic chromosome that mediates microtubule attachment and the spindle checkpoint (for a review, see Cleveland et al20 ). Many proteins localizing to these structures are involved in the regulation of chromosomal alignment on the mitotic spindle and the spindle checkpoint. Proteins recruited to the microtubules and cortex composing the midzone between the separating chromosomes in late anaphase are generally thought to be required for completion of cytokinesis.21,22 Aurora-B kinase (also known as AIM1) localizes to the centromere and later to the midzone and is proposed to function in the spindle checkpoint and in cytokinesis. Type C aurora kinase appears to be specific to the testis.23
Aurora-B kinase is a member of a growing family of chromosomal passenger proteins that also includes survivin and inner centromere protein (INCENP; for a review, see Adams et al15 ). These proteins share a characteristic pattern of association with chromatin in prophase, centromeres in metaphase and early anaphase, and then the midzone and midbody in late anaphase and telophase, respectively.24-26 Studies in several organisms have established that these proteins form a complex and that the appropriate localization of each member of the complex is dependent on the presence of the others.27-32 In addition, studies have demonstrated that kinetochore localization of checkpoint proteins including centromere protein E (CENP-E), BubR1, and Mad2 requires Aurora-B kinase activity.33-35 Disruption of any of these 3 chromosomal passengers results in a similar phenotype, including chromosome segregation defects, failure of cytokinesis, and polyploidy.28,36-41 Inhibition of the function of Aurora-B kinase using various strategies is associated with chromosome missegregation, abnormal cytokinesis, and polyploidy.29,34,35,42 Growing numbers of Aurora-B kinase substrates have been identified, including the yeast kinetochore protein Ndc10p, the kinesin-related protein Eg5, CENP-A, the myosin II regulatory light chain, survivin, INCENP, mitotic centromere-associated kinesin (MCAK), and histone H3.32,43-51 Recently, small-molecule inhibitors with specificity for aurora kinase have been developed and used to probe the function of Aurora-B kinase in mitotic cells using phosphorylation of histone H3 as a convenient marker of kinase activity.35,42
Because of the role of the chromosomal passenger proteins in anaphase and cytokinesis, we examined the expression, localization, and function of Aurora-B kinase in EnM MKs and compared them to those seen in diploid cells. Through metaphase we consistently observed a similar pattern of localization of these proteins in EnM and mitotic cells, with Aurora-B kinase present at centromeres. In human MKs, most of which were of low ploidy, we also observed centromere localization of the associated chromosomal passengers survivin and INCENP. In addition, histone H3 is phosphorylated at Ser10 in EnM MKs and ablated by incubation with the aurora kinase inhibitor ZM447439,35 suggesting that Aurora-B kinase function at least toward this substrate is intact. Further, CENP-E is appropriately localized at kinetochores in EnM cells, again supporting the function of Aurora-B kinase in early stages of EnM. The related kinase aurora-A is also appropriately localized to the multiple centrosomes present in polyploid MKs. Although Aurora-B kinase (as well as the other passenger proteins examined) was readily detected localized at centromeres up through EnM metaphase, it was not clear whether Aurora-B kinase consistently translocates to a bundled midzone structure in anaphase. Although this could be demonstrated in polyploid human MKs identified in late anaphase, it was not detected in MKs of murine origin. These data suggest that the simple absence of Aurora-B kinase activity is unlikely to be the underlying cause of EnM. These data suggest that the defect leading to the failure of cytokinesis in EnM MKs does not involve failure at the level of the chromosomal passenger proteins.
Materials and methods
Cells
Primary MKs were prepared both from murine bone marrow and from human CD34+ cells. Briefly, for murine MKs, 3 to 5 mice were killed according to UCSD animal care program guidelines, and marrow cells from femurs and tibiae were flushed into Iscove modified Dulbecco medium (IMDM) containing 2% fetal bovine serum (FBS). A single-cell suspension was obtained by passing the cells twice through a 22-gauge needle and then a 70-μm filter. Cells were centrifuged and then resuspended in Optimem (Invitrogen, Carlsbad, CA) containing 5% thrombopoietin (TPO) supernatant (prepared from a secreting BHK cell line). After 2 to 3 days of culture, MKs were purified on a discontinuous albumin gradient as previously described.52 For human MKs, CD34+ cells isolated from peripheral blood from granulocyte colony-stimulating factor (G-CSF)–stimulated donors were purchased (Cambrex, East Rutherford, NJ, or AllCells, Berkley, CA). Cells were thawed according to the manufacturer's directions and cultured for 10 to 14 days in StemPro (Invitrogen) containing recombinant human TPO (rhTPO; 50 ng/mL), human stem cell factor (hSCF; 10 ng/mL), and human interleukin 6 (hIL-6; 10 ng/mL) to promote MK differentiation. Because the starting material for the human cells was already somewhat purified and because human MKs do not get as large as murine MKs, they were not subjected to an albumin gradient for further purification; analysis for the megakaryocytic marker CD41 showed that approximately half of the cells differentiated into MKs. For comparison of antibody staining and optimization of inhibitors, 3T3 cells, HeLa cells, a human megakaryocytic leukemia cell line (UT7/TPO), and a murine lymphoblastic leukemia cell line (BaF3/Mpl), were also used.
Fixation
MKs were fixed in suspension by the addition of an equal volume of 4% paraformaldehyde prepared in phosphate-buffered saline (PBS, pH 7.4) for a final concentration of 2% paraformaldehyde. Cells were fixed at room temperature for 45 minutes and then centrifuged and washed twice with PBS. Cells were resuspended in PBST (PBS containing 0.5% Triton-X100) with 3% bovine serum albumin (BSA) and incubated at room temperature for 15 to 30 minutes to provide blocking against nonspecific antibody reactivity.
Antibodies
All staining of MKs was performed in suspension in PBST. Primary antibodies used in this work include anti–Aurora-B kinase (1:100, murine monoclonal; Transduction Laboratories, San Diego, CA), anti–Aurora-A kinase (1:100, murine monoclonal; Transduction Laboratories), anti–phospho-Ser10 histone H3 (1:200, rabbit; Upstate Biotechnology, Waltham, MA); biotinylated anti–α-tubulin (1:100, murine monoclonal; Molecular Probes, Eugene, OR), anti–α-tubulin (1:1000, rat; Serotec, Raleigh, SC), anti–CENP-E (1:200, rabbit; gift of Donald Cleveland), anti-INCENP (1:250, rabbit; gift of Changjun Zhu), antisurvivin (1:100, rabbit sc-10811; Santa Cruz Biotechnology, Santa Cruz, CA), and anti-human CD41 (1:100, murine monocloncal; Immunotech, Marseille, France). Secondary antibodies were conjugated to Alexa Fluor 488, Alexa Fluor 568, or Alexa Fluor 647 and were obtained from Molecular Probes. After final staining, cells were applied to a slide, allowed to dry, and mounted in Vectashield with DAPI (4,6 diamidino-2phenylindole; Vector Laboratories, Burlingame, CA).
Inhibitors
ZM447439 has been described35 and was the gift of AstraZeneca (Wilmington, DE). Stocks were prepared in dimethyl sulfoxide (DMSO) and it was used at a final concentration of 5 μM, which inhibited phosphorylation of histone H3 and promoted polyploidy in 3T3 cells (data not shown). Monastrol has been described53 and was the gift of Tarun Kapoor. Stocks were prepared in DMSO and it was used at a final concentration of 100 μM, which resulted in the formation of monopolar spindles and mitotic arrest in 3T3 cells.
Microscopy and image processing
Images were captured with a DeltaVision Deconvolution Microscope (Applied Precision, Issaquah, WA). The system includes a Sony Coolsnap HQ CCD camera mounted on a Nikon TE-200 inverted epifluorescence microscope. All images were obtained using a × 60 objective. Exposure times were set such that the camera response was in the linear range for each fluorophore. The data sets were deconvolved and analyzed using SoftWorx 2.5 software (Applied Precision) on a Silicon Graphics Octane workstation (Mountainview, CA).
Western blotting
Mature murine MKs were isolated by albumin gradient, pelleted by centrifugation, and lysed in protein lysis buffer (200 mM Tris [tris(hydroxymethyl)aminomethane], 137 mM NaCl, 10% glycerol, 1% NP-40, 10 mM EDTA [ethylenediaminetetraacetic acid, 100 mM NaF) containing protease inhibitors (phenylmethylsulfonyl fluoride [PMSF], aprotinin, leupeptin, sodium orthovanadate). Eight to 10 μg total cell lysate was boiled with Western sample loading buffer and size-fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) according to the method of Laemmli.54 As a diploid control, an equivalent amount of total cell lysate from BaF3/Mpl cells was run in an adjacent lane. Proteins were eletrophoretically transferred to a nitrocellulose membrane and then blotted with monoclonal anti–Aurora-B kinase at a dilution of 1:500. Secondary antimouse antibody conjugated to horseradish peroxidase was applied (BioRad, Hercules, CA) and antibody detection was performed using chemiluminescence (LumiGlo, Cell Signaling Technology, Beverly, MA).
RT-PCR
Total RNA was prepared from either gradient-purified murine MKs from a 3-day culture or BaF3/Mpl cells according to the manufacturer's instructions (Rneasy, Qiagen, Santa Clarita, CA). One μg total RNA was reverse transcribed (RT; Superscript II, Invitrogen) and 2 μL cDNA was amplified by polymerase chain reaction (PCR) using HotStart Taq polymerase (Qiagen). Primers were synthesized by Invitrogen as follows: Aurora-B kinase (5′-GCTCAAAGACGTCTCA and 5′-CGAAGCTGGTGCTCTA, or 5′-CGATCATGGAGGAACT and 5′-CTTGACAATCCGACGA); survivin (5′-CCCTTCCTGGGGACT and 5′-GGCATGTCACTCAGGT); INCENP (5′-CTGGACTTTGTCTGCA and 5′-GCCATTCTCCTCAACA).
Aurora-B kinase assay
Mature murine MKs were isolated by albumin gradient and a total cell lysate was prepared as described for Western blotting. BaF3/Mpl cell lysate was similarly prepared as a diploid control. An equivalent amount of protein from each sample was immunoprecipitated using anti–Aurora-B kinase antibody followed by the addition of protein A/G beads (Santa Cruz Biotechnology). Beads were washed 3 times in kinase buffer (50 mM Tris, pH 8.0, 10 mM MgCl2, 1 mM dithiothreitol [DTT], 100 mM NaCl, 100 μM adenosine triphosphate [ATP]), then incubated in 30 μL kinase buffer to which 1 μg purified histone H3 had been added (Upstate Biotechnology) for 20 minutes at room temperature. The reaction was stopped by the addition of 5 × Western sample loading buffer. Control samples lacked either substrate or immunoprecipitated kinase. Samples were electrophoresed on a 12% SDS-PAGE gel and blotted for phosphorylation of histone H3 using anti–phospho-Ser10 histone H3. The blots were reprobed to confirm substrate presence using anti–histone H3 antibody (Cell Signaling Technology).
Results
Aurora-B kinase is present and active in EnM MKs
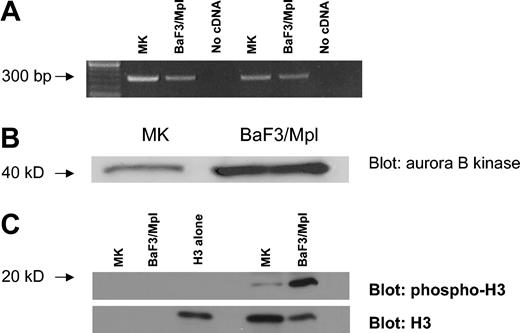
The expression of Aurora-B kinase was investigated by RT-PCR. Total RNA was prepared from murine bone marrow MKs purified on a discontinuous albumin gradient to isolate large polyploid cells. BaF3/Mpl cells, a murine lymphoblastic leukemia cell line, served as a diploid control.55 An equal amount of RNA was reverse transcribed and the cDNA was amplified using 2 independent pairs of primers for Aurora-B kinase. Both RT-PCRs yielded an appropriate product indicating the presence of Aurora-B kinase transcripts in murine endomitotic MKs (Figure 1A).
Aurora-B kinase is expressed in EnM MKs. (A) RT-PCR assay for Aurora-B kinase using RNA isolated from polyploid murine MKs and unsynchronized BaF3/Mpl cells. Two independent primer pairs were designed to yield products of about 300 base pair (bp). (B) Western blot comparing Aurora-B kinase expression in polyploid murine MKs and diploid cells. Gradient-purified MKs compared to unsynchronized BaF3/Mpl cells. Equivalent amounts of total cell protein were loaded in each lane. Aurora-B kinase is 41 kDa. (C) Kinase assay comparing Aurora-B kinase activity in polyploid murine MKs and diploid cells. Aurora-B kinase was immunoprecipitated from mature MK or BaF3/Mpl lysates, and kinase activity was assayed using purified histone H3 as a substrate. Phosphorylation was detected by blotting with phospho-specific histone H3 antibody. Blots were reprobed with antibody to histone H3. Lanes 1 and 2 contain no substrate, lane 3 contains purified histone H3 alone, lane 4 is empty, and lanes 5 and 6 show the kinase assay of MK and BaF3/Mpl immunoprecipitates against the purified histone H3 (19 kDa).
Aurora-B kinase is expressed in EnM MKs. (A) RT-PCR assay for Aurora-B kinase using RNA isolated from polyploid murine MKs and unsynchronized BaF3/Mpl cells. Two independent primer pairs were designed to yield products of about 300 base pair (bp). (B) Western blot comparing Aurora-B kinase expression in polyploid murine MKs and diploid cells. Gradient-purified MKs compared to unsynchronized BaF3/Mpl cells. Equivalent amounts of total cell protein were loaded in each lane. Aurora-B kinase is 41 kDa. (C) Kinase assay comparing Aurora-B kinase activity in polyploid murine MKs and diploid cells. Aurora-B kinase was immunoprecipitated from mature MK or BaF3/Mpl lysates, and kinase activity was assayed using purified histone H3 as a substrate. Phosphorylation was detected by blotting with phospho-specific histone H3 antibody. Blots were reprobed with antibody to histone H3. Lanes 1 and 2 contain no substrate, lane 3 contains purified histone H3 alone, lane 4 is empty, and lanes 5 and 6 show the kinase assay of MK and BaF3/Mpl immunoprecipitates against the purified histone H3 (19 kDa).
To show that Aurora-B kinase protein is present in EnM MKs we again used murine MKs from bone marrow cultures that were gradient-purified. BaF3/Mpl cells were used as a diploid control. Equal amounts of total cell protein were size-fractionated by SDS-PAGE and the proteins were analyzed by Western blotting for the presence of Aurora-B kinase. As shown in Figure 1B, a 41-kDa protein is recognized by the anti–Aurora-B kinase antibody corresponding to the expected size of the kinase. This band is of lesser intensity in the MKs than the BaF3/Mpl cells; however, this is to be expected because the fraction of mitotic cells in the BaF3/Mpl sample is much greater than that in maturing MKs. Therefore it is apparent that Aurora-B kinase protein is present in EnM MKs.
To confirm the activity of Aurora-B kinase in EnM MKs, the kinase was immunoprecipitated from equivalent amounts of total cell lysate prepared from gradient-purified polyploid murine MKs or diploid BaF3/Mpl cells and assayed for kinase activity using purified histone H3. As shown in Figure 1C, Aurora-B kinase activity is present in both cell types, as demonstrated by phosphorylation of histone H3. Although the activity of the immunoprecipitated Aurora-B kinase from MKs is less than that in an equivalent amount of lysate from BaF3/Mpl cells, the activity is proportionate to the relative amounts of the Aurora-B kinase observed in Western blotting (Figure 1B). This suggests that the Aurora-B kinase present has the expected activity.
One disadvantage of using whole-cell lysates is that it is difficult to correct for differences in the mitotic fraction between various cell populations; in addition, information regarding the subcellular localization of proteins is lost. To examine the localization and function of Aurora-B kinase in actively EnM cells, we prepared primary MKs for immunostaining both from murine bone marrow and from human peripheral blood CD34+ cells, because some antibodies do not recognize murine antigens well especially in immunofluorescence applications. Over 25 separate preparations of murine MKs were examined, as well as 3 preparations of human MKs. For comparison and to verify antibody staining patterns and species reactivity, 3T3 cells, HeLa cells, a human megakaryocytic leukemia cell line (UT7/TPO),56 and BaF3/Mpl cells, were also examined (data not shown). Fixed cells were labeled with primary antibody against Aurora-B kinase alone and in combination with a second antibody recognizing related mitotic proteins. Cells were then incubated with appropriate fluoresceinated secondary antibodies and DNA dye and examined by deconvolution microscopy.
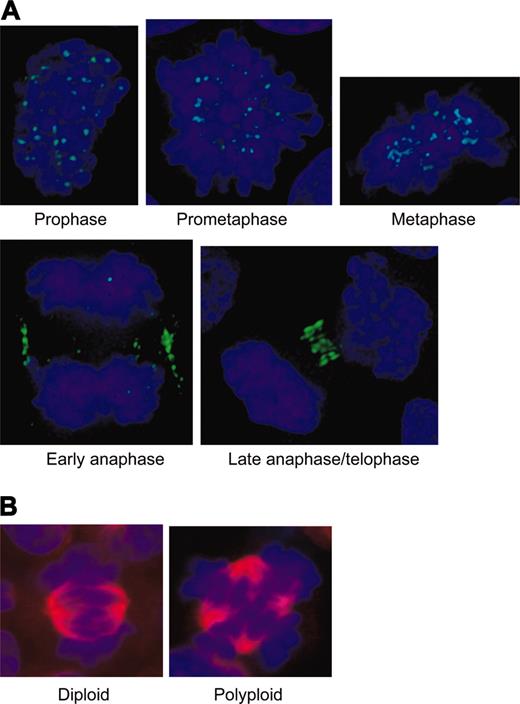
The expected localization pattern of Aurora-B kinase (and other chromosomal passenger proteins) in diploid cells is shown in Figure 2A. The protein localizes to the centromere in prometaphase and metaphase, then transfers to the cortex and spindle midzone in anaphase, and finally stains the compacted midbody in telophase. Diploid mitotic cells were recognized by a bipolar spindle and the alignment of their chromosomes onto a single metaphase plate. In contrast, EnM MKs have a multipolar spindle with poles arranged in a sphere, and any individual spindle pole often forms microtubule associations with more than one other pole (Figure 2B). Depending on the number of spindle poles, EnM cells align their chromosomes in multiple metaphase plates. Stages of the EnM cycle were inferred primarily from the condensation and organization of the chromosomes as either prometaphase, metaphase, or anaphase.
Diploid controls for Aurora-B kinase localization in mitosis. (A) Diploid CD34+ cells showing expected pattern of Aurora-B kinase localizing to centromeres in prophase through metaphase, then transferring to the midzone cortex and the condensing midbody in anaphase. Note chromosomal organization defining the different stages with condensation beginning at prophase, chromosomal alignment occurring at metaphase, sister chromatid separation in early anaphase with spindle elongation, and the beginning of chromosome decondensation in late anaphase and telophase. (B) Comparison of a diploid metaphase (bipolar spindle, single metaphase plate) with a polyploid metaphase (more than 2 spindle poles and more than a single metaphase plate).
Diploid controls for Aurora-B kinase localization in mitosis. (A) Diploid CD34+ cells showing expected pattern of Aurora-B kinase localizing to centromeres in prophase through metaphase, then transferring to the midzone cortex and the condensing midbody in anaphase. Note chromosomal organization defining the different stages with condensation beginning at prophase, chromosomal alignment occurring at metaphase, sister chromatid separation in early anaphase with spindle elongation, and the beginning of chromosome decondensation in late anaphase and telophase. (B) Comparison of a diploid metaphase (bipolar spindle, single metaphase plate) with a polyploid metaphase (more than 2 spindle poles and more than a single metaphase plate).
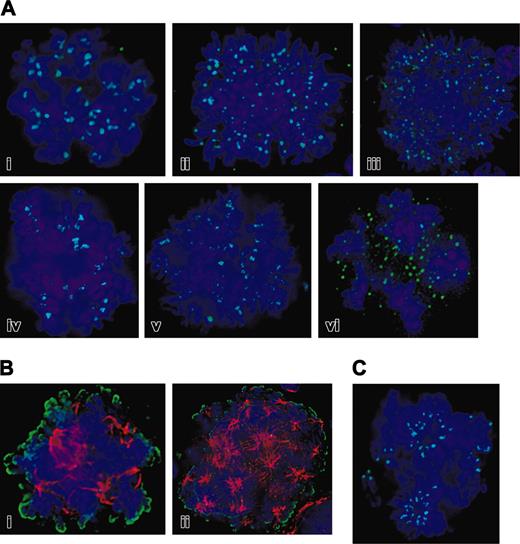
Thus defined, Aurora-B kinase was detected in association with centromeres through metaphase in both mitotic and EnM cells (Figure 3A). In murine cells, the most commonly observed EnM cells appeared to be in prometaphase or metaphase (approximately 80%), with another 15% in early anaphase, but occasional cells were seen with chromosomal separation consistent with late anaphase. Clear localization to midzone microtubules in murine MKs captured in anaphase was generally not seen. In human MKs, late anaphases with pronounced spindle elongation were more readily observed, and in at least 5 examples of these cells we could detect the transfer of Aurora-B kinase to the spindle midzone (Figure 4B). Compact midbodies were not seen, although these could be readily demonstrated in polyploid cells of a human megakaryocytic leukemia cell line, UT7 (data not shown). The activity of Aurora-B kinase in EnM is demonstrable in even highly polyploid cells as histone H3 is phosphorylated on Ser10 (Figure 3B), as has been described for Aurora-B kinase and as was demonstrated in Figure 1. However, centromere localization of Aurora-B kinase did not require the activity of the kinase because it was retained after treatment with the aurora kinase inhibitor ZM447439 (Figure 3C), nor was it dependent on intact microtubules because it actually intensified at centromeres on treatment of cells with nocodazole (data not shown). Therefore, Aurora-B kinase is present in early EnM and localizes similarly to the centromere in early EnM and mitotic cells.
Functional Aurora-B kinase is expressed in EnM MKs. (A) Deconvolution images of a series of murine polyploid MKs showing localization of Aurora-B kinase (green) to centromeres. DNA is blue. Cells shown are in prometaphase with chromosomes not yet completely aligned, metaphase with chromosomal alignment evident to several plates, and anaphase. Note centromere localization of Aurora-B kinase in up through metaphase with indistinct localization in anaphase. (B) Histone H3 is phosphorylated on Ser10 in EnM murine MKs. Phospho-histone H3 is shown in green and tubulin in red; left image shows a cell with chromosomal alignment suggesting metaphase, whereas the very large cell on the right appears to be in early anaphase with chromosomal separation. (C) Localization of Aurora-B kinase (green) to centromeres does not require intact kinase activity. Purified mouse MKs were incubated for 2 hours with 5 μM ZM447439 prior to fixation and immunostaining for Aurora-B kinase (green) and phospho-histone H3 (red). Phosphorylation of histone H3 is absent due to inhibition of aurora kinase.
Functional Aurora-B kinase is expressed in EnM MKs. (A) Deconvolution images of a series of murine polyploid MKs showing localization of Aurora-B kinase (green) to centromeres. DNA is blue. Cells shown are in prometaphase with chromosomes not yet completely aligned, metaphase with chromosomal alignment evident to several plates, and anaphase. Note centromere localization of Aurora-B kinase in up through metaphase with indistinct localization in anaphase. (B) Histone H3 is phosphorylated on Ser10 in EnM murine MKs. Phospho-histone H3 is shown in green and tubulin in red; left image shows a cell with chromosomal alignment suggesting metaphase, whereas the very large cell on the right appears to be in early anaphase with chromosomal separation. (C) Localization of Aurora-B kinase (green) to centromeres does not require intact kinase activity. Purified mouse MKs were incubated for 2 hours with 5 μM ZM447439 prior to fixation and immunostaining for Aurora-B kinase (green) and phospho-histone H3 (red). Phosphorylation of histone H3 is absent due to inhibition of aurora kinase.
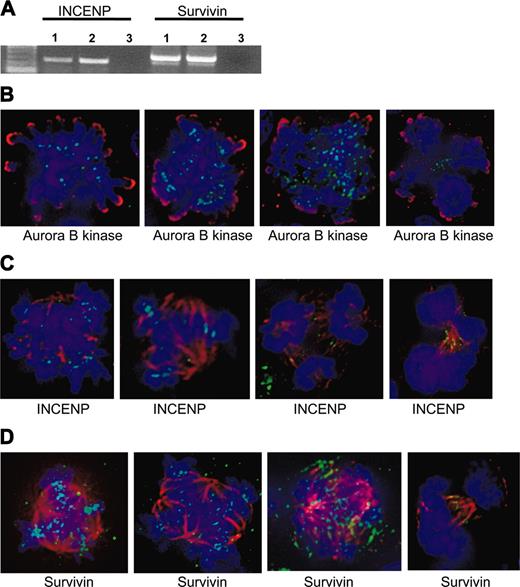
The chromosomal passengers, INCENP and survivin, are present at centromeres in EnM MKs. (A) RT-PCR showing expression of INCENP and survivin in polyploid murine MKs. RNA was isolated from gradient-purified MKs or BaF3/Mpl cells as a diploid control. Lanes 1, MKs; lanes 2, BaF3/Mpl; lanes 3, negative control. (B) Aurora-B kinase is present and functional in human MKs. Aurora-B kinase is shown in green and phospho-histone H3 is shown in red. Note the localization to midzone structures in the two rightmost images shown in anaphase. (C-D) INCENP and survivin (green) are present and properly localized in human MKs. Tubulin is shown in red, and DNA is blue. Note the localization to centromeres in the leftmost 2 images of metaphase cells, then midzone structures in the next 2 images shown in anaphase.
The chromosomal passengers, INCENP and survivin, are present at centromeres in EnM MKs. (A) RT-PCR showing expression of INCENP and survivin in polyploid murine MKs. RNA was isolated from gradient-purified MKs or BaF3/Mpl cells as a diploid control. Lanes 1, MKs; lanes 2, BaF3/Mpl; lanes 3, negative control. (B) Aurora-B kinase is present and functional in human MKs. Aurora-B kinase is shown in green and phospho-histone H3 is shown in red. Note the localization to midzone structures in the two rightmost images shown in anaphase. (C-D) INCENP and survivin (green) are present and properly localized in human MKs. Tubulin is shown in red, and DNA is blue. Note the localization to centromeres in the leftmost 2 images of metaphase cells, then midzone structures in the next 2 images shown in anaphase.
The chromosomal passenger proteins INCENP and survivin are properly localized at the kinetochore in EnM MKs
Expression of the chromosomal passenger proteins INCENP and survivin was determined by RT-PCR. As performed for Aurora-B kinase, equivalent amounts of total RNA from gradient-purified murine EnM MKs or BaF3/Mpl cells were reverse transcribed and amplified using primers designed to be specific for either INCENP or survivin. Both RT-PCRs yielded products of the expected size, indicating the presence of INCENP and survivin transcripts in these cells (Figure 4).
Because the chromosomal passenger proteins are dependent on each other for their localization,27-32 we used deconvolution fluorescence microscopy to detect whether INCENP and survivin are appropriately localized in EnM MKs. These proteins could only be examined in human MKs due to the species reactivity of our antibodies. In particular, despite reactivity by Western blot, several commercially available antibodies to survivin did not give us the accepted pattern of centromere localization in diploid murine cells such as NIH3T3 or BaF3/Mpl (including rabbit and mouse antibodies from Novus Biologicals [Littleton, CO], 500-201 and 3258, and rabbit and mouse antibodies from Santa Cruz Biotechnology, sc-10811 and 17779, all tested at dilutions of 1:50-1:500). Human CD34+ cells purified from peripheral blood were cultured with TPO for 10 days at which point approximately 50% of the cells expressed CD41. As noted, the polyploid cells in these preparations were mostly of low ploidy (8-16N). These cells were fixed and stained for immunofluorescence. Microscopic examination of this population stained for CD41 demonstrated that nearly all of the cells exhibiting polyploid mitotic figures were positive for CD41 confirming that they are MKs (data not shown). As shown in Figure 4B-D, deconvolution microscopy clearly shows the presence of INCENP, survivin, and Aurora-B kinase at the centromeres of EnM human MKs. As discussed for Aurora-B kinase, polyploid cells in anaphase could be identified in which Aurora-B kinase, INCENP, and survivin were seen on midzone structures. In the human MKs examined, more than half of the polyploid MKs identified in late anaphase (approximately 30 cells identified) showed clear midzone localization of the passenger proteins. The appropriate localization of survivin and INCENP provides indirect support for the presence of functional Aurora-B kinase. These findings indicate that aurora kinase and the associated passenger proteins are present in polyploid MKs and their absence is not necessary at least for the early stages of EnM.
Aurora-B kinase is functional in EnM MKs and is required to maintain the multipolar spindle
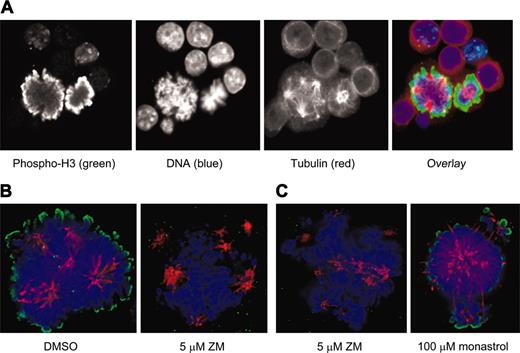
Because of the discrepancies between the mitotic fractions in unsynchronized populations of polyploid MKs and diploid cells, we again used immunofluorescence for phosphorylated histone H3 to compare the function of Aurora-B kinase in these cell types. Evaluation of murine MKs stained with a primary antibody specific for histone H3 that is phosphorylated on Ser10 showed a pattern of bright peripheral staining on condensed mitotic chromosomes that was of similar intensity to that seen in diploid mitotic cells (Figure 5A). Incubation of MKs with an inhibitor of aurora kinase, ZM447439, ablated the phosphorylation of histone H3 within 2 hours, indicating that this phosphorylation is due to an aurora kinase, and also resulted in some collapse of the multipolar spindle and loss of kinetochore-associated microtubules (Figure 5B). The spindle collapse did not appear to be due to the loss of Eg5 activity (a known target of Aurora-B kinase) because incubation with the Eg5 inhibitor monastrol resulted in a very different pattern of spindle reorganization with less overall microtubule loss (Figure 5C). Note, however, that the collapse of the microtubule spindle in the presence of monastrol did not affect phosphorylation of histone H3. The relatively high specificity of ZM447439 for aurora kinases (both A and B) has been described35 ; in particular it has been shown to have little activity against PLK1, CHK1, CDK1, CDK2, and CDK4, and it has approximately 20-fold less activity against MEK1 than against the aurora kinases.
Phosphorylation of histone H3 is similar in EnM and mitosis and is ablated by an inhibitor of aurora kinase. (A) EnM murine MK and mitotic cell adjacent to each other allowing direct comparison of phosphorylation of histone H3. Phospho-histone H3 is shown in green; tubulin is stained in red. (B) Murine MKs were purified and incubated for 2 hours with either vehicle (DMSO) or 5 μM ZM447439 prior to fixation and immunostaining for phospho-histone H3 (green). (C) Spindle structure in MKs treated with an aurora inhibitor is not the same as that in MKs treated with an Eg5 inhibitor. Murine MKs were incubated for 2 hours with either 5 μM ZM447439 or 100 μM monastrol prior to fixation and immunostaining for tubulin and phospho-histone H3.
Phosphorylation of histone H3 is similar in EnM and mitosis and is ablated by an inhibitor of aurora kinase. (A) EnM murine MK and mitotic cell adjacent to each other allowing direct comparison of phosphorylation of histone H3. Phospho-histone H3 is shown in green; tubulin is stained in red. (B) Murine MKs were purified and incubated for 2 hours with either vehicle (DMSO) or 5 μM ZM447439 prior to fixation and immunostaining for phospho-histone H3 (green). (C) Spindle structure in MKs treated with an aurora inhibitor is not the same as that in MKs treated with an Eg5 inhibitor. Murine MKs were incubated for 2 hours with either 5 μM ZM447439 or 100 μM monastrol prior to fixation and immunostaining for tubulin and phospho-histone H3.
Aurora-related proteins are correctly localized in EnM MKs
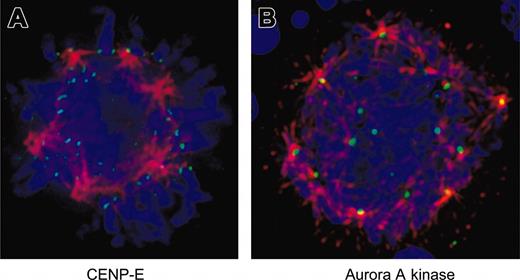
Because inhibition of Aurora-B kinase activity has been shown to disrupt localization of CENP-E in other systems,34,35 we examined the localization of this checkpoint protein in EnM MKs. Figure 6A demonstrates that CENP-E localizes appropriately to kinetochores on the multipolar spindle of a murine MK.
Aurora-related proteins are correctly localized in EnM MKs. (A) Murine MKs were fixed and stained for CENP-E (green) and α-tubulin (red). DNA is shown in blue. Note that CENP-E is localized to kinetochores and bi-orientation of chromosomes is visible at top of the spherical arrangement of spindle poles. (B) Murine MKs were fixed and stained for aurora-A kinase (green) and α-tubulin (red). DNA is shown in blue. Note localization of aurora-A kinase to spindle poles. Image is shown as a projection to display multiple spindle poles in the same view.
Aurora-related proteins are correctly localized in EnM MKs. (A) Murine MKs were fixed and stained for CENP-E (green) and α-tubulin (red). DNA is shown in blue. Note that CENP-E is localized to kinetochores and bi-orientation of chromosomes is visible at top of the spherical arrangement of spindle poles. (B) Murine MKs were fixed and stained for aurora-A kinase (green) and α-tubulin (red). DNA is shown in blue. Note localization of aurora-A kinase to spindle poles. Image is shown as a projection to display multiple spindle poles in the same view.
Similar to Aurora-B kinase, the deficiency of aurora-A kinase activity has also been associated with defects in anaphase and chromosome segregation,17 and previous studies have concluded that aurora-A kinase is deficient in polyploid megakaryocytic cells.14 We examined the localization of this kinase in EnM MKs. Immunofluorescence staining clearly showed aurora-A kinase localized to the poles of multipolar spindles in both human and murine MKs (Figure 6B). These data provide additional evidence that the aurora kinases are expressed normally in MKs.
Discussion
Our studies using RT-PCR and Western blotting confirm that Aurora-B kinase is present in EnM MKs. Using immunofluorescence and deconvolution microscopy, we have shown that Aurora-B kinase is appropriately localized at centromeres through metaphase, as are the chromosomal passenger proteins INCENP and survivin. The localization of Aurora-B kinase does not depend on its kinase activity or on the presence of intact microtubules. Additionally, Aurora-B in MKs is a functional kinase because histone H3, a known target of Aurora-B kinase, is specifically phosphorylated on Ser10 and that phosphorylation is ablated by an accepted inhibitor of aurora kinases. Eg5, another downstream target of Aurora-B kinase, would also appear to be active because the cells exhibit spindle collapse in response to its inhibition by monastrol. Additional evidence supporting the activity of Aurora-B kinase is provided by the proper localization of CENP-E, because it has been shown that disruption of Aurora-B kinase activity using either a dominant-negative kinase34 or a small-molecule inhibitor results in the dispersion of CENP-E as well as BubR1 and Mad2.35
The difficulty in which we detected transfer of the passenger proteins to an organized midzone or midbody structure, especially in murine MKs, is consistent with the current model of EnM in which MKs abort normal mitosis in late anaphase before a prominent midzone would be formed.1 However, we have observed multiple examples of EnM human MKs with chromosome separation consistent with late anaphase and clear midzone localization of the chromosomal passengers. These discrepant observations prevent us from concluding whether Aurora-B kinase is specifically mislocalized in anaphase. The situation in murine cells is made more difficult by the lack of reagents that identify the midzone, because many antibodies to known midzone markers such as the other chromosomal passengers (including survivin and INCENP), PRC1, or MKLP1 are not reactive in mouse by immunofluorescence. It is not clear whether defects in midzone formation could be a cause or a result of aborted anaphase and subsequent cytokinesis. Alternatively, it is possible that the midzone in these cells is a transient or unstable structure and thus difficult to capture in the static images viewed in deconvolution microscopy, especially when one considers the relatively low frequency of EnM figures found in unsynchronized cultures of polyploid MKs. We are pursuing live cell imaging of MKs undergoing EnM to gain additional insights into the subcellular redistribution of Aurora-B kinase during EnM.
Our data are in contrast to recent studies12-14 in which it was concluded that Aurora-B kinase is either deficient or absent in polyploidizing megakaryocytic cell lines or in MKs. There are several possible explanations for the discrepant findings between our data and these reports. First, these studies evaluated mRNA levels of Aurora-B kinase in unsynchronized populations of polyploidizing megakaryocytic cell lines or human MKs. Our observation of unsynchronized populations of maturing MKs by deconvolution microscopy reveals a relatively small fraction of cells with condensed chromosomes, and work by others indicates that the G2 and M phases of the EnM cycle comprise only 3% of the duration of an EnM cycle.57 Additional studies have shown that as MKs mature they drop out of the cell cycle, accompanied by overexpression of p21 and p27.58 Because the expression of Aurora-B kinase is regulated by the cell cycle,37,59 it is reasonable to hypothesize that as MKs become more polyploid, the proportion that are actively cycling and in G2/M phase would decline and thus the amount of detectable Aurora-B kinase mRNA would also decline. Given the practical difficulties in synchronizing primary MKs, we believe that microscopy is a more accurate method for evaluating these proteins in this heterogeneous cell population. In the work by Kawasaki et al,14 Aurora-B kinase was exogenously expressed in a megakaryocytic cell line with a consequent reduction in polyploidization. However, cell lines may behave differently than primary cells; for example, polyploid cells of the megakaryocytic line UT7/TPO routinely progress well into telophase and form compact midbodies, whereas this is not true of primary MKs (our data not shown). In addition, the data in these studies do not rule out the possibility that the cells are arrested rather than cycling as diploid cells. The effects of unscheduled overexpression of several aurora kinases have been shown in several studies to be detrimental and in some cases similar to the effects of expressing a dominant-negative form of the same protein, suggesting that the timing and degree of expression of these proteins is carefully regulated and cannot be simply manipulated.18,59
While this manuscript was in preparation, additional work was published by Zhang et al60 that indicated that although Aurora-B kinase is present in EnM murine MKs, it is inappropriately degraded in early anaphase and hence not seen at the midzone. The result that Aurora-B kinase is present at centromeres in early EnM murine MKs is consistent with our conclusions here. However, in that work the authors concluded that survivin is not expressed in EnM murine MKs, based on its absence by immunofluorescence. Due to our poor experience with multiple survivin antibodies in immunofluorescence applications in murine cells, we believe that this is possibly an artifact of the antibody reactivity. When the same antibodies are used in low-ploidy MKs of human origin, survivin staining at the kinetochore and midzone can be demonstrated (Figure 4C). This finding, in conjunction with the detection of survivin in murine MKs by RT-PCR (Figure 4A), is more consistent with the literature from other cell types in which the chromosomal passenger proteins Aurora-B kinase, INCENP, and survivin are dependent on each other for proper localization.27,32,51,61 However, we cannot exclude the possibility that a minor component of contaminating marrow cells contributed to the amplification of survivin and INCENP, and we did not examine the possibility of alternate spliceoforms in our primer design. In addition, in that work, as in ours, Aurora-B kinase was not seen to translocate to the midzone in murine MKs. However, as discussed, our observations in human MKs differ in that the chromosomal passenger proteins are shown to translocate in at least some EnM MKs. Although in general the MKs obtained from culture of CD34+ cells are of lower ploidy that those obtained from murine bone marrow, they represent a valid model of EnM as a primary MK that clearly develops polyploidy; it could in fact be argued that the processes that underlie the onset of EnM must be present from the first aborted anaphase and that therefore the lowest polyploid cell that can be clearly identified is the most informative model.
The function of Aurora-B kinase in mitosis is proposed to include roles in chromosome congression, chromosome segregation, and cytokinesis.28,29,34,35,42,45,62 In polyploid MKs, chromosomes appear to congress relatively normally albeit to multiple metaphase plates; however, defects in chromosome segregation have been demonstrated.10 Importantly, in addition to cytokinesis, karyokinesis fails in MKs, indicating that chromosome separation is insufficient to result in the formation of separate nuclei. Aurora-B kinase phosphorylates the kinesin Eg5, which helps to maintain the bipolar spindle, and inhibition of Eg5 with the small-molecule inhibitor monastrol results in collapse of the spindle and arrest in prometaphase with a monopolar spindle.53,63 However, in MKs the pattern of spindle abnormalities in response to inhibition of Eg5 using monastrol was strikingly different from those observed after treatment with ZM447439. It is possible that Aurora-B kinase has targets other than Eg5 that are important for spindle stability, such as its recently described regulation of the microtubule depolymerizing activity of MCAK.50
In conclusion, Aurora-B kinase is present and functional in EnM MKs, as determined by detection of the protein by immunofluorescence at centromeres and phosphorylation of its substrate histone H3, as well as appropriate centromere localization of the passengers INCENP and survivin and of the checkpoint protein CENP-E, whose localization has been shown to depend on intact Aurora-B kinase activity. The riddle of EnM remains unsolved; however, the events leading to failure of cytokinesis do not seem to entail abnormal expression of the chromosomal passenger proteins. Further investigation into the formation and function of the spindle midzone is warranted.
Prepublished online as Blood First Edition Paper, May 6, 2004; DOI 10.1182/blood-2004-02-0419.
Supported in part by National Institutes of Health grants K08 HL0442001 and RO1CA1615.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors would like to thank Jim Feramisco and Steve McMullen at the UCSD Cancer Center Digital Imaging Shared Resource for their invaluable help with deconvolution microscopy and image processing. Donald Cleveland and members of his laboratory including Jagesh Shaw and Beth Weaver, Kevin Sullivan, Sam Zeitlan, Changjun Zhu, and Paul Maddox provided helpful discussions and technical advice.