Using a combination of molecular cytogenetic and large-scale expression analysis in human T-cell acute lymphoblastic leukemias (T-ALLs), we identified and characterized a new recurrent chromosomal translocation, targeting the major homeobox gene cluster HOXA and the TCRB locus. Real-time quantitative polymerase chain reaction (RQ-PCR) analysis showed that the expression of the whole HOXA gene cluster was dramatically dysregulated in the HOXA-rearranged cases, and also in MLL and CALM-AF10-related T-ALL cases, strongly suggesting that HOXA genes are oncogenic in these leukemias. Inclusion of HOXA-translocated cases in a general molecular portrait of 92 T-ALLs based on large-scale expression analysis shows that this rearrangement defines a new homogeneous subgroup, which shares common biologic networks with the TLX1- and TLX3-related cases. Because T-ALLs derive from T-cell progenitors, expression profiles of the distinct T-ALL subgroups were analyzed with respect to those of normal human thymic subpopulations. Inappropriate use or perturbation of specific molecular networks involved in thymic differentiation was detected. Moreover, we found a significant association between T-ALL oncogenic subgroups and ectopic expression of a limited set of genes, including several developmental genes, namely HOXA, TLX1, TLX3, NKX3-1, SIX6, and TFAP2C. These data strongly support the view that the abnormal expression of developmental genes, including the prototypical homeobox genes HOXA, is critical in T-ALL oncogenesis.
Introduction
T-cell acute lymphoblastic leukemias (T-ALLs) are highly malignant tumors representing 10% to 15% of pediatric and 25% of adult ALLs in humans.1,2 T-ALL cells derive from partially differentiated thymocytes. These cells originate from pluripotent progenitors that are progressively committed to the αβ or γδ T-cell lineages in the thymus.3 Somatic V(D)J recombination rearranges the T-cell-receptor (TCR) gene segments, generating protein products that allow cells to pass through several cellular processes, so-called β-selection and selection.4,5 Hence, V(D)J recombination both directly compromises genome integrity and indirectly controls cellular functions considered to be critical for oncogenesis, such as cell cycle, proliferation, and apoptosis.6 However, T-ALLs remain rare, consistent with efficient control mechanisms that can only be overcome by accumulation of oncogenic events. Several classes of proto-oncogenes can be activated by chromosomal rearrangements or epigenetic mechanisms in T-ALL.7-9 The resulting oncoproteins include basic helix-loop-helix proteins (TAL1/SCL, TAL2, LYL1), homeodomain-containing proteins (TLX1/HOX11, TLX3/HOX11L2), and LIM only proteins (LMO1, LMO2), which are likely to be involved in transcription factor complexes.8,10 In addition, chromosomal alterations or DNA mutations can activate genes involved in signal transduction and thymus differentiation, such as NOTCH1 and LCK,11-13 or can lead to gene fusion (MLL-ENL, CALM-AF10, NUP214-ABL1).14-16 Moreover, the CDKN2A/INK4A/ARF tumor suppressor gene is inactivated by deletion or mutation in most T-ALL cases.17,18 Recently, T-ALL classifications based on selected oncogene expression and corresponding microarray gene expression patterns have been reported.19
In the present study, a combination of molecular cytogenetic and large-scale expression analyses allowed us to identify and characterize a new recurrent chromosomal translocation directly targeting and dysregulating the HOXA locus. This new rearrangement extends the catalog of T-cell oncogenes to these prototypical homeobox genes and strongly suggests that HOX dysregulation can be not only a consequence of cancer in humans but also a cause.20 Moreover, we show that HOXA genes can be included in biologic networks that associate oncogenesis and thymocyte differentiation processes. Our data suggest that inappropriate use or perturbation of specific biologic pathways involved in thymic maturation, and ectopic expression of genes, often developmental genes, are critical for T-ALL oncogenesis.
Patients, materials, and methods
Patients, leukemic cells, and annotations
T-ALL was diagnosed in 92 patients who were treated at Saint Louis Hospital (Paris, France). Seven patients were studied at diagnosis and relapse (total 99 T-ALL samples). There were 56 children (median age, 9 years; range, 1-16 years) and 36 adults (median age, 27 years; range, 17-66 years). Informed consent was obtained from the patients or relatives. The study was approved by the Review Board of the Féderation d'Hématologie, Hôpital Saint-Louis, Paris, France. T-ALL diagnosis was based on morphologic and immunophenotypical criteria using flow cytometry and an extended monoclonal antibody panel. Bone marrow samples from healthy donors (n = 4), unpurified thymus cells, and MOLT4, HSB2, CEM, and Jurkat T-cell lines were also included in the study. DNA and RNA were extracted from the cryopreserved leukemic cells. Expression of the main T-ALL oncogene transcripts, TAL1, TLX1, TLX3, LMO1, LMO2, and LYL1, and the SIL-TAL1, CALM-AF10, and NUP214-ABL1 transcripts were analyzed by real-time quantitative polymerase chain reaction (RQ-PCR; shown in Supplemental Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article), as described.21 Because TAL1 is highly expressed in erythroblasts,22 10 cases with high expression of erythroid-associated genes (Table S1) were excluded from the final analysis of TAL1 expression to avoid spurious results. MLL and CDKN2A/INK4A/ARF genomic abnormalities were searched for in the HOXA-expressing cases by fluorescent in situ hybridization (FISH; LSI MLL DualColor, VYSIS/ABOTT, Rungis, France, and RP11-149I2 BAC probe, respectively). MLL partners were identified in the 3 MLL-translocated cases using the MLL fusionChip kit (IPSOGEN, Marseille, France). BMI1, NKX3-1, SIX6, TFAP2C, RAG1, and CD8A gene expression was tested by RQ-PCR (see “PCR systems for gene expression analysis” in the Supplemental Materials and methods).
Isolation of normal human thymocyte subsets
Postnatal thymocytes were obtained from thymus samples removed during corrective cardiac surgery in 3 patients aged 1 month to 3 years old.
Thymocyte subpopulations (for a review, see Carrasco et al4 ) were isolated and analyzed by fluorescence-activated cell sorting (FACS) for purity as previously described.23-26 Briefly, after rosetting to remove CD2bright thymocytes, CD34+ thymocytes were magnetically sorted and then treated with CD1a microbeads to isolate the CD34+CD1a+ cells (Pre-T1, 96.5% purity). The most immature CD34brightCD33lowCD1a- cells (early thymic progenitors [ETPs]), were sorted from the CD1a-depleted fraction with anti-CD33-phycoerythrin (PE) and anti-PE microbeads (94.6% purity).26 The remaining CD34+ CD33-depleted thymocytes corresponded to CD34+CD1a- Pro-T thymocytes (99.4%). Other thymic subpopulations were purified after centrifugation of total thymus cells on stepwise Percoll density gradients as previously described.4,24 Characteristics of these subsets were: Pre-T2 and Pre-T3(E) (not separated here and further referred as Pre-T2/Pre-T3[E]), 99% CD4+CD8αβ-CD3-TCRαβ-; Pre-T3(L), 98% CD4+CD8αβ+CD3-TCRαβic; immature DPs (immature double-positive cells), 98% CD4+CD8αβ+CD3-TCRαβ-; DP-TCRαβlow, 63.2% CD4+CD8αβ+CD3lowTCRαβlow; DP-TCRαβint, CD8αβ+CD4+ CD3intTCRαβ+, 99% purity with 74% CD3int; DP-TCRαβbr, 99.9% CD8αβ+CD4+CD3brTCRαβbr; CD4-SP (CD4 single-positive cells), 97.2% CD4+CD3brTCRαβbr; and CD8-SP, 99.8% CD8αβ+CD3brTCRαβbr; see “Reagents used for human thymocyte subsets isolation” in the Supplemental Materials and methods.
Genomic analysis of the TCRB and HOXA loci
TCRB-flanking FISH probes were prepared from the CTD-3092H9 P1 artificial chromosome (PAC; 5′ probe, centromeric) and RP11-368I15 bacterial artificial chromosome (BAC; 3′ probe, telomeric). HOXA-flanking FISH probes were prepared from the RP11-167F23 and RP5-1200I23 BACs (telomeric and centromeric of the HOXA locus, respectively). FISH mapping within the HOXA locus was performed using the RP11-163M21, RP11-1132K14, RP11-1025G19, and RP11-1148E13 BACs. Images were captured and analyzed using a DMRXA epifluorescence microscope (Leica), a Plan Apo 63 ×/1.32 oil immersion objective lens (0.132 mm/pixel; Leica), a CCD camera (Cohu), and Q-FISH software (Leica). Southern blot analysis of the HOXA locus was performed using a panel of probes shown in Supplemental Materials and methods. Genome information was collected in the UCSC Genome Bioinformatics site (http://genome.ucsc.edu).
For inverse PCR amplification of the TCRB-HOXA breakpoint regions, leukemic gDNAs were digested by the MaeI and MseI restriction enzymes (cases TL43 and TL46, respectively) and ligated at low concentration. The circularized breakpoint-containing fragments were amplified by nested PCR. Primers were 5′-ATACCCCACCACCAACTTG-3′ and 5′-ATGTCTTTGTAAGGCTGTTTG-3′ (first round PCR), and 5′-CGCGGATCCATGTCACCCACCCTCCA-3′ and 5′-CGCGGATCCCAGTACCCACCCCTCTCC-3′ (nested PCR). Reciprocal breakpoint sequences were long-range PCR amplified using the forward HOXA primers 5′-GAGCCTCGTGTCTTTCTCC-3′ (case TL43) and 5′-GTTCTAGCTCTATTTACTTCTA-3′ (case TL46), and a set of reverse Jβ primers, including 5′-CTGCTCAGCTCTTTTTGCC-3′ (case TL43), and 5′-CCTTCCCACCGCTGAGAG-3′ (case TL46). Translocation breakpoints and flanking sequences were sequenced both directly and after cloning in the PCR-Script vector (Stratagene, La Jolla, CA).
RQ-PCR analysis of genes from the HOXA locus
Leukemic and control cDNAs were analyzed for expression of the 11 HOXA and EVX1 genes using SYBRgreen reagents (Applied Biosystems, Courtaboeuf, France). Primers are indicated in Supplemental Materials and methods. Results were expressed in Ct of the amplification curve and normalized on the basis of the expression of 2 housekeeping genes, ABL1 and TBP. Because the HOX paralogs share high sequence similarity, primers were carefully designed to avoid cross-priming. The specificity of the amplified fragments was further checked by analyzing the dissociation curves, migration on polyacrylamide gels, and by direct sequencing of the PCR products for at least one product of each type. In most cases, the primers overlap exon boundaries to prevent contamination by gDNA.
Microarray experiments and data analysis
Large-scale expression and analyses were performed on quality-controlled RNAs using the Affymetrix U133A technology, according to standardized procedures at the Strasbourg Genopole national facility. See “Microarray experiments and data analysis” in the Supplemental Materials and methods for details on RNA control and data normalization.
Microarray analyses were performed on the quantile-normalized data using dChip1.3 (http://biosun1.harvard.edu/complab/dchip/) and BRB (http://linus.nci.nih.gov/BRB-ArrayTools.html) software, as described in the Supplemental Materials and methods. LocusLink symbols and ID (http://www.ncbi.nih.gov/entrez/query.fcgi?db=gene) were used when the information was available to label genes corresponding to the probe sets.
Results
A new recurrent chromosomal translocation in T-ALL targets the HOXA locus
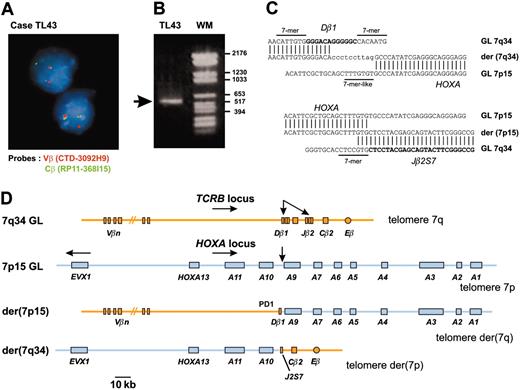
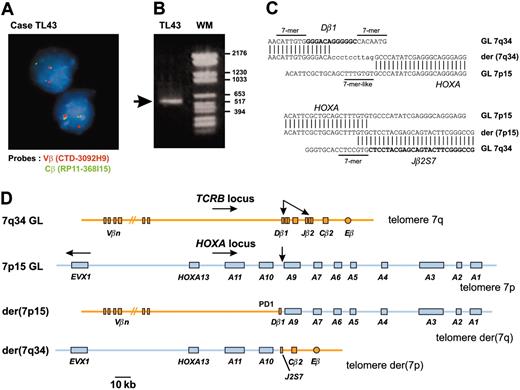
In an attempt to identify new oncogenic abnormalities in T-ALL, we carried out an interphasic FISH screening in 15 T-ALL cases that did not show expression of the 3 oncogenes most commonly involved in this type of leukemia (SIL-TAL1, TLX1, and TLX3). We used FISH probes of the T-cell-receptor β genes locus (TCRB) because this locus is often involved in oncogenic rearrangements of T-ALL and considering that the near-telomeric location of TCRB could lead to cryptic translocations. In this way, we identified 2 T-ALL cases (TL43, TL44) with a TCRB translocation (Figure 1A). Cloning of the TL43 translocation identified the HOXA locus as the TCRB partner sequence (Figure 1B-D). The HOXA locus was also involved in the second case (TL44), as demonstrated by FISH and Southern blot (data not shown).
We addressed the hypothesis that this new recurrent translocation could define a homogeneous T-ALL subgroup by analyzing global gene expression on a series of unselected T-ALL samples.
A new chromosomal translocation involves the TCRB and HOXA loci. (A) A TCRB chromosomal translocation was identified in leukemic cells of patient TL43 by dissociation of TCRB-flanking FISH probes. The centromeric Vβ-flanking probe (CTD-3092H9) and the telomeric Cβ-flanking probe (RP11-168I15) were labeled with Texas red and fluorescein isothiocyanate (FITC), respectively. Original magnification, × 63. (B) A 541-bp fragment containing the Dβ1 breakpoint was produced by circled nested PCR (left lane, the rearranged band is shown by an arrow; right lane, molecular weight marker). (C) Breakpoint and flanking sequences of both derivative chromosomes of the TCRB-HOXA translocation TL43. Recognition sequence signal (RSS) heptamer and putative heptamer-like sequence are indicated according to consensus.27 Untemplated nucleotides (N-diversity) are typed in lowercase. GL indicates germline; der, derivative chromosomes. (D) Schematic representation of the TCRB-HOXA translocation TL43, drawn to scale according to data from the IMGT Information System (http://imgt.cines.fr) and the UCSC Genome Bioinformatics site (http://genome.ucsc.edu/), and to the DNA sequences of the breakpoint regions of the 2 derivative chromosomes. Chromosomes are depicted according to the direction of the TCRB and HOXA gene transcription (except for the EVX1 gene, which is transcribed in the opposite direction as indicated by arrow). Breakpoints are indicated with arrows.
A new chromosomal translocation involves the TCRB and HOXA loci. (A) A TCRB chromosomal translocation was identified in leukemic cells of patient TL43 by dissociation of TCRB-flanking FISH probes. The centromeric Vβ-flanking probe (CTD-3092H9) and the telomeric Cβ-flanking probe (RP11-168I15) were labeled with Texas red and fluorescein isothiocyanate (FITC), respectively. Original magnification, × 63. (B) A 541-bp fragment containing the Dβ1 breakpoint was produced by circled nested PCR (left lane, the rearranged band is shown by an arrow; right lane, molecular weight marker). (C) Breakpoint and flanking sequences of both derivative chromosomes of the TCRB-HOXA translocation TL43. Recognition sequence signal (RSS) heptamer and putative heptamer-like sequence are indicated according to consensus.27 Untemplated nucleotides (N-diversity) are typed in lowercase. GL indicates germline; der, derivative chromosomes. (D) Schematic representation of the TCRB-HOXA translocation TL43, drawn to scale according to data from the IMGT Information System (http://imgt.cines.fr) and the UCSC Genome Bioinformatics site (http://genome.ucsc.edu/), and to the DNA sequences of the breakpoint regions of the 2 derivative chromosomes. Chromosomes are depicted according to the direction of the TCRB and HOXA gene transcription (except for the EVX1 gene, which is transcribed in the opposite direction as indicated by arrow). Breakpoints are indicated with arrows.
Large-scale expression analysis identifies a HOXA-expressing subgroup within HOX-related T-ALL samples
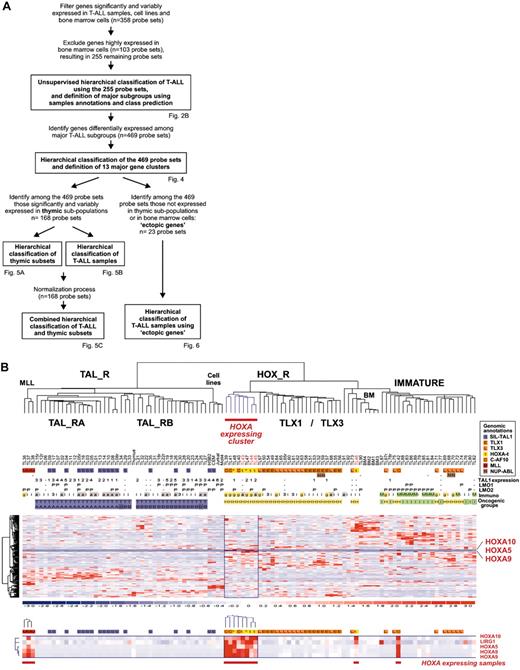
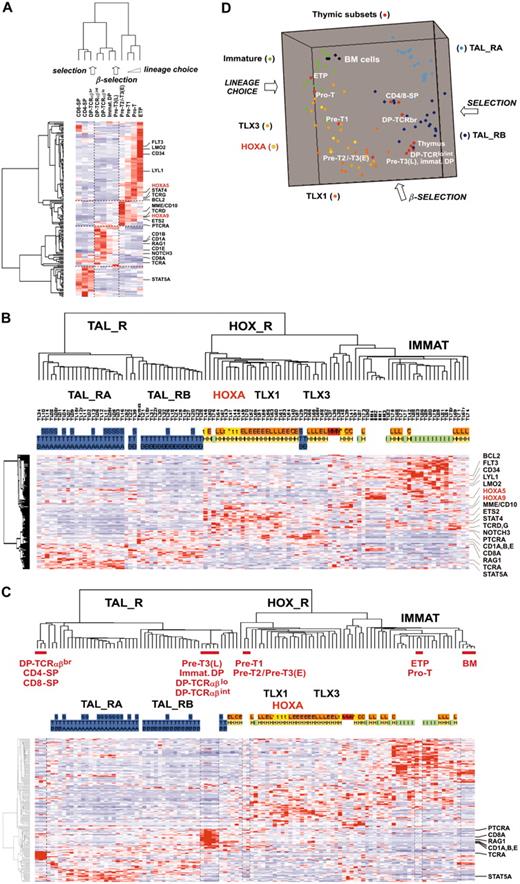
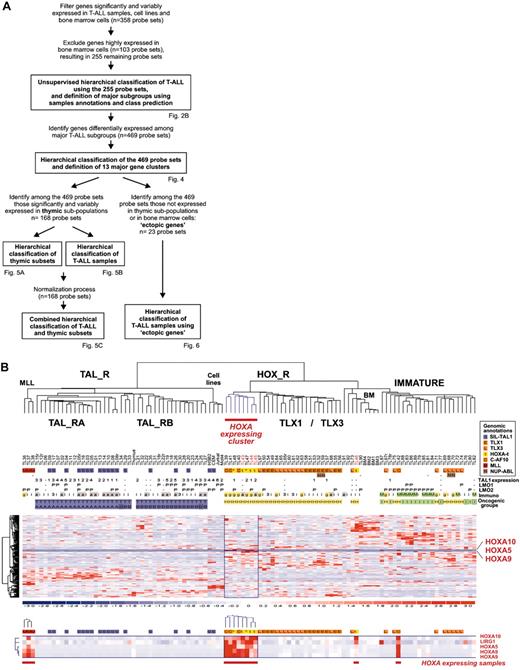
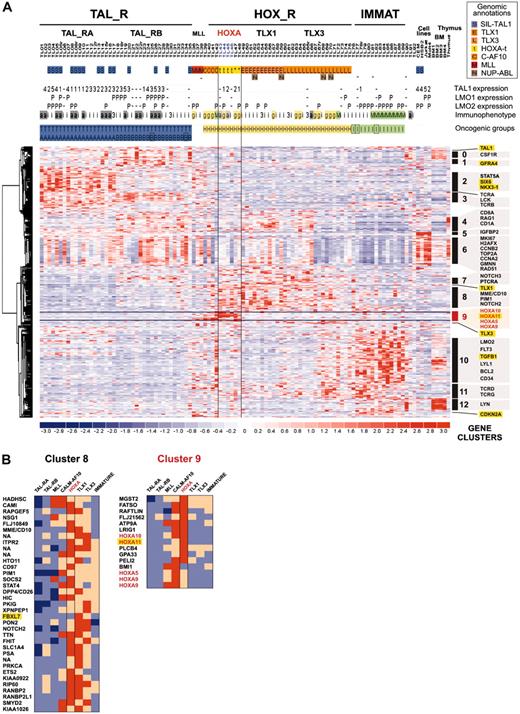
Leukemic cells from a series of 92 patients, including the 2 patients with the newly identified TCRB-HOXA rearrangement, were studied by transcriptome analysis of tumor cells using Affymetrix U133A microarrays. Sample and gene classifications were performed with unsupervised hierarchical methods using a list of 255 probe sets selected on the basis of expression variability and reliability among samples (the general microarray strategy is shown in Figure 2A; see also “Microarray experiments and data analysis” in the Supplemental Materials and methods for probe-set selection and methods). Major clusters of T-ALL were obtained, and significant correlations with the immunologic and oncogene expression annotations allowed definition of major T-ALL oncogenic groups (Figure 2B).
The first branch of the hierarchy (labeled as TAL1-related, TAL_R) includes all samples with SIL-TAL1 rearrangement (P < .001, 16 cases). Cases of this branch are associated with strong TAL1 expression (P < .001, 33 of 40 T-ALL cases with significant TAL1 expression are found in this branch; see also Figure S1). The cell-surface TCRαβ receptor is frequently expressed (P < .001, 16 of 20 αβ T-ALL cases are included in this branch but no γδ cases), in line with previous work connecting SIL-TAL1 rearrangement with αβ T-cell differentiation.19,28,29 Furthermore, this analysis delineates 2 stable subgroups (TAL_RA and TAL_RB) not identified by previous works.
The second branch of the hierarchy includes all cases with TLX1 and TLX3 expression (P < .001 in both cases). All the γδ T-ALL cases are included in this branch (P < .001, 14 cases). In addition to the TLX1- and TLX3-expressing cases, 2 clusters of samples are found in this branch. A first cluster contains 8 cases, including the 2 HOXA-translocated cases. Stability of this cluster was demonstrated by sampling analysis (Figure S2). The genes that mainly define this cluster are HOXA5, HOXA9, HOXA10, and LRIG1, and accordingly, this subgroup of samples was provisionally named the “HOXA-expressing cluster” (Figure 2B). These cases, and the TLX1- and the TLX3-expressing samples, were collectively named the HOX_R group. Another cluster of samples is characterized by strong expression of genes expressed in immature cells (eg, CD34 or LMO2) and frequent lymphoid and myeloid CD13 or CD33 surface antigen coexpression (P < .001, 9 of 14 cases, excluding the TLX3 samples). Considering that oncogenic events are uncharacterized in these cases but probably occurred at an early stage of differentiation (with lymphoid and myeloid potential), this group was provisionally named “Immature group” and constitutes the third major group of T-ALL. Half the TLX3-expressing samples cocluster with cases of this group, due to expression of immature genes.
Unsupervised hierarchical classification of samples and genes based on expression microarray data. (A) General flowchart describing the sequence of microarray analysis procedures throughout the study. (B) This bidimensional classification was performed using expression data of probe sets selected on expression variability and reliability through the samples (n = 358 probe sets), and exclusion of probe sets highly expressed in bone marrow cells; the resulting list of 255 probe sets is shown in Table S1. The top panel shows a hierarchical tree of samples (columns), sample subgroups, and sample annotations. Major subgroups corresponding to main hierarchy branches are indicated on the tree. MLL indicates cases with MLL rearrangements; TAL_R, TAL1-related cases, with 2 TAL_R subgroups (TAL_RA and TAL_RB); HOX_R, HOX-related cases; the HOXA-expressing cluster of cases is highlighted in red, and the main TLX1- and TLX3-expressing subgroup is indicated; IMMATURE, subgroup of cases characterized by strong expression of genes expressed in immature cells and frequent coexpression of myeloid genes; BM, normal human bone marrow cells. TL is the unique identification number for samples. Sample group annotations: for genomic annotations, S indicates cases with SIL-TAL1 transcripts; E, TLX1-expressing cases, L, TLX3-expressing cases (both also quoted as genomic annotations for simplicity); t, HOXA-rearranged cases; C, cases with CALM-AF10 transcripts; M, cases with MLL rearrangement as detected in these cases by FISH and Southern blot; and N, cases with NUP214-ABL transcripts. *HOXA_R cases without identified rearrangement. For TAL1, LMO1, and LMO2 gene expression, RQ-PCR evaluations of expression are indicated; - indicates cases not annotated to avoid bias due to erythroid contamination (n = 9 cases, including the HOXA-rearranged cases TL46), or not available (n = 2 cases). TAL1 expression was scored for significant increased levels compared to normal thymus level, evaluated from 1 to 5 (moderate to highest levels), and significant LMO1 and LMO2 expressions compared to normal thymus levels are indicated as P (positive); see Figure S1. Immuno indicates immunologic markers: i, immature; 3, cCD3 expression; g, γδ expression; a, αβ expression; and M, myeloid markers (CD13 or CD33 or both). For oncogenic groups, T indicates TAL_R cases; H, HOX_R cases; I, immature cases without TLX3 expression. For TAL_R subgroup annotations, A indicates TAL_RA; and B, TAL_RB. These labels were assigned based on sample annotations and microarray analyses (Figure S2). Group assignment by prediction models was questionable in 2 TAL_RB cases and 3 immature cases and these cases are indicated by boxes (TL25, TL34, TL76, TL77, and TL82; Figure S2D). The “Immature group” label is provisional because oncogenic events are unidentified in this group. In the middle panel, left, hierarchical tree of genes (rows); center, for all samples (columns), relative expression levels are indicated according to the color scale shown on the bottom of the figure (from deep blue, lower expression, to deep red, higher expression); right, rows corresponding to the HOXA genes are indicated. The bottom panel shows magnification of the cluster of genes defining the HOXA-expressing cluster. These cases, and the 5 additional samples with HOXA expression, are indicated by red boxes (HOXA-expressing samples).
Unsupervised hierarchical classification of samples and genes based on expression microarray data. (A) General flowchart describing the sequence of microarray analysis procedures throughout the study. (B) This bidimensional classification was performed using expression data of probe sets selected on expression variability and reliability through the samples (n = 358 probe sets), and exclusion of probe sets highly expressed in bone marrow cells; the resulting list of 255 probe sets is shown in Table S1. The top panel shows a hierarchical tree of samples (columns), sample subgroups, and sample annotations. Major subgroups corresponding to main hierarchy branches are indicated on the tree. MLL indicates cases with MLL rearrangements; TAL_R, TAL1-related cases, with 2 TAL_R subgroups (TAL_RA and TAL_RB); HOX_R, HOX-related cases; the HOXA-expressing cluster of cases is highlighted in red, and the main TLX1- and TLX3-expressing subgroup is indicated; IMMATURE, subgroup of cases characterized by strong expression of genes expressed in immature cells and frequent coexpression of myeloid genes; BM, normal human bone marrow cells. TL is the unique identification number for samples. Sample group annotations: for genomic annotations, S indicates cases with SIL-TAL1 transcripts; E, TLX1-expressing cases, L, TLX3-expressing cases (both also quoted as genomic annotations for simplicity); t, HOXA-rearranged cases; C, cases with CALM-AF10 transcripts; M, cases with MLL rearrangement as detected in these cases by FISH and Southern blot; and N, cases with NUP214-ABL transcripts. *HOXA_R cases without identified rearrangement. For TAL1, LMO1, and LMO2 gene expression, RQ-PCR evaluations of expression are indicated; - indicates cases not annotated to avoid bias due to erythroid contamination (n = 9 cases, including the HOXA-rearranged cases TL46), or not available (n = 2 cases). TAL1 expression was scored for significant increased levels compared to normal thymus level, evaluated from 1 to 5 (moderate to highest levels), and significant LMO1 and LMO2 expressions compared to normal thymus levels are indicated as P (positive); see Figure S1. Immuno indicates immunologic markers: i, immature; 3, cCD3 expression; g, γδ expression; a, αβ expression; and M, myeloid markers (CD13 or CD33 or both). For oncogenic groups, T indicates TAL_R cases; H, HOX_R cases; I, immature cases without TLX3 expression. For TAL_R subgroup annotations, A indicates TAL_RA; and B, TAL_RB. These labels were assigned based on sample annotations and microarray analyses (Figure S2). Group assignment by prediction models was questionable in 2 TAL_RB cases and 3 immature cases and these cases are indicated by boxes (TL25, TL34, TL76, TL77, and TL82; Figure S2D). The “Immature group” label is provisional because oncogenic events are unidentified in this group. In the middle panel, left, hierarchical tree of genes (rows); center, for all samples (columns), relative expression levels are indicated according to the color scale shown on the bottom of the figure (from deep blue, lower expression, to deep red, higher expression); right, rows corresponding to the HOXA genes are indicated. The bottom panel shows magnification of the cluster of genes defining the HOXA-expressing cluster. These cases, and the 5 additional samples with HOXA expression, are indicated by red boxes (HOXA-expressing samples).
Therefore, this analysis identifies a “HOXA-expressing cluster” of T-ALL that includes the 2 HOXA-rearranged samples. These cases cluster in the same branch as TLX1 and half the TLX3-related cases, suggesting a biologic link between these leukemias.
Genomic and expression analysis links HOXA expression to TCRB-HOXA, MLL, and CALM-AF10 chromosomal rearrangements
We investigated the 8 cases of the HOXA-expressing cluster defined by the unsupervised hierarchy, and 5 additional samples with detectable HOXA expression (HOXA-expressing samples, n = 13; Figure 2B). FISH, Southern blot, and reverse transcription-PCR (RT-PCR) were performed. Clinical, immunologic, and genetic features of the corresponding patients are indicated in Table 1.
In line with previous work,30,31 MLL rearrangements were found in 3 cases, which clustered as an independent homogeneous group (Figure 2B). In addition and unexpectedly, 4 HOXA-expressing cases demonstrated a CALM-AF10 translocation, as detected by RT-PCR.
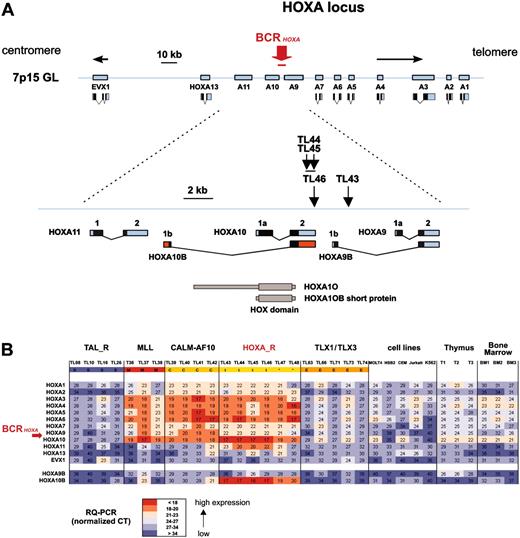
Considering that the HOXA-expressing cases included 2 cases (TL43 and TL44) with a TCRB-HOXA translocation, we searched for additional HOXA rearrangements in this subgroup (Table 1). Two more TCRB-HOXA translocations were identified using FISH (cases TL45 and TL46). The HOXA breakpoint location was mapped by Southern blot analysis in the 4 cases, and the TL46 translocation was fully characterized by sequencing of the 2 derivative breakpoint regions, in addition to case TL43 (Figure S3). Strikingly, the 4 breakpoints clustered in a genomic region of 2.6-kb length located between the HOXA10 and HOXA9 genes (BCRHOXA; Figure 3A). In the final 2 HOXA-expressing cases (TL47 and TL48), no HOXA rearrangement was detected, suggesting additional mechanisms of HOXA activation, as has also been reported for TAL1 and TLX1 activation.35-37 The 4 TCRB-HOXA cases were collectively named the HOXA-related (HOXA_R) subgroup, along with cases TL47 and TL48, because these 2 cases most frequently clustered with HOXA-rearranged cases in clustering experiments (Figure S2).
To analyze the consequence of the translocations on the expression of the HOXA locus, all 11 HOXA genes, as well as the non-HOX homeobox gene EVX1 located in the vicinity of the HOXA locus, were analyzed for expression using specific RQ-PCR. All cases with TCRB-HOXA, CALM-AF10, and MLL rearrangements consistently demonstrated global overexpression of most HOXA genes (but no EVX1 deregulation), compared to levels in normal thymus and control T-ALLs (Figure 3B). Strikingly, HOXA genes located on both sides of the TCRB-HOXA breakpoints were overexpressed. In addition, whereas we failed to amplify any potential TCRB-HOXA fusion transcript (data not shown), we found that a HOXA10 gene transcript, referred to as HOXA10B, was present at high levels in all the 4 HOXA-rearranged cases and in the 2 additional HOXA_R cases (Figure 3B). By contrast, no consistent expression of this HOXA10B transcript was detected in the thymus and other samples.
Therefore, the combination of large-scale expression, molecular cytogenetic, and gene-specific expression analyses allowed detailed description of the new recurrent HOXA chromosomal rearrangement. This rearrangement strongly deregulates the HOXA multigene cluster. In addition, a new association was detected between the CALM-AF10 rearrangement and expression of HOXA genes.
Inclusion of HOXA-expressing cases in a general molecular portrait of T-ALL
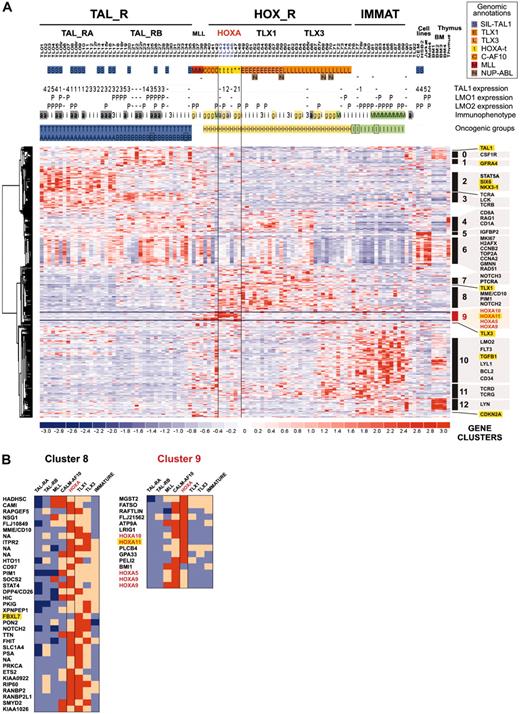
To analyze gene pathways differentially involved in T-ALL (including those of the HOXA_R subgroup), we studied expression profiles of the oncogenic T-ALL groups. First, a compendium of 469 probe sets differentially expressed between the previously defined T-ALL groups was obtained (see “Microarray experiments and data analysis” in the Supplemental Materials and methods). Then, a supervised hierarchical classification of the genes was performed, allowing definition of 13 gene clusters based on correlated expression in the T-ALL groups (Figure 4; Table S2 provides a list of genes and clusters). Tentative examination of the known biologic function of the genes shows that a high number of them are linked to thymocyte differentiation or general cellular functions like signaling, cell cycle, DNA repair, and antiapoptotic pathways.
As expected, the HOXA_R, CALM-AF10, and MLL cases express a cluster of genes containing HOXA5, HOXA9, HOXA10, and HOXA11 (cluster C9; Figure 4B). HOXA_R cases are also characterized by high average expression of genes, which have been involved in T-cell differentiation or oncogenesis (or both), including NOTCH2, NOTCH3, PTCRA (Pre-Tα gene), TCRG, TCRD, and PIM1 (clusters C7, C8, and C11), also shared with the TLX1 and TLX3 subgroups. Together with the fact that HOXA-rearranged, TLX1-, and most of the TLX3-expressing cases cluster in the unsupervised hierarchy (Figure 2B), these data reinforce the view that these subgroups share biologic similarities by using common pathways involved in thymocyte differentiation and, most likely, in T-cell oncogenesis.
Clustering of the breakpoints and dysregulated expression of the HOXA genes in leukemias with TCRB-HOXA rearrangement. (A) The 4 breakpoints cluster in a 2.6-kb breakpoint cluster region (BCRHOXA) as indicated. The direction of transcription is shown by arrows (centromere to telomere for the HOXA genes, and opposite direction for EVX1). Transcription products of the HOXA genes are drawn according to the data of the UCSC site. The HOXA10B exons are shown as red boxes. The rearrangements disrupt the HOXA locus at the 3′ end of HOXA10 gene (cases TL44, TL45, and TL46), and within the intron Ib region of HOXA9 (TL43), as indicated. Open reading frames are shown as black boxes. The 2 isoforms of the HOXA10 protein (HOXA10A, quoted HOXA10, and HOXA10B) are drawn according to previous reports, and the homeobox DNA-binding domain is indicated.32-34 (B) Analysis of expression of the HOXA genes by specific real-time RT-PCR in T-ALL samples. The HOXA and EVX1 expression values are shown in the 4 HOXA-rearranged cases (marked t), and the 2 HOXA-related cases without detected chromosomal rearrangement (TL47 and TL48, marked *), which constitute the HOXA_R group. Values in the 3 MLL-rearranged cases, the 4 CALM-AF10 cases, a panel of TAL_R and TLX1/TLX3 samples, T-cell lines, normal human thymus, and normal bone marrow samples are shown. HOXA9B and HOXA10B transcripts were also analyzed (bottom rows). Median normalized Ct values are indicated and a color scale was used as shown.
Clustering of the breakpoints and dysregulated expression of the HOXA genes in leukemias with TCRB-HOXA rearrangement. (A) The 4 breakpoints cluster in a 2.6-kb breakpoint cluster region (BCRHOXA) as indicated. The direction of transcription is shown by arrows (centromere to telomere for the HOXA genes, and opposite direction for EVX1). Transcription products of the HOXA genes are drawn according to the data of the UCSC site. The HOXA10B exons are shown as red boxes. The rearrangements disrupt the HOXA locus at the 3′ end of HOXA10 gene (cases TL44, TL45, and TL46), and within the intron Ib region of HOXA9 (TL43), as indicated. Open reading frames are shown as black boxes. The 2 isoforms of the HOXA10 protein (HOXA10A, quoted HOXA10, and HOXA10B) are drawn according to previous reports, and the homeobox DNA-binding domain is indicated.32-34 (B) Analysis of expression of the HOXA genes by specific real-time RT-PCR in T-ALL samples. The HOXA and EVX1 expression values are shown in the 4 HOXA-rearranged cases (marked t), and the 2 HOXA-related cases without detected chromosomal rearrangement (TL47 and TL48, marked *), which constitute the HOXA_R group. Values in the 3 MLL-rearranged cases, the 4 CALM-AF10 cases, a panel of TAL_R and TLX1/TLX3 samples, T-cell lines, normal human thymus, and normal bone marrow samples are shown. HOXA9B and HOXA10B transcripts were also analyzed (bottom rows). Median normalized Ct values are indicated and a color scale was used as shown.
In addition to the expression of HOXA genes, the CALM-AF10- and MLL-related cases are characterized by a high expression of the BMI1 gene (Figure 4B; BMI1 expression was confirmed by specific RQ-PCR, not shown). Bmi1 has been implicated in experimental murine B-cell leukemias38 and in self-renewal of normal and leukemic stem cells.39 This gene is considered a repressor of the HOX genes,40 which contrasts intriguingly with the high expression of HOXA genes in the MLL- and CALM-AF10-related cases (Figure 4B).
By comparison with the HOXA_R samples, and more generally with the HOX_R group, samples from the immature group strongly express the LMO2, LYL1, CD34, BCL2, FLT3, and TGFB1 genes (cluster C10, “hematopoiesis”) but have low expression of genes from other clusters, particularly from cluster C6, which appears linked to “proliferation and mitosis,” whereas there is a variable expression of this cluster throughout samples in the other groups (Figure 4A). Half the TLX3-expressing cases coexpress genes characteristic of both the immature and HOX_R groups, and accordingly these cases cluster with immature cases. Samples from the TAL_R group express genes involved in TCRB signaling (cluster C3), particularly in the TAL_RB samples. In addition, the TAL_RB samples share with HOX_R cases (particularly TLX1 cases) a high expression of T-cell differentiation genes (cluster C4 and C7), including NOTCH3 and PTCRA. Samples from the TAL_RA subgroup specifically express the signaling genes STAT5A and STAM1 (cluster C2). Interestingly, numerous genes not normally expressed in thymus have ectopic expression in TAL_R samples, including 2 homeobox genes, NKX3-1 and SIX6.
Analysis of oncogenic T-ALL subgroups with respect to normal T-cell differentiation
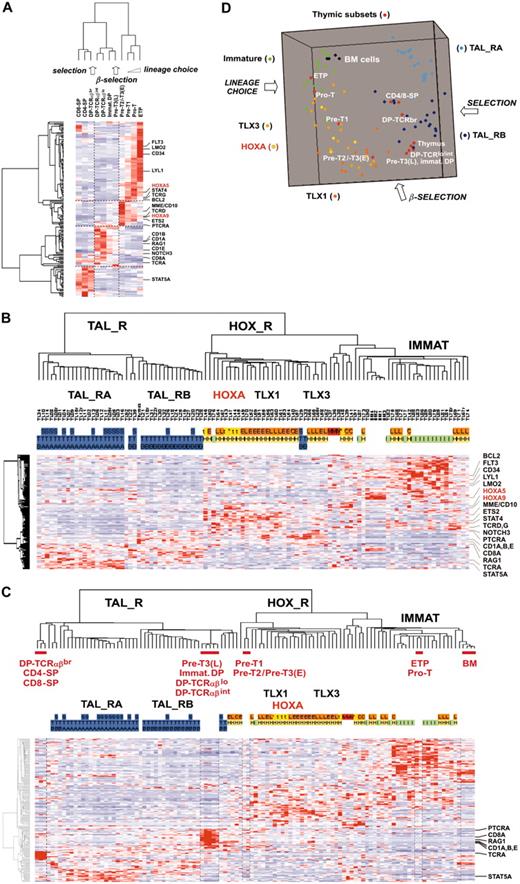
At this point, we analyzed gene expression in T-ALL samples with respect to normal thymic differentiation. For this purpose, 11 distinct thymic subpopulations were purified from normal human thymus, and global gene expression analysis using U133A microarrays was performed. The 469 probe sets differentially expressed between oncogenic groups (Figure 4A) were filtered for thymic expression and proliferation genes were excluded, resulting in 168 genes (see “Microarray experiments and data analysis” in the Supplemental Materials and methods).
First, an unsupervised hierarchy of the thymic subpopulations and the 168 genes was performed. Importantly, the sample classification recapitulates the sequential steps of thymocyte differentiation (Figure 5A). Moreover, examination of the gene clusters of normal thymic subpopulations frequently shows similar patterns compared to gene clusters of T-ALL (compare Figures 4A and 5A), pinpointing networks whose coordinated expression is conserved in leukemic cells (ie, FLT3-LMO2-CD34-LYL1 and CD1A-CD8A-RAG1). In rarer cases, different gene clustering suggests that abnormal coexpression in T-ALL samples could be important in the oncogenic process. For instance, PTCRA and NOTCH3 are coexpressed in subgroups of T-ALL samples, including HOX_R samples (Figure 4A), whereas in normal thymic subpopulations PTCRA is expressed at the highest level before the β-selection process and NOTCH3 afterward (Figure 5A).
Supervised clustering of genes differentially expressed among major sample groups. (A) This hierarchical classification was performed using 469 probe sets detecting differential expression among sample groups (Figure 2A). Samples were preclassified according to oncogenic group assignment as defined in Figure 2B. The same conventions for sample annotations are used. The HOXA_R subgroup is indicated (HOXA-rearranged, n = 4, and HOXA-related, n = 2). Clusters of genes defined by this supervised clustering are indicated (clusters 0-12). Genes of particular interest are labeled using Locuslink symbol, and genes without significant thymic expression are highlighted in yellow (ectopic thymic expression). The cluster C9, which is defined by HOXA-expressing samples and contains HOXA genes, is highlighted in red. (B) List of genes from clusters C8 and C9 and relative expression in oncogenic subgroups. Genes from cluster C8 are frequently highly expressed in subsets from the HOX_R group; cluster C9 is defined mainly by the HOXA-expressing samples (HOXA_R, CALM-AF10, and MLL) and includes HOXA genes (in red); “ectopic” genes with respect to thymus expression are highlighted in yellow. NA indicates Locuslink symbol not available. Relative expression in each T-ALL group is shown by color scale.
Supervised clustering of genes differentially expressed among major sample groups. (A) This hierarchical classification was performed using 469 probe sets detecting differential expression among sample groups (Figure 2A). Samples were preclassified according to oncogenic group assignment as defined in Figure 2B. The same conventions for sample annotations are used. The HOXA_R subgroup is indicated (HOXA-rearranged, n = 4, and HOXA-related, n = 2). Clusters of genes defined by this supervised clustering are indicated (clusters 0-12). Genes of particular interest are labeled using Locuslink symbol, and genes without significant thymic expression are highlighted in yellow (ectopic thymic expression). The cluster C9, which is defined by HOXA-expressing samples and contains HOXA genes, is highlighted in red. (B) List of genes from clusters C8 and C9 and relative expression in oncogenic subgroups. Genes from cluster C8 are frequently highly expressed in subsets from the HOX_R group; cluster C9 is defined mainly by the HOXA-expressing samples (HOXA_R, CALM-AF10, and MLL) and includes HOXA genes (in red); “ectopic” genes with respect to thymus expression are highlighted in yellow. NA indicates Locuslink symbol not available. Relative expression in each T-ALL group is shown by color scale.
Then, by analyzing leukemic samples using the 168 “thymic genes,” we found that a consistent hierarchy of T-ALL could be obtained (Figure 5B; compare with Figure 2B). Therefore, to define the differentiation stages of leukemic cells according to biologic pathways, T-ALL and thymic subpopulations data were combined. Hierarchical analysis and multidimensional scaling visualization were performed and informative coclustering of T-ALL subgroups and thymic subsets was observed (Figure 5C-D). The HOXA_R samples cluster with the Pre-T1 and Pre-T2/Pre-T3(E) subpopulations, together with the TLX1- and half the TLX3-expressing samples. Therefore, these samples appear arrested at a stage corresponding to T-lineage engagement preceding the β-selection process. By contrast, the TAL_R samples cluster with subpopulations corresponding to steps following β-selection. Samples from the immature group, as well as the CALM-AF10, MLL, and half the TLX3-expressing leukemic samples, cluster with the most immature ETP and Pro-T subpopulations, which correspond to the lineage choice process. Notably, further examination of gene expression in leukemic samples with respect to normal subpopulations reveals numerous abnormalities in the differentiation process (in addition to the differentiation arrest itself). For instance, a few HOX_R samples, including 2 TCRB-HOXA cases, unexpectedly coexpress CD10/MME and TCRαβ heterodimer (Figure 4B). Strikingly, numerous T-cell differentiation-associated genes are expressed at dramatically lower levels in T-ALL samples compared with the relevant normal thymic subpopulations, such as CD8A, RAG1, CD1s, PTCRA, and TCRA in the TAL-R cases (Figure 5C; CD8A and RAG1 expression was confirmed in thymic subsets and T-ALL samples using RQ-PCR, not shown). Conversely, several genes are expressed at higher levels in T-ALL than in normal thymic subsets, such as STAT5A in TAL_RA cases.
Oncogenic groups of T-ALL can be classified using “ectopic” gene expression
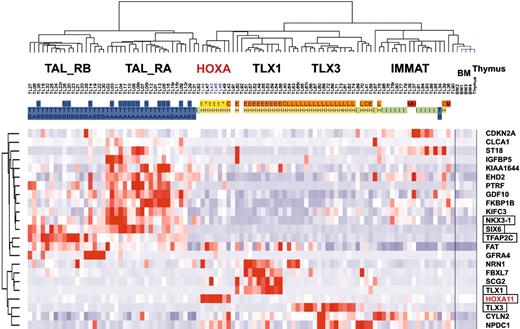
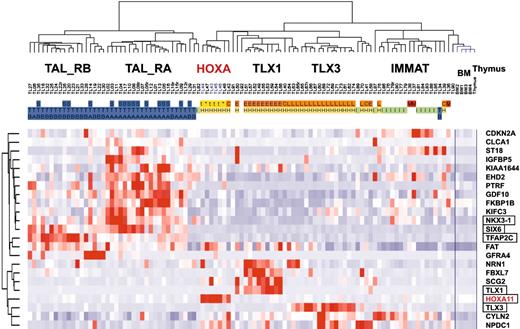
Because subgroups of T-ALL can be easily classified by genes normally expressed during thymic differentiation, we wondered whether the unsupervised clustering of T-ALL samples defined in this work merely reflects the differentiation status of the cells, or whether it may also specifically highlight oncogenic events. The 469 probe sets differentially expressed between oncogenic groups (Figure 4A) were filtered for ectopic expression with reference to expression in normal thymic subpopulations and bone marrow (considering that a fraction of T-ALL cells coexpress immature and myeloid-associated genes), resulting in a working list of 23 “ectopic” genes (see “Microarray experiments and data analysis” in the Supplemental Materials and methods). An unsupervised classification of the leukemic samples was performed using these genes. Strikingly, the resulting hierarchy properly classifies the major T-ALL oncogenic subgroups (Figure 6).
Importantly, this gene-filtering strategy was able to select the TLX1 and TLX3 genes and a HOXA gene (HOXA11), all genes that can be targeted by chromosomal translocations as demonstrated here for the HOXA locus. Note, however, that some genes linked to oncogenesis were excluded due to their expression in certain thymic subpopulations or in bone marrow, such as TAL1, LMO2, and the HOXA genes except HOXA11. Interestingly, 2 homeobox genes, NKX3-1 and SIX6, are included in the list of ectopic genes, and they are expressed in TAL_R cases. Notably, a gene related to NKX3-1, namely NKX2-5, has been involved in chromosomal translocations in T-cell lines.41 Additional genes involved in development or morphogenesis were also selected, including TFAP2C and GDF10 (BMP family). Expression level of NKX3-1, SIX6, and TFAP2C was confirmed by specific RQ-PCR in representative T-ALL samples. It therefore appears that ectopic expression of at least one developmental gene is found in most leukemic cases from the major HOX_R and TAL_R groups.
These results demonstrate that the subgroups of T-ALL are not only defined by their differentiation status but also by ectopic expression of genes, including developmental genes.
Discussion
The HOXA gene cluster is an oncogenic target in T-ALL
By combining molecular cytogenetic and expression analysis approaches, we have identified and characterized a new recurrent molecular rearrangement involving the HOXA and TCRB loci in T-ALL. Interphasic FISH patterns and breakpoint sequencing were consistent with chromosomal inversion inv(7)(p15q34) in cases TL44, TL45 and TL46, and translocation t(7;7)(p15;q34) in case TL43 (no karyotypic data were available). Notably, such chromosomal rearrangements have been recurrently reported in T-ALL,42 (see also http://cgap.nci.nih.gov/Chromosomes/Mitelman), but it was probably assumed that they would always reproduce in T-ALL the TCRG-TCRB rearrangements frequently found in T cells in patients with ataxia-telangiectasia.43,44
Unsupervised clustering of human thymocyte subpopulations and leukemic cells. (A) Unsupervised classification of 11 subpopulations purified from normal human thymus and subsequently analyzed by global gene expression on the U133A array. For human thymic subsets, ETP indicates early thymocyte precursor with lymphomyeloid potential25,26 ; the Pro-T, Pre-T1, Pre-T2/Pre-T3(E) (ie, Pre-T2 and Pre-T3(E), not distinguished here), Pre-T3(L), immature DP (immature double-positive cells), DP-TCRαβlo, DP-TCRαβint and DP-TCRαβbr, CD4-SP (CD4 single-positive cells), and CD8-SP, have been described previously (for a review, see Carrasco et al4 ). The lineage choice, β-selection, and selection processes as reported in normal thymus differentiation are indicated. The 469 probe sets differentially expressed between oncogenic groups (Figure 4A) were filtered for variable and reliable thymic expression. In addition, “proliferation genes” defined by the “proliferation-mitosis” cluster (cluster C6 in Figure 4A) were discarded from this analysis to avoid spurious sample clustering due to proliferation rate. The resulting final list of 168 “thymic” genes is shown in Table S3. Genes of particular interest are shown, and HOXA genes are indicated in red. (B) Unsupervised classification of T-ALL samples using the same 168 “thymic” gene list. The same conventions are used as in Figure 2. The main oncogenic subgroups are indicated. The same genes of interest as in Figure 5A are shown. (C) Unsupervised analysis of combined T-ALL samples, normal human thymic subpopulations, and bone marrow (BM) cells. A selection of genes with a strongly altered expression level compared with the corresponding normal thymic subsets is indicated (RAG1 and CD8A gene expression was also directly analyzed by RQ-PCR in normal thymic samples and T-ALL, giving consistent data; not shown). (D) Three-dimensional representation of a multidimensional scaling analysis performed using normal thymic subsets, normal bone marrow, and T-ALL samples of the main oncogenic subgroups (TAL_RA, TAL_RB, HOXA, TLX1, TLX3, immature). The lineage choice, β-selection, and selection processes are indicated.
Unsupervised clustering of human thymocyte subpopulations and leukemic cells. (A) Unsupervised classification of 11 subpopulations purified from normal human thymus and subsequently analyzed by global gene expression on the U133A array. For human thymic subsets, ETP indicates early thymocyte precursor with lymphomyeloid potential25,26 ; the Pro-T, Pre-T1, Pre-T2/Pre-T3(E) (ie, Pre-T2 and Pre-T3(E), not distinguished here), Pre-T3(L), immature DP (immature double-positive cells), DP-TCRαβlo, DP-TCRαβint and DP-TCRαβbr, CD4-SP (CD4 single-positive cells), and CD8-SP, have been described previously (for a review, see Carrasco et al4 ). The lineage choice, β-selection, and selection processes as reported in normal thymus differentiation are indicated. The 469 probe sets differentially expressed between oncogenic groups (Figure 4A) were filtered for variable and reliable thymic expression. In addition, “proliferation genes” defined by the “proliferation-mitosis” cluster (cluster C6 in Figure 4A) were discarded from this analysis to avoid spurious sample clustering due to proliferation rate. The resulting final list of 168 “thymic” genes is shown in Table S3. Genes of particular interest are shown, and HOXA genes are indicated in red. (B) Unsupervised classification of T-ALL samples using the same 168 “thymic” gene list. The same conventions are used as in Figure 2. The main oncogenic subgroups are indicated. The same genes of interest as in Figure 5A are shown. (C) Unsupervised analysis of combined T-ALL samples, normal human thymic subpopulations, and bone marrow (BM) cells. A selection of genes with a strongly altered expression level compared with the corresponding normal thymic subsets is indicated (RAG1 and CD8A gene expression was also directly analyzed by RQ-PCR in normal thymic samples and T-ALL, giving consistent data; not shown). (D) Three-dimensional representation of a multidimensional scaling analysis performed using normal thymic subsets, normal bone marrow, and T-ALL samples of the main oncogenic subgroups (TAL_RA, TAL_RB, HOXA, TLX1, TLX3, immature). The lineage choice, β-selection, and selection processes are indicated.
Unsupervised clustering of T-ALL samples based on `ectopic' genes. The 469 probe sets differentially expressed between oncogenic groups (Figure 4A) were filtered for ectopic expression with reference to expression in normal thymic subpopulations and bone marrow, resulting in a working list of 23 ectopically expressed genes. Genes known to be involved in development are boxed. The main T-ALL oncogenic subgroups and annotations are indicated as in other figures.
Unsupervised clustering of T-ALL samples based on `ectopic' genes. The 469 probe sets differentially expressed between oncogenic groups (Figure 4A) were filtered for ectopic expression with reference to expression in normal thymic subpopulations and bone marrow, resulting in a working list of 23 ectopically expressed genes. Genes known to be involved in development are boxed. The main T-ALL oncogenic subgroups and annotations are indicated as in other figures.
The HOXA gene cluster is one of the 4 major homeobox gene clusters. Each cluster displays 9 to 11 paralogous HOX genes, with a conserved structural organization and a tightly regulated expression.45-48 We found that the disruption of the HOXA locus by TCRB chromosomal rearrangements was associated with a global overexpression of the HOXA genes (Figure 3B). This may be due to the juxtaposition of TCRB regulatory elements in the vicinity of the HOXA genes7,49 and also to the separation of the locus into 2 parts, which may disrupt the normal regulatory elements of the cluster. Alternatively, nonconventional deregulation mechanisms involving miRNA might also be involved50 or, at a higher level, the new chromosome structure resulting from the rearrangement might induce global changes in the nucleus and chromatin organization.51 Interestingly, the 4 HOXA breakpoints cluster tightly at the vicinity of the HOXA10 and HOXA9 genes (Figure 3A). Whereas we could not detect a TCRB-HOXA fusion transcript, strong expression of an alternative transcript known as the HOXA10B transcript was detected in addition to the global HOXA gene overexpression (Figure 3A-B). This transcript, also called the HOXA10 short transcript, has been found in embryonic cells and myeloid cell lines but not in normal hematopoiesis.32,33,52 It encodes a short protein that contains the HOXA10 homeobox DNA-binding domain but lacks the N-terminal regulatory region and that may have distinct functional properties.33,34 The TCRB-HOXA translocation is, to our knowledge, the first example of a recurrent chromosomal rearrangement associated with such a direct and major dysregulation of a multigenic locus.
MLL-translocated cases of the present series demonstrate a strong expression of the HOXA genes, consistent with previous reports.30,31 The direct targeting of the HOXA locus by chromosomal rearrangement strongly supports the hypothesis that dysregulation of HOXA gene expression, triggered by MLL fusion proteins, is causal in MLL-related oncogenesis, consistent with in vitro properties of MLL proteins.53-56 The fact that MLL and HOX_R groups do not cluster in unsupervised hierarchy may suggest that additional genes targeted by the MLL fusions could play an important role in oncogenesis. A major oncogenic role of HOXA genes in T-ALL with CALM-AF10 translocation is also suggested by our results.
The HOX gene network is involved in embryogenesis, stem cell renewal, cell fate, and control of cell differentiation in many tissue types including hematopoietic system and thymus.45,57-60 The evidence increasingly suggests that HOX genes might play a central role in oncogenesis, including in leukemia.2,20,58,61-65 In myeloid leukemias, several HOX genes including HOXA9 can be involved in gene fusion with NUP98, leading to chimeric proteins.66-68 The present identification of the TCRB-HOXA rearrangement in human leukemia, involving the prototypical homeobox genes HOXA, strongly reinforces the view that abnormal expression of developmental genes can be causal in human cancer.
Large-scale expression analysis defines complete typology of T-ALL
Unsupervised classifications allow definition of the overall typology of samples, without a priori knowledge of the sample annotations.69 New homogeneous subgroups can be identified using these methods, as shown here in the HOXA-rearranged cases. Importantly, comparison of the sample annotations with the distinct oncogenic groups in our series of 92 cases suggests that a complete model of T-ALL has been here delineated. Major stable groups have been defined by unsupervised analysis on a large number of samples and genes, and this classification makes sense because it pinpoints links between these groups and genetic rearrangements, ectopic expression of genes including known oncogenes, and critical stages of thymus differentiation. It is therefore likely that potential new subgroups in T-ALL would represent discreet variations within these major groups or rare distinct subgroups.
Using a supervised approach based on oncogene expression, Ferrando et al identified a set of genes differentially expressed in T-ALL samples.19 An unsupervised classification of our samples using a list of genes based on this set properly classifies the major oncogenic groups (Figure S4). It is likely that the LYL1 group defined in this previous work partially overlaps with our provisionally named immature group (note, however, that LYL1 is expressed in the corresponding normal thymic immature subpopulation). Importantly, 3 cases here identified as HOXA-translocated cocluster using this former set of genes, within a branch containing the vast majority of TLX1 and TLX3 cases (Figure S4). Comparison of these 2 studies demonstrates the reliability of the gene expression approach and emphasizes the view that TCRB-HOXA-rearranged cases represent a homogeneous T-ALL subgroup.
HOXA genes are included in biologic networks that connect oncogenesis, thymic differentiation pathways, and ectopic gene expression
Important data were obtained by analyzing expression profiles of leukemic samples with respect to those of normal human thymocyte subpopulations. Although oncogenic processes dramatically alter levels of gene expression, we found that conserved gene expression networks allow pertinent coclustering of leukemic and thymic samples (Figure 5C). T-ALL cells of the major groups (TAL_R, HOX_R, and immature) appear arrested at distinct stages of differentiation, as previously suggested,19,28,29,70 corresponding to critical events in thymic maturation. Specifically, the HOXA-rearranged and TLX1-expressing cases closely match differentiation stages of Pre-T1 to Pre-T2/Pre-T3(E) subsets, whereas TLX3-expressing cases extend from ETP to Pre-T2/Pre-T3(E), all stages preceding the β-selection process. This correlation between oncogene expression and arrest at precise stages of differentiation may suggest that expression of distinct oncogenes is associated with the inhibition of distinct differentiation pathways. For instance, it has been reported that ectopic expression of TAL1 oncoprotein may perturb T-cell differentiation by disturbing the expression of E2A target genes.71-73 Consistently, we found in TAL_R samples that many genes characteristic of steps later than β-selection have dramatic underexpression, although these samples cocluster with thymic subpopulations corresponding to these late stages (Figure 5C). It therefore appears that these samples do not display a complete β-selection pattern. One can hypothesize that protection mechanisms have been selected in evolution to avoid the generation of tumors at this stage normally associated with pre-TCR signaling and a resulting high proliferation rate. For instance, RAG2 phosphorylation inhibits V(D)J recombination in cycling thymocytes,74 and this may protect highly proliferative cells in β-selection from a major mechanism of oncogenic chromosomal rearrangement.
Once arrested, leukemic cells might continuously activate certain biologic pathways normally transiently used at these stages, like antiapoptotic pathways. In addition, we found a distortion in expression of certain genes critical for thymic differentiation. For instance, NOTCH2, NOTCH3, and PTCRA genes are coexpressed in most human T-ALL cases (Figure 4A), as reported.75 Interestingly, these genes cocluster in T-ALL samples but not in normal thymic subpopulations, suggesting that an abnormal coexpression of these genes could be specifically associated with oncogenesis. Consistent with a major oncogenic role of the activated NOTCH genes, NOTCH1 is frequently targeted by activating mutations (or rare chromosomal translocation) in human T-ALL,11,13 and expression of a constitutively active intracellular domain of Notch3 favors murine T-cell leukemia in a Ptcra-dependent manner.75 Interestingly, 2 of the 4 HOXA-rearranged cases here identified also have prototypical NOTCH1 DNA mutation (leading to L1601P and H1602L+1603insP mutated proteins in cases TL43 and TL45, respectively, data not shown), demonstrating that HOXA and NOTCH1 genomic abnormalities can be associated and probably cooperate in these cases.
Finally, it appears from our data that a number of genes, including developmental genes, have abnormal, frequently ectopic, expression in certain T-ALL samples compared with normal thymic subpopulations. These include HOXA, TLX1, and TLX3 genes, which can be targeted by recurrent chromosomal translocation as demonstrated here for the HOXA genes, and CDKN2A/INK4A/ARF, frequently inactivated by DNA mutation or deletion as a tumor suppressor gene.17,18 Ectopic expression in T-ALL samples of other developmental genes, such as NKX3-1, SIX6, and TFAP2C suggests that these genes might also be involved in T-ALL oncogenesis (Figure 6). Interestingly, expression of some of these genes has been previously linked to cancer.76,77 Notably, both the NKX2-5 gene (homeobox gene paralogous to NKX3-1) and the TAL1 gene are targeted by chromosomal rearrangements in the T-ALL cell line CEM-CCRF, suggesting potential oncogenic cooperation.41 Altogether, our results are consistent with a model in which developmental genes, including the prototypical HOXA genes in some leukemia subgroups, play a central role in T-ALL oncogenesis.63
Prepublished online as Blood First Edition Paper, March 17, 2005; DOI 10.1182/blood-2004-10-3900.
Supported by grants from the Ligue Nationale contre le Cancer (`Programme Cartes d'Identité des Tumeurs'), the Ministère de la Recherche (`Programme Génomique'), the Ministère de la Santé (`Programme Tumorothèques'), and INSERM.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
This paper reports data of the pilot project of the Carte d'Identité des Tumeurs program from the Ligue National Contre le Cancer.
We thank all physicians who sent leukemic samples to the Hematology Laboratory, and especially Thierry Leblanc and Emmanuel Raffoux. We gratefully thank F.A.B. members Georges Flandrin and Marie-Thérèse Daniel for expert morphologic evaluation of leukemic samples. We thank the Xavier Fund for RNA extraction and contribution to oncogene expression analysis, Marie Romeo for CALM-AF10 and BMI1 transcript evaluation, Claire Pichereau for contribution to FISH analysis, and Antoine Crinquette for contribution to Southern blot experiments. We thank Philippe Dessen (Institut Gustave Roussy, Villejuif, France) for updates of gene annotations. We also thank Christelle Thibault, Philippe Kastner, and Doulaye Dembelé (Strasbourg Genopole National Facility, Illkirch, France), Fabien Patel (CIT program, Ligue Nationale contre le Cancer), and Elisabeth Savariau and Robin Nancel (Service d'Infographie, Institut Universitaire d'Hématologie, Paris). We thank Catharine M. Green, Jacques Haiech, Alain Aurias, and Didier Auboeuf for critical comments on the manuscript.