Abstract
Clinical observations and experimental evidence link bone marrow failure in myelodysplastic syndrome (MDS) with a T cell–dominated autoimmune process. Immunosuppressive therapy is effective in improving cytopenias in selected patients. Trisomy 8 is a frequent cytogenetic abnormality in bone marrow cells in patients with MDS, and its presence has been associated anecdotally with good response to immunotherapy. We studied 34 patients with trisomy 8 in bone marrow cells, some of whom were undergoing treatment with antithymocyte globulin (ATG). All had significant CD8+ T-cell expansions of one or more T-cell receptor (TCR) Vβ subfamilies, as measured by flow cytometry; expanded subfamilies showed CDR3 skewing by spectratyping. Sorted T cells of the expanded Vβ subfamilies, but not of the remaining subfamilies, inhibited trisomy 8 cell growth in short-term hematopoietic culture. The negative effects of Vβ-expanded T cells were inhibited by major histocompatibility complex (MHC) class 1 monoclonal antibody (mAb) and Fas antagonist and required direct cell-to-cell contact. Sixty-seven percent of patients who had de novo MDS with trisomy 8 as the sole karyotypic abnormality responded to ATG with durable reversal of cytopenias and restoration of transfusion independence, with stable increase in the proportion of trisomy 8 bone marrow cells and normalization of the T-cell repertoire. An increased number of T cells with apparent specificity for trisomy 8 cells is consistent with an autoimmune pathophysiology in trisomy 8 MDS.
Introduction
Myelodysplastic syndrome (MDS) is a diverse group of hematologic disorders characterized by dysplastic bone marrow (BM) and defective hematopoiesis, producing anemia, leukopenia, and thrombocytopenia.1 In MDS, blood counts are low despite normal or increased BM cellularity. An important mechanism of marrow failure in these syndromes is increased apoptosis in hematopoietic progenitors and their progeny.2-6 We and others7,8 have identified increased T-cell inhibitory activity that may suppress hematopoiesis in MDS. Cytokine deregulation, particularly increased levels of tumor necrosis factor alpha (TNF-α),2,9,10 has been described. Clonal expansion of T cells, identified by their selective use of the T-cell receptor (TCR) variable β chain, and lymphocyte-mediated suppression of hematopoiesis by CD8+ cytotoxic T cells (CTL) in coculture suggest a mechanism of progenitor and stem cell inhibition similar to that encountered in aplastic anemia (AA).11 Like AA, MDS can be successfully treated with cyclosporine (CsA) and antithymocyte globulin (ATG).12-15 De novo MDS with trisomy 8 especially often shows hematologic improvement after immunosuppressive therapy (IST). In patients in whom MDS has evolved from AA, trisomy 8 is associated with a relatively good prognosis, and cytopenias show sustained response to IST. In contrast, patients who evolve to monosomy 7 do not usually respond to IST and often die of progression to leukemia or of refractory cytopenia.16
We recently found evidence that MDS with trisomy 8 differs in its pathophysiology from other types of MDS.17 Trisomy 8 cells were more sensitive to Fas-mediated apoptosis and more likely to express activated caspase, Fas, and tumor necrosis factor receptor-1 (TNFR-1) than were karyotypically normal cells. These findings contrasted with the relative resistance to Fas-mediated apoptosis in monosomy 7 and normal Fas sensitivity of 5q–-associated MDS. Several mechanisms might underlie the high Fas expression and increased apoptotic markers in trisomy 8 disease. Fas expression could be an intrinsic property of the trisomy 8 cell, related to genetic changes resulting from aneuploidy of chromosome 8. Alternatively, extrinsic up-regulation of apoptotic markers could occur as a result, for example, of a T-cell–mediated immune response to an antigen presented by MDS cells. In the current study we used TCR-Vβ analysis, colony inhibition assays,18,19 and fluorescence in situ hybridization (FISH) to define cytotoxic T cells with specificity for trisomy 8 progenitors.
Patients, materials, and methods
Patient population characteristics
After informed consent, using protocols approved by the Institutional Review Board of the National Heart, Lung, and Blood Institute, BM samples were obtained from 52 patients with MDS, as categorized by French-American-British criteria (FAB),20 and from 19 healthy volunteers (Table 1). Patients were selected for study based on results of routine cytogenetic analysis. They were part of a larger cohort of MDS patients enrolled in Hematology Branch trials at the National Institutes of Health Clinical Center between 1995 and 2003. Patients with MDS and the FAB classification refractory anemia (RA) were enrolled to receive treatment with ATG,21 ATG/CsA,22 or CsA in protocols approved by the Institutional Review Board of the National Heart, Lung and Blood Institute. We treated consecutive qualifying patients with MDS older than 17 years of age.23 To qualify, patients had to have at least 1 of the following criteria: anemia requiring transfusion support with at least 1 U packed red blood cells per month for 2 months or longer; thrombocytopenia (platelet count less than 50 000/μL); or neutropenia (absolute neutrophil count less than 500/μL). Patients with excess blasts (more than 20%) with a matched related donor who were eligible to undergo transplantation were not eligible. Patients with a history of initial failure to respond to ATG, antilymphocyte globulin (ALG), or another immunosuppressive agent were also excluded. Complete hematologic remission was defined as normalization of all affected cell lines and less than 5% marrow blasts. Partial response was defined as greater than 50% improvement from baseline to normal levels of all cell counts in 2 serial blood values obtained 1 month apart. Response was determined on the basis of hematologic characteristics 3 months after treatment.
Cell preparation
BM mononuclear cells (BMMCs) were prepared after aspiration from the posterior iliac crest into syringes containing media supplemented 1:10 with heparin (O'Neill and Feldman, St Louis, MO) and separated by density gradient centrifugation with lymphocyte separation medium (Organon, Durham, NC).
Fluorescence in situ hybridization
At days 0, 7, and 14, cells were treated with hypotonic buffer consisting of KCl, HEPES (N-2-hydroxyethypiperazine-N′-2-ethanesulfonic acid), EGTA (ethyleneglycotetraacetic acid), and NaOH to expose the nucleus at interphase and were fixed onto slides using methanol/acetic acid (3:1). FISH was performed with probes for chromosomes 5q, 7, and 8 (Vysis Inc, Downers Grove, IL). Slides were denatured by immersion in formamide/20 × SSC solution for 5 minutes at 73°C, followed by several washes in 70%, 85%, and 100% ethanol at room temperature for 1 minute in each wash. Probes were similarly denatured and applied to cells on the slides to hybridize at 42°C overnight. After hybridization, the slides were washed in prewarmed 0.4 × SSC at 73°C for 2 minutes, 2 × SSC/0.1% NP-40 at room temperature for 1 minute, allowed to dry in the dark, and counterstained with DAPI-II for enumeration using a fluorescence microscope. Percentage positive staining was based on 400 cells scored. Three different observers, blinded with respect to sample identity, examined 3 different sets of slides. Scores were averaged, and the mean of the 3 was recorded. A healthy positive control and a trisomy 8–positive control were hybridized and scored with each separate run.
Characterization of the TCR of patients with trisomy 8
We used flow cytometry to analyze the TCR repertoire in MDS patients; in experimental models, flow cytometry has been used as a surrogate for an antigen-driven immune response.24-27 As controls, we used a normal range of the TCR previously obtained from 23 healthy controls by flow cytometry with the same panel of Vβ subfamily mAbs used in these experiments,28 and we added 19 healthy persons as controls in our test cohort. We determined the use of the different TCR-Vβ variable regions in CD4+ and CD8+ T cells and in their effector subset as determined by the low expression of CD28. Fresh peripheral blood (PB) was stained with a mixture of 4 mAbs: energy-coupled dye (ECD)–conjugated CD4, phycoerythrin cytochrome 5 (PECY5)–conjugated CD8, and phycoerythrin (PE)–, or when appropriate fluorescein isothiocyanate (FITC)–, conjugated CD28 (all from Beckman-Coulter, Fullerton, CA). For the variable region of the TCR-Vβ), a pool of 22 mAbs (either PE- or FITC-conjugated) was used to quantitate the individual contribution of TCR-Vβ subfamilies to the CD4+ and CD8+ lymphocyte pool (all from Immunotech [Beckman-Coulter], except mAb anti–Vβ6.7 [Endogen, Woburn, MA]). Samples stained with appropriate isotypic PE-, FITC-, PECY-, and ECD-conjugated mAbs served as controls to establish the fluorescence limits of background staining. After 20 minutes of incubation at room temperature, erythrocytes were lysed; the remaining cells were fixed with a Q-prep apparatus (Beckman-Coulter) and were further analyzed by a Coulter XL flow cytometer equipped with Epics Elite software. A 4-color protocol was used: the lymphocyte gate was set according to their size and forward scatter properties. TCR-Vβ use was determined within CD4+CD28low, CD8+CD28low, and total CD4+ and CD8+ populations, by using appropriate gates. In addition, FITC-conjugated αβ mAb (Becton Dickinson, Mountain View, CA) was used to determine the contribution of each Vβ subfamily to the total α/β TCR repertoire. Values obtained for individual Vβ families were expressed as a percentage of α/β TCR-expressing CD4+ or CD8+ cells. The 19 healthy, age-matched controls were run and compared with the 23 healthy samples from previous studies.28 Values greater than 2 SD of the mean control TCR-Vβ percentage were designated abnormal. All samples were tested in duplicate, and positive samples were verified by staining an additional sample.
Polymerase chain reaction and complementarity-determining region 3 size distribution analyses
Details of the complementarity-determining region 3 (CDR3) size distribution assay have been reported, including reaction conditions and primer sequences.29 After reverse transcription using an oligo d (T)16 primer, cDNA was amplified by the polymerase chain reaction (PCR) with a TCR-Vβ family-specific primer and an antisense TCR common primer. A cocktail mix of 10 × buffer (Takara Biomedicals, Shiga, Japan) containing 15 μM MgCl2, 2.5 mM dTNP each, and 20 μM each Vβ primer was mixed with the cDNA and 5 U/μL TakaRaEx Taq (Takara Biomedicals) in a final volume of 20 μL. PCR was performed in a Peltier Thermal Cycler-200 (MJ Research, Waltham, MA) under the following conditions: 15 cycles of initial touch-down by denaturation at 94°C for 1 minute, followed by annealing of primers at 60°C for 1 minute with a 0.5° gradient reduction of annealing temperature for the subsequent cycles to 53°C and extension at 72°C for 1 minute. Subsequently, 20 additional amplification cycles (denaturation at 94°C for 1 minute, followed by annealing at 53°C for 1 minute and extension at 72°C for 1 minute) were performed with a final extension of the primers at 72°C for 10 minutes. Subsequently, 1 μL amplification products was mixed with 12.5 μL deionized formamide (Sigma, St Louis, MO) and 0.5 μL size standard (Genescan-400 ROX, ABI 310; Perkin-Elmer, Norwalk, CT), heated at 90°C for 2 minutes, chilled on ice, and applied to an ABI 310 sequencer to analyze CDR3 size distribution.
Analysis of spectratype
Because of the recombination events occurring during TCR generation, the size of the amplicon varies in length. In a normal population of T cells, CDR3 length analysis produces approximately 5 to 10 identifiable peaks spaced by 3 nucleotides, with fluorescence intensity following a quasi-Gaussian distribution. Spectratypes were analyzed in 3 different ways. First, the spectratype pattern was visually assessed by 3 independent observers. A normal spectratype profile was defined as showing an approximated Gaussian bell-shaped distribution, with discrete peaks spaced by 3 nucleotides. If discrete peaks were observed but did not have the Gaussian profile, the spectratype was classified as skewed; if discrete peaks were not present, skewing was as absent. Second, spectratypes were scored mathematically, as previously described. Evidence of oligoclonal expansion or skewing was assessed by calculating the relative fluorescence intensity (RI) of each peak (RI [%] = 100 × clonal peak area/total peak area). A skewed profile was determined if a single peak was observed and the RI of the dominant peak was greater than 35% of total peak area; if 2 dominant peaks were observed and each peak's RI was greater than 25% of total peak area; or if multipeaks were observed, the dominant peaks differed from a Gaussian pattern, and the RI of the peaks exceeded 25% of total peak area. Finally, overall complexity within a Vβ subfamily was determined by counting the number of discrete peaks per Vβ subfamily, with each subfamily graded on a scale of 0 to 5.30 Spectratypes containing more than 5 peaks were given a score of 5; no spectratype signal was given a score of 0; and spectratypes with 1, 2, 3, or 4 peaks were given a score of 1, 2, 3, or 4, respectively. Overall spectratype complexity score per sample was calculated as the sum of the scores for each subfamily, with a maximum complexity score for any one patient of 110 (22 Vβ×5). Nineteen age-matched healthy controls were analyzed for comparison.
CDR3 cloning and sequencing
PCR-amplified CDR3 fragments were directly ligated into a pCR 2.1-TOPO cloning vector using the TOPO-TA cloning system (Invitrogen, Carlsbad, CA) and cloned in bacteria, according to the manufacturer's instructions. Single colonies were randomly selected, picked, and grown in Luria-Bertani broth; after purification of plasmid DNA (miniprep kit; Qiagen, Valencia, CA), the inserted DNA fragments were sequenced using an ABI PRISM Big Dye Terminator version 3.0 Cycle Sequencing Ready Reaction Kit and an ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA). Sequences of the CDR3 motif were analyzed with a DNA* software source, and the amino acid sequences were deduced. At least 20 single colonies from each cloned CDR3 amplicon were analyzed; as controls, colonies from CDR3 fragments amplified from healthy controls were analyzed to establish the normal range of diversity within a given Vβ family.
Trisomy 8 cell colony formation
To assess the effect of autologous T cells on trisomy 8 hematopoiesis, we performed short-term colony culture after 4-hour incubation of BMMCs with autologous lymphocytes obtained from PB by Ficoll-Hypaque separation. BMMCs were preincubated with a variety of lymphocyte preparations before being placed in short-term culture. Samples were centrifuged before incubation to permit close contact between effector and target cells; incubations between lymphocytes and BM were at 37°C for 4 hours. After incubation, lymphocytes were removed from all samples using immunomagnetic beads, and the BMMCs were placed in methylcellulose with growth factors as previously described.31 After 2 weeks, colonies were counted, and individual erythroid and myeloid cells were plucked, pooled, and assessed by FISH. A number of conditions were compared: (1) BMMCs alone; (2) lymphocyte-depleted BMMCs (CD3d-BMMC) (depletion performed using C3 micromagnetic beads [MACS; Miltenyi Biotech, Auburn, CA]); (3) BMMCs plus autologous lymphocytes at an effector-target ratio of 2:1; (4) BMMCs plus selected CD4 cells; and (5) BMMCs plus selected CD8 cells of the expanded Vβ subfamily at a ratio of 1:6. After these experiments, dependence on HLA class 1 recognition was evaluated by preincubation of CD3d-BMMCs with anti–class 1 mAb (1:20) for 20 minutes at 37°C before incubation with T cells. To determine whether membrane-associated Fas-L, present on Vβ CD8 cells, was responsible for cross-linking Fas on trisomy 8 progenitors, we incubated Vβ-expanded cells with a Fas antagonist (ZB4) for 20 minutes before incubation of the CD3d-BM with T cells. To determine whether contact between effector and target cells was essential for colony inhibition, we placed CD3d-BMMCs in culture separated from lymphocytes by a 0.4-μm pore filter. HLA-matched and mismatched allogeneic lymphocytes from an unrelated healthy donor, at an effector-target ratio of 1:2, were also added in a separate experiment to assess the specificity of patients' lymphocytes for trisomy 8 cells.
Results
Patients
Thirty-four patients with MDS and trisomy 8 (28 with trisomy 8 as the sole abnormality, 2 with an additional 5q–-clone, 3 with an additional monosomy 7 clone, and 1 with an additional 20q–-clone) were studied. In 4 patients with trisomy 8, MDS had evolved from AA that had previously been treated successfully with immunosuppressive drugs, and 2 of these patients were receiving CsA at the time of sampling. No patient had radiation- or chemotherapy-related secondary MDS. Five patients with MDS but without cytogenetic abnormalities also were studied. A subset of 19 patients underwent treatment for MDS with ATG alone or with ATG and CsA, as previously described.21,32
Preferential Vβ use by effector CD4+ and CD8+ T-cell subpopulations
Our previous findings of increased Fas expression on trisomy 8 cells and the consistent functional effects of Fas agonist and Fas antagonist suggested that the destruction of cells with trisomy 8 might be immune mediated. To explore this hypothesis, we studied CD28–CD4+ and CD28–CD8+ effector cells from patients with trisomy 8 or monosomy 7 for evidence of T-cell selection and expansion. During the course of an antigen-driven immune response, clonal expansion of antigen-specific T cells leads to increased representation of specific Vβ families.27 We analyzed CDR3 size distribution within the overrepresented Vβ families.
Previous work from our laboratory and by others has demonstrated that clonal T-cell expansion in patients with MDS and AA are best detected in effector lymphocyte subsets.33-35 CD28 downmodulation was used as a marker of the effector phenotype (CD4+CD28dim and CD8+CD28dim). In controls, effector cells were irregularly distributed within CD4+ or CD8+ Vβ families. For example, within CD4+CD28dim cells, Vβ1, Vβ2, Vβ5.1, and Vβ16 were underrepresented (P < .05; Student t test), whereas Vβ14 was relatively expanded. In CD8+CD28dim cells, Vβ5.2, Vβ6.7, Vβ9, Vβ13.1, Vβ13.6, and Vβ17 were all underused in the effector pool compared with total CD8+ T cells.28
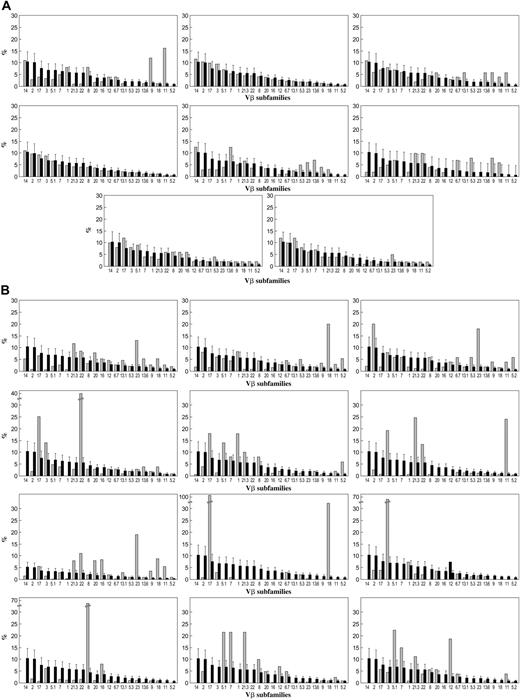
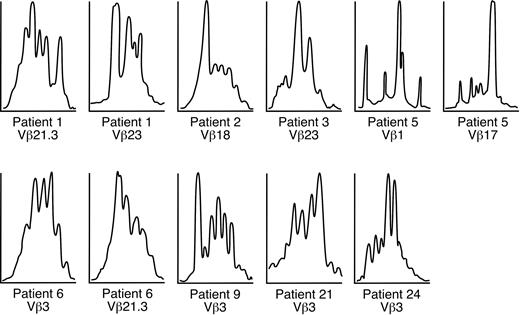
T-cell Vβ expansions were observed in all 33 patients with trisomy 8 (representative sample seen in Figure 1A) who underwent Vβ testing (including 5 patients with complex cytogenetics) but in 1 of 11 patients with monosomy 7 (as the sole cytogenetic abnormality) (representative sample seen in Figure 1B), none of the patients with 5q–, and 1 of 5 patients with MDS without cytogenetic abnormalities. Patient samples that were spectratyped showed skewing in some of the expanded Vβ subfamilies (Table 2; example seen in Figure 2). Values (mean ± 2 SD) for 19 age-matched healthy controls tested concurrently with patient samples are displayed at the bottom of Table 2; these were not significantly different from the validation cohort of 23 controls or from our previously published values using a different group of healthy donors. Expansions of Vβ subfamilies occurred within CD4 and CD8 subsets and affected a median of 3 Vβ families. In 8 patients, massive Vβ expansion within the effector T-cell subset was observed, with 1 or 2 Vβ families constituting more than half the total effector T-cell population. Vβ3 expansion appeared to be more frequent among HLA-A2 patients, but this association did not reach statistical significance. TCR-Vβ expansion was observed in CD4+ and CD8+ cells in PB but only in CD8 cells in BM (n = 5). CD8 cell expansion in BM was limited to 1 or 2 Vβ subfamilies, which almost always were also overrepresented in the PB (data not shown); 1 patient had expanded BM Vβ not present in blood. Based on our previously published experience in AA10,28,36 and our current flow cytometry results, all Vβ families overrepresented by more than 2 SD from the mean of control in the CD28dim compartment were considered likely to represent oligoclonal or monoclonal expansion of disease-specific CD4+ or CD8+ T cells. CDR3 spectratyping in the expanded Vβ subfamilies was performed in 9 patients with trisomy 8, and skewing of the repertoire was seen in many of the expanded Vβ subfamilies (Figure 2).
CDR3 cloning and sequencing of Vβ TCR of expanded T-cell populations
In 2 patients tested, CDR3 cloning showed oligoclonal rather than polyclonal T-cell expansion, with predominance of a few sequences within the repertoire. In both cases, within the expanded Vβ3 CD8 T cells, 2 predominant clonotypes were demonstrated (CASSDFRGAGYEQYFGPGTRLTVT and CASSGGLEQYFGPGTRLTVT). Both clonotypes had a JB2.7 joining segment.
Effect of preincubation of BMMCs with T cells on trisomy 8 and normal cell colony formation
The ability of autologous lymphocytes to affect the proliferation of hematopoietic progenitors of trisomy 8 and of normal karyotype was studied in 20 patients with trisomy 8, 4 patients with monosomy 7, 2 patients with 5q–, and 4 healthy donors. On average, incubation with lymphocytes before short-term culture significantly decreased the number of cells with trisomy 8 or normal karyotype, with preferential and sometimes complete loss of trisomy 8 cells (Figure 3A). Although colony size appeared to be relatively homogeneous, we looked at individual colonies (which were plucked and in which FISH was performed) to determine whether the numbers of trisomy colonies were preferentially decreased. When 1 patient with trisomy 8 was studied in this way, there was a 35% reduction in the number of trisomy 8 colonies (n = 20) and a 42% reduction in trisomy 8 cells in the lymphocyte-treated fraction.
TCR repertoire in trisomy 8 and monosomy 7. Thirty-three patients (light bars) with trisomy 8 (including 5 with complex cytogenetics), 11 patients with monosomy 7, and 19 age-matched healthy donors (dark bars) were studied. PBMC preparations were stained with CD28 FITC- and PE-conjugated mAbs directed against individual TCR-Vβ subfamilies and subjected to flow cytometry. Patients were compared with 19 age-matched controls. (A) Vβ subfamily distributions for a selection of patients with monosomy 7, only one of whom showed evidence of T-cell expansion. Values for each patient sample (light bars) are superimposed on mean of normal cohort (dark bars). (B) Examples of the Vβ subfamily distributions of patients with trisomy 8, all of whom showed expansion of Vβ subfamilies.
TCR repertoire in trisomy 8 and monosomy 7. Thirty-three patients (light bars) with trisomy 8 (including 5 with complex cytogenetics), 11 patients with monosomy 7, and 19 age-matched healthy donors (dark bars) were studied. PBMC preparations were stained with CD28 FITC- and PE-conjugated mAbs directed against individual TCR-Vβ subfamilies and subjected to flow cytometry. Patients were compared with 19 age-matched controls. (A) Vβ subfamily distributions for a selection of patients with monosomy 7, only one of whom showed evidence of T-cell expansion. Values for each patient sample (light bars) are superimposed on mean of normal cohort (dark bars). (B) Examples of the Vβ subfamily distributions of patients with trisomy 8, all of whom showed expansion of Vβ subfamilies.
Spectratyping performed on trisomy 8 samples. Spectratyping demonstrated skewing of the CDR3 distribution in most expanded CD8+ Vβ subfamilies shown in Table 2.
Spectratyping performed on trisomy 8 samples. Spectratyping demonstrated skewing of the CDR3 distribution in most expanded CD8+ Vβ subfamilies shown in Table 2.
When unmanipulated BMMCs from patients with trisomy 8 were divided into 2 aliquots and 1 was depleted of CD3 cells using magnetic beads, there was a significant increase in the absolute number and in the percentage of trisomy 8 colonies as a result of lymphocyte removal (P < .05; Figure 3B). No specific activity of lymphocytes against cells bearing other cytogenetic abnormalities was seen in 4 patients with monosomy 7, 2 patients with 5q–, or 4 healthy controls subjected to the same in vitro manipulations.
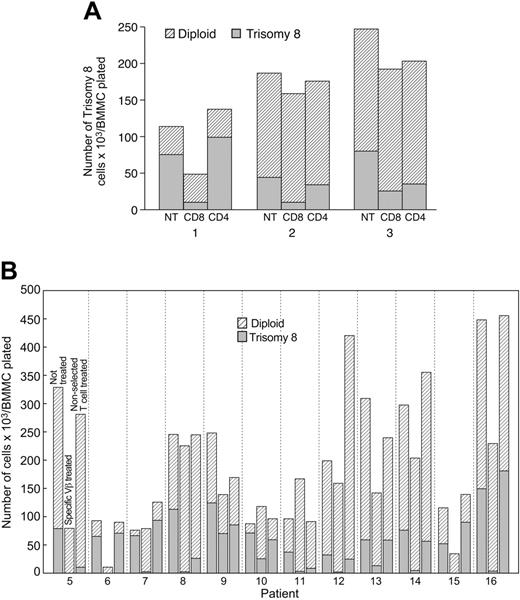
Effect of expanded trisomy 8 patient–derived CD8 T cells compared with normal HLA-matched allogeneic T cells on trisomy 8 hematopoiesis. We next characterized the contributions of CD4 and CD8 cells to the inhibition of BMMCs in patients with trisomy 8. In the 4 patients examined, CD8 cells inhibited colony formation but CD4 cells showed little or no influence on cell growth (Figure 4A). CD8 cells from specific Vβ subfamilies, identified as expanded by flow cytometry, were isolated by cell sorting and incubated with BMMCs for 4 hours, followed by cell culture. These T cells produced significant suppression of erythroid and myeloid trisomy 8 cells. In contrast, little or no inhibition occurred after coculture with the control population of nonexpanded Vβ subfamily lymphocytes, even when they were present at 3-fold greater numbers compared with the specific Vβ expanded cells (Figure 4B-C). Suppression by Vβ-expanded T cells appeared to be partially dependent on Fas because the addition of a Fas antagonist to the mixture trisomy 8 cells partially abrogated the inhibition. When a Fas antagonist (ZB4 mAb) was added to 3 patients' unmanipulated BMMCs and the samples were placed in long-term culture, trisomy 8 colonies proliferated at a faster rate than did normal cells (Figure 3C). Fas antagonist had no effect on CD3d-BMMCs, nor did it have an effect on unmanipulated BMMCs obtained from 3 patients receiving CsA (data not shown). Soluble factors released from CD8 cells not in direct contact with BMMC because of separation by a filter did not influence cell growth in trisomy 8. Blocking experiments performed by preincubating CD3d-BM with anti-HLA class 1 mAb were able to ameliorate the effect of Vβ cells on trisomy 8 cells, implicating a role for MHC class 1. The inhibitory effect appeared to be specific to the patients' CD8 cells because allogeneic CD8 cells matched at class 1 and added in excess (2:1 ratio of lymphocytes to BMMCs) did not produce any change in the proportion of trisomy 8 cells or inhibit normal cell growth (Table 3). The inhibitory effect of unmatched allogeneic lymphocytes also did not preferentially affect trisomy 8 cells.
Autologous lymphocytes decrease the proportion of BM trisomy B cells while Fas antagonist increases their growth. Fas agonist increases the proportion of trisomy 8 cells in short-term culture. BMMCs were obtained from 10 patients with trisomy 8, 4 patients with monosomy 7, and 4 healthy controls. Samples were divided into 2 aliquots, one of which was incubated with autologous PBLs for 4 hours, as described in “Patients, materials, and methods.” Samples were subsequently depleted of lymphocytes and placed in semisolid media with growth factors for short-term culture. Colonies were counted, and FISH was performed. The number of trisomy 8 cells and the percentage of trisomy 8 cells were decreased by lymphocyte cocultivation, with little effect seen in diploid cells (A). When one aliquot was depleted of T cells using CD3-specific magnetic beads before short-term culture and compared with the non–T cell–depleted sample, T-cell cultures consistently showed increased trisomy 8 progenitor–derived cell growth (B, left). T-cell depletion had little effect on the growth of cytogenetically normal cells (B, right). When samples of BM were plated in long-term culture with autologous lymphocytes, with and without Fas antagonist (ZB4 mAb), trisomy 8 cells increased compared to karyotypically normal cells (C).
Autologous lymphocytes decrease the proportion of BM trisomy B cells while Fas antagonist increases their growth. Fas agonist increases the proportion of trisomy 8 cells in short-term culture. BMMCs were obtained from 10 patients with trisomy 8, 4 patients with monosomy 7, and 4 healthy controls. Samples were divided into 2 aliquots, one of which was incubated with autologous PBLs for 4 hours, as described in “Patients, materials, and methods.” Samples were subsequently depleted of lymphocytes and placed in semisolid media with growth factors for short-term culture. Colonies were counted, and FISH was performed. The number of trisomy 8 cells and the percentage of trisomy 8 cells were decreased by lymphocyte cocultivation, with little effect seen in diploid cells (A). When one aliquot was depleted of T cells using CD3-specific magnetic beads before short-term culture and compared with the non–T cell–depleted sample, T-cell cultures consistently showed increased trisomy 8 progenitor–derived cell growth (B, left). T-cell depletion had little effect on the growth of cytogenetically normal cells (B, right). When samples of BM were plated in long-term culture with autologous lymphocytes, with and without Fas antagonist (ZB4 mAb), trisomy 8 cells increased compared to karyotypically normal cells (C).
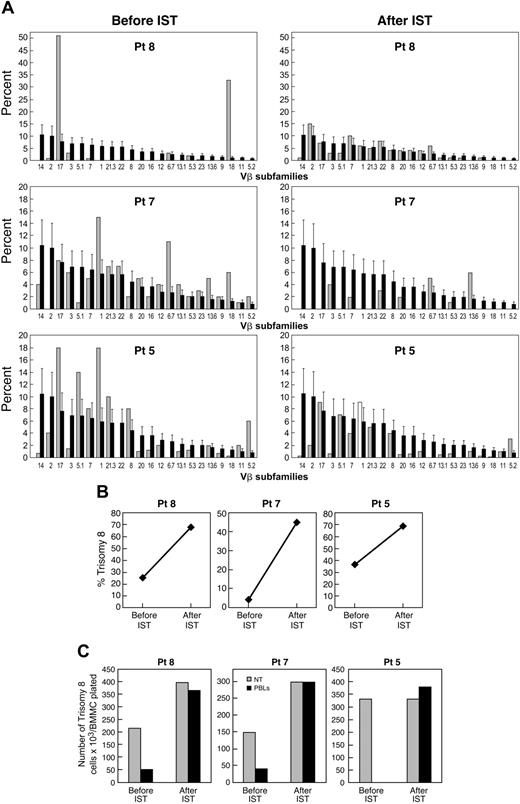
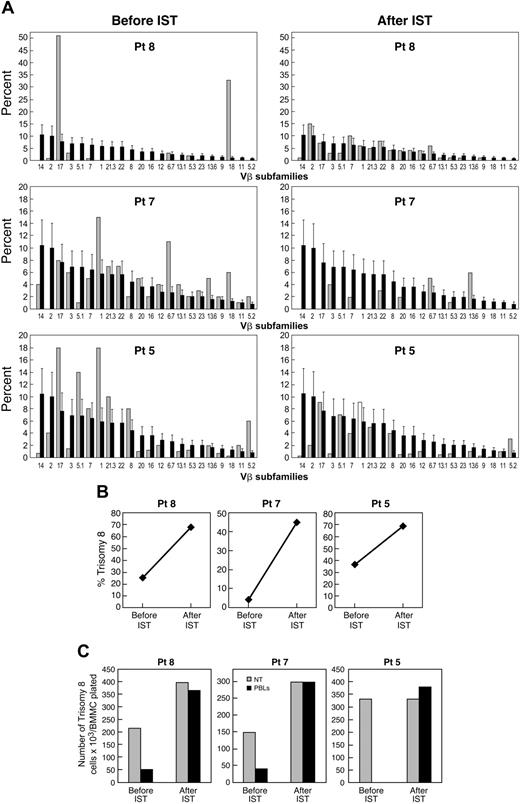
Effect of immunosuppressive therapy on the number of trisomy 8 cells. Eighteen MDS patients with trisomy 8 as the sole cytogenetic abnormality underwent treatment with an ATG regimen: 10 received ATG and 8 received ATG and CsA. Eight of 13 (61%) patients with de novo MDS responded to immunosuppression, and all 5 patients with a history of AA responded to immunosuppression. BM cytogenetics and FISH were performed before and 6 months after therapy to assess the size of the trisomy 8 clone (Table 4). After immunosuppressive therapy (IST), we found an increase in the number of trisomy 8 cells in all 21 patients. The clinical response to immunosuppression was associated with a loss of expansion of T cells from a specific Vβ TCR subfamily on flow cytometry, such that the pattern resembled that in healthy controls (example seen in Figure 5). All responding patients have stable disease or are in partial or complete hematologic remission; leukemia has not developed in any of the patients with trisomy 8 as the sole karyotypic abnormality. Deaths from complications related to pancytopenia occurred in 6 of 21 treated patients. Among 3 patients in whom stored samples were available before the development of trisomy 8, none retrospectively showed evidence of Vβ expansion by flow cytometry (Table 2).
None of the patients with complex cytogenetic abnormalities responded to IST; 1 patient with monosomy 7 improved, as evidenced by significant decreases in the proportion of his cytogenetically abnormal clone. Although expansion of the Vβ subfamilies was not observed in any of the nonresponding patients with monosomy 7, the sole responding patient experienced T-cell clonal expansion that regressed after treatment with ATG. Patients with complex cytogenetic abnormalities had increased proportions of trisomy 8 and little change in the other cytogenetic clones. In none of these patients was there a demonstration of expansion of any Vβ subfamily.
Preferential inhibition of trisomy 8 cell colony growth related to suppression of growth by Vβ subfamily expanded CD8+ cells. Autologous column-purified CD4+ or CD8+ cells were incubated with 3 BMMC aliquots for 4 hours and placed in semisolid media for short-term culture in an additional 3 patients. Suppression of trisomy 8 progenitor–derived cell growth is seen for the BM cells previously incubated with CD8+ cells only (A). Similar experiments performed with Vβ-selected CD8+ cells (B) from an additional 7 patients showed preferential inhibition by selected, but not unselected, cells of trisomy 8 but not karyotypically normal cells. Erythroid and myeloid cells were affected.
Preferential inhibition of trisomy 8 cell colony growth related to suppression of growth by Vβ subfamily expanded CD8+ cells. Autologous column-purified CD4+ or CD8+ cells were incubated with 3 BMMC aliquots for 4 hours and placed in semisolid media for short-term culture in an additional 3 patients. Suppression of trisomy 8 progenitor–derived cell growth is seen for the BM cells previously incubated with CD8+ cells only (A). Similar experiments performed with Vβ-selected CD8+ cells (B) from an additional 7 patients showed preferential inhibition by selected, but not unselected, cells of trisomy 8 but not karyotypically normal cells. Erythroid and myeloid cells were affected.
Discussion
IST improves bone marrow function in selected patients with MDS, though it does not generally eliminate the cytogenetically abnormal clone.37 Patients who are younger, are HLA DR15 positive, and have more recent onset of disease have a high response rate.22 This high-responder group includes most patients with trisomy 8 (E.M.S., unpublished data, 2005).
In our experiments, clonally expanded CD8 cells showed apparent cytotoxicity specifically for autologous trisomy 8 hematopoietic progenitors. MDS with 5q– and monosomy 7 have neither skewing of the T-cell repertoire nor T-cell effector cytotoxicity for karyotypically abnormal progenitors. Intrinsic sensitivity of trisomy 8 cells to lymphocytes of their cytokine products, while not rigorously excluded, seems unlikely because of the following: (1) alloactivated T lymphocytes do not produce the same effect as autologous T cells; (2) adding interferon-γ (IFN-γ) to cell culture did not decrease the proportion of trisomy 8 cells (data not shown); (3) Fas antagonist did not preferentially conserve trisomy 8 cells in CD3-depleted marrow; (4) lymphocytes from patients on CsA therapy did not diminish the proportion of trisomy 8 cells; and (5) there was a reciprocal relationship between trisomy 8 clone expansion after IST and trisomy 8–specific T-cell reduction.
Recently, CD8+ T-cell responses against a number of antigens presented by myeloid cells have been discovered in patients with myeloid malignancies, including overexpressed self-proteins such as proteinase-3 (PR-1)38 and Wilms tumor (WT-1)39 or neoantigens created by chromosomal translocations such as breakpoint-cluster region/Abelson leukemia (BCR-ABL).40 Central memory and effector memory T cells recognizing WT-1, PR-1, and BCR-ABL were measured in patients with chronic myelogenous leukemia (CML) in higher frequencies than they were encountered in healthy persons.41 The presence of circulating, high-avidity, PR1-specific T cells was correlated with response to IFN-γ in patients with CML, whereas reduced numbers of these cells was associated with disease progression.42 Our results in patients with trisomy 8 MDS are consistent with an immune response against the trisomy 8 clone.
In the hypothesized immune mechanism of MDS, CD8 T cells of a specific Vβ subfamily might expand in response to a neoantigen, to a quantitatively up-regulated antigen, or to an aberrantly expressed normal protein, presented by the trisomy 8 clone through MHC class 1 molecules.43 Patients with MDS overexpress WT-1 and PR-1 and can have circulating PR-1– and WT-1–specific T cells (personal communication, K. Rezvani, April 2005). However, the antigens recognized by the clonally expanded T cells in trisomy 8 have not yet been identified. Immortalized T-cell clones generated from the expanded Vβ subfamilies44 might be used to determine peptides involved in MDS by screening them against a universal combinatorial peptide library. Alternatively, overexpressed proteins could be deduced from microarray differences between cytogenetically abnormal and normal CD34 cells.45
An immunologic description of the pathophysiology of trisomy 8 MDS must also explain the myelosuppression encountered in this condition. During the immune response to trisomy 8 cells, normal hematopoietic cells could be damaged as “bystanders,” leading to generalized hematopoietic cell destruction and pancytopenia. Such a bystander effect has been described in a mouse model of immune-mediated marrow failure46 : donor lymphocytes, activated in response to H2 differences in an F1 hybrid recipient, were cytotoxic to hematopoietic stem cells genetically matched at H2 to the effector cells. Bystander killing may result from cytokines released by activated T cells47 or by the cross-recognition of targets through molecular mimicry or epitope spreading.48-50
Cells expressing an autoantigen elicit T-cell responses through TCR engagement with self-peptides bound to HLA molecules.51 An effective CTL response requires T-cell engagement with the costimulatory molecules CD86 and CD28.52-54 Colony inhibition by CTL in trisomy 8 MDS (which express CD86 on CD34 cells; data not shown) indicates that progenitors from these patients are competent antigen-presenting cells and susceptible CTL targets.
The factors leading to the persistence of clones of cytogenetically abnormal hematopoietic cells in patients with BM failure disorders are unclear. Clinically detectable MDS may derive from a preclinical stage, when an abnormal stem cell with genomic instability develops successive cytogenetic abnormalities, altering the transcription of genes governing growth and resistance to apoptosis (genes such as p53, FLT3,or RAS).55-57 The occurrence in the marrow of small numbers of trisomy 8 cells, identified by FISH, long before cytogenetic conversion suggests that trisomy 8 aneuploidy is an early event in MDS. These small clonal populations could nevertheless induce a CTL response to trisomy 8 cells, resulting in the destruction of normal cells as bystanders and causing cytopenia. The persistence of trisomy 8 cells, despite immune attack, would be attributed to their increased proliferation relative to normal cells, failure to fully undergo programmed cell death,58 or immune escape.
The absence of expanded Vβ T cells that suppress trisomy 8 colony formation after IST in patients with MDS was not associated with disease progression or with the development of leukemia. In our MDS patients, trisomy 8 appeared to be a relatively benign chromosomal abnormality (when it was the sole cytogenetic finding; patients with complex cytogenetic abnormalities did poorly59 ), compatible with normal hematopoiesis. In our experience, patients responding to IST remain clinically stable, despite increases in their trisomy 8 populations,16 and no patients with trisomy 8 have developed leukemia. Nevertheless, underlying clonal instability in trisomy 8 MDS could increase the risk for further mutations that might lead to disease progression and malignancy.
Our findings may have clinical implications. Flow cytometry with quantitation of individual Vβ subfamilies may provide a useful means for monitoring the immune response in patients with trisomy 8, because loss of Vβ expansion appears to correlate with clinical response. These results, our microarray data,60 and our clinical data on AA patients and 133 patients with de novo MDS treated and observed for 10 years59,61 indicate that MDS with different chromosomal abnormalities are distinct entities requiring disease-specific treatments. In patients with trisomy 8, IST may be more effective in improving hematopoiesis than is similar therapy in patients with MDS and other cytogenetic abnormalities.
Clinical development or disappearance of trisomy 8 associated with presence or absence of Vβ expansion. (A) Flow cytometric examination of Vβ subfamilies from patients 5, 7, and 8 before and after IST. Values for each patient (light bars) are superimposed on mean values of normal cohort (dark bars). (B) FISH data showing percentage of trisomy 8 before and after IST. (C) Effect of lymphocytes on trisomy 8 cell growth before and after IST. Paired lymphocytes and BMMCs obtained before and after IST were incubated in close contact with effector cells (with effector-target ratios of 2:1) for 2 hours before placement in short-term culture for 14 days. When FISH was performed, the number of trisomy 8 cells was decreased by cocultivation with the lymphocytes obtained before IST only.
Clinical development or disappearance of trisomy 8 associated with presence or absence of Vβ expansion. (A) Flow cytometric examination of Vβ subfamilies from patients 5, 7, and 8 before and after IST. Values for each patient (light bars) are superimposed on mean values of normal cohort (dark bars). (B) FISH data showing percentage of trisomy 8 before and after IST. (C) Effect of lymphocytes on trisomy 8 cell growth before and after IST. Paired lymphocytes and BMMCs obtained before and after IST were incubated in close contact with effector cells (with effector-target ratios of 2:1) for 2 hours before placement in short-term culture for 14 days. When FISH was performed, the number of trisomy 8 cells was decreased by cocultivation with the lymphocytes obtained before IST only.
Prepublished online as Blood First Edition Paper, April 12, 2005; DOI 10.1182/blood-2004-05-2017.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.