Abstract
A major barrier in hematopoietic gene function studies is posed by the laborious and time-consuming generation of knockout mice with an appropriate genetic background. Here we present a novel lentivirus-based strategy for the in situ generation of hematopoietic knockdowns. A short hairpin RNA (shRNA) was designed targeting murine CC-chemokine receptor 2 (CCR2), which was able to specifically blunt CCR2 expression at the mRNA, protein, and functional levels in vitro. Reconstitution of irradiated recipient mice with autologous bone marrow that had been ex vivo transduced with shRNA lentivirus led to persistent down-regulation of CCR2 expression, which translated into a 70% reduction in CCR2-dependent recruitment of macrophages to an inflamed peritoneal cavity without noticeable side effects on related chemokine receptors or general inflammation status. These findings clearly demonstrate the potential of shRNA lentivirus–infected bone marrow transplantation as a rapid and effective method to generate hematopoietic knockdowns for leukocyte gene function studies.
Introduction
Bone marrow transplantation studies have greatly facilitated the identification of macrophage genes that play a significant role in inflammatory disorders1 ; for example, atherogenesis.2-4 However, this experimental setup essentially depends on the availability of transgenic mice or knockouts of the gene of interest for providing bone marrow. Thus, progress is considerably hampered by the laborious construction of, in particular, knockout mice and the subsequent backcrossing to an appropriate disease background for analysis of gene function. Moreover, the development of knockouts is even more difficult when deletion results in embryonic lethality as is the case for tissue factor (TF)5 or vascular endothelial growth factor (VEGF).6
Efficient and stable gene transfer to hematopoietic stem cells has been a major challenge in gene therapy. For several years, a range of vectors of onco-retroviral origin, which integrate into the host-cell genome, has been developed.7 A major drawback of these retroviral systems, mostly based on Moloney leukemia viruses, is their inability to integrate in the genome of nonmitotic cells. Since hematopoietic stem cells are rather quiescent, their retroviral transduction generally is inefficient. While transduction efficiency can be improved by cytokine supplementation,8 by reducing cyclin-dependent kinase inhibitor levels9 or by fibronectin coating to induce stem-cell proliferation,10,11 these manipulations may likely affect stem-cell differentiation. Unlike onco-retroviral vectors, lentivirus offers the advantage of stably transducing not only mitogenic but also quiescent cells.12-15 Indeed, efficient lentiviral transduction of bone marrow stem cells using lentiviral vectors has been reported.16 Such transduction is not only required to obtain required gene expression levels in vivo, but also obviates the use of time-consuming preselection strategies.
We argued that the high levels of lentiviral gene transfer to bone marrow may be exploited to knockdown hematopoietic genes in vivo via RNA interference (RNAi), defined as sequence-specific posttranscriptional gene silencing.17-24 The advent of RNAi or small interfering RNA (siRNA) has instigated a true revolution in functional genomics and allowed the in situ knockdown of genes. Synthetic short double-strand (ds) DNA sequences of 21 to 25 nucleotides in length25 were reported to be efficient in inducing selective mRNA degradation and to suppress gene expression.26,27 Recently, Brummelkamp et al28 have generated a mammalian expression vector, encoding a short hairpin dsRNA transcript under the control of the RNA polymerase III–dependent H1 gene promoter for sustained silencing of a transgene. At present, several groups reported successful lentiviral delivery of short hairpin RNA (shRNA) to eukaryotic cells in vitro. Tiscornia et al29 showed decreased green fluorescent protein (GFP) levels after transduction with a lentiviral shRNA construct. Likewise, successful lentiviral delivery of shRNAs to human cells30,31 and even to human hematopoietic stem cells was reported.32
In this study we have addressed the potential of lentivirus-aided gene silencing of genes in bone marrow for the in situ generation of chimeras with blunted gene expression in the hematopoietic-cell lineage in vivo. As a model gene we chose murine CC-chemokine receptor 2 (Ccr2), which was previously shown to play a central role in the recruitment of monocytes to sites of inflammation and during atherogenesis.33-35 An shRNA construct was developed against CCR2 (shCCR2), which proved highly efficient in down-regulating CCR2 expression both at an mRNA as well as at a protein level in CCR2-transfected HEK 293 cells. Furthermore, we have efficiently transduced bone marrow cells and reconstituted irradiated C57Bl/6 mice with shCCR2-transduced bone marrow cells, which considerably attenuated macrophage recruitment to the site of inflammation to a level comparable with that of mice reconstituted with Ccr2–/– bone marrow.
Materials and methods
Cloning and expression of murine CCR2-GFP in HEK 293 cells
Total RNA was extracted from the murine monocytic cell line WEH1, reverse transcribed using M-MuLV reverse transcriptase (RevertAid; MBI Fermentas, Leon-Roth, Germany) and the cDNAs were amplified by polymerase chain reaction (PCR) to obtain full-length cDNA for murine CCR2 using the following primers: forward: 5′-AGCCTCGAGATGGAAGACAATAATATGTT-3′ and reverse: 5′-ATAAAGCTTTTACAACCCAACCGAGACCTCT-3′. Primers contained extra XhoI and HindIII restriction sites to facilitate cloning (underlined). Purified PCR products were subsequently cloned into pCR2.1 (Invitrogen, Breda, The Netherlands) and subcloned into pEGFPN-1 (Clontech, Palo Alto, CA). Murine CCR2 and GFP were fused at the GFP N-terminus via removal of the stop codon, affording the pEGFPN-1/CCR2 fusion vector. A CCR2 expression vector was generated by cloning full-length Ccr2 into pcDNA3.1(-) (Invitrogen), designated as pcDNA3.1(-)/CCR2. All constructs were sequenced to confirm the identity of murine Ccr2.
Design and cloning of shRNA directed against murine CCR2
A pair of 64-nucleotide oligonucleotides encoding a 19-nucleotide CCR2 shRNA (5′-GATCCCCCTGTGTGATTGACAAGCACTTCAAGAGAGTGCTTGTCAATCACACAGTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAACTGTGTGATTGACAAGCACTCTCTTGAAGTGCTTGTCAATCACACAGGGG-3′) were designed, which contained extra BglII and HindIII restriction sites to facilitate cloning. The position of the core 19–nucleotide sequence (underlined) targeted nucleotides 867 to 885 of the murine Ccr2. Blast search was performed using the National Center for Biotechnology Information (NCBI) Expressed Sequence Tags (EST) database to ensure that the shRNA construct was targeting only murine CCR2. The 64-nucleotide oligonucleotides were annealed and cloned into the BglII and HindIII sites of the pSUPER vector (kindly provided by Dr Agami, The Netherlands Cancer Institute, Amsterdam, The Netherlands) and the resulting vector, designated pSUPER-H1.shCCR2, was subsequently sequenced to confirm identity.
Silencing effect of pSUPER-H1.shCCR2
HEK 293 cells, grown at 37°C and 5% CO2 in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS), 100 μg/mL streptomycin, 100 U/mL penicillin, and 2 mM l-glutamine (all from Cambrex Bio Science, Verviers, Belgium), were transiently cotransfected with a constant amount of pEGFPN-1/CCR2 (0.5 μg) and 0.1 μg, 0.5 μg, and 1.5 μg pSUPER-H1.shCCR2, supplemented to 1.5 μg with pSUPER-H1. Empty vector using Lipofectamine 2000 (Invitrogen). As controls, the effects of an shRNA designed against luciferase (pSUPER-H1.shLuc, CGTACGCGGAATACTTCGA36 ) and a nonsense sequence (pSUPER-H1.shInert, AGGCTGCTTGCACGATCTA) cloned into pSUPER on CCR2 expression levels were determined. After 48 hours, total RNA was extracted from the HEK 293 cells and reverse transcribed as described under “Cloning and expression of murine CCR2-GFP in HEK 293 cells.” Quantitative analysis of CCR2 expression was performed using an ABI PRISM 7700 Taqman apparatus (Applied Biosystems, Foster City, CA) as described previously37 (Table 1). The amplicon encompassed the shCCR2 core sequence (nucleotides 851-991), enabling the analysis of sequence-specific degradation of murine CCR2 mRNA and excluding artifacts due to the presence of shRNA. Human hypoxanthine phosphoribosyltransferase (hHPRT) was used as a standard housekeeping gene and non–reverse transcribed RNA samples were used as controls to determine genomic DNA contamination. Murine CCR2 mRNA expression was calculated relative to that of hHPRT on the basis of ΔCt values.
After transfection with pEGFPN-1/CCR2 and pSUPER-H1.Empty, pSUPER-H1.shLuc, pSUPER-H1.shInert, or different amounts of pSUPER-H1.shCCR2, the cells were harvested, resuspended in phosphate-buffered saline (PBS), and analyzed using fluorescence activated cell sorting (FACS; FACScalibur, BD Biosciences) for GFP expression. For Western blotting, transfected cells were washed gently with PBS, scraped off, and subsequently lysed in sodium dodecyl sulfate (SDS) sample buffer supplemented with a protease inhibitor cocktail (Roche, Mannheim, Germany). Equal amounts of protein were applied to a polyacrylamide gel (12.5%) and transferred to nitrocellulose. Visualization of GFP with a mouse anti-GFP monoclonal antibody and a peroxidase-conjugated rat anti–mouse IgG as a secondary antibody (1:2000 and 1:1000 dilution, respectively; Santa Cruz Biotechnology, Santa Cruz, CA) was performed via the enhanced chemiluminescence (ECL) technique (Amersham Pharmacia Biotechnology, Piscataway, NJ). Blots were simultaneously probed with a mouse anti–β-tubulin antibody (Sigma, Zwijndrecht, The Netherlands) to verify equal loading of the samples.
Functional loss of CCR2 by shCCR2
The effect of the shRNA vector on CCR2 function was determined by analyzing calcium influx in response to the endogenous ligand for murine CCR2, CC-chemokine ligand 2 (CCL2), or JE. HEK 293 cells, expressing murine CCR2, were transfected with 0.1 μg pSUPER-H1.shCCR2 or with pSUPER-H1.Empty control vector. After 48 hours, cells were seeded onto poly-lysine–coated glass coverslips to measure transient intracellular calcium influx. Cells were loaded with 20 μM Fura-2-AM (Invitrogen) for 60 minutes and after washing with Hank balanced salt solution, the coverslips were mounted in an incubation chamber. JE (Santa Cruz Biotechnology) was added to a final concentration of 1 nM and real-time digital fluorescence imaging was performed using a fluorescence microscope (excitation: 340 nm and 380 nm; emission: 495 nm) for 20 minutes. The mean fluorescence intensity was determined to calculate intracellular calcium concentrations.
Lentivirus vector construction and production
The lentiviral expression vectors pRRl-cPPt-H1.PreSIN (H1.Empty) and pRRl-cPPt-H1.shCCR2-PreSIN (H1.shCCR2) were constructed by removing the cytomegalovirus (CMV) promotor from the expression vector pRRl-cPPt-CMV-PreSIN15 using ClaI and PstI digestion, followed by insertion of the H1 promoter or the complete H1.shCCR2 construct. Virus was produced as described38 using transient calcium phosphate cotransfection of 293T cells with the H1.Empty or H1.shCCR2 vector together with pMDL/RRE, pRSV-REV, and pVSV-G. Viral supernatants were passed through a 0.45 μm filter and concentrated by ultracentrifugation (41 000 g, 2 hours, 4°C). Viral titers were determined essentially as described by Sastry et al.39 In short, HEK 293 cells were transduced with serially diluted viral supernatant, and 48 hours after transduction total genomic DNA was isolated from these cells and the number of vector DNA copies was determined using PCR analysis with pRRl-cPPt-H1.PreSIN vector as the calibration standard (forward primer: GTGCAGCAGCAGAACAATTTG; reverse primer: CCCCAGACTGTGAGTTGCAA).
Animals
All animal work was approved by the regulatory authority of Leiden and performed in compliance with the Dutch government guidelines. C57Bl/6 mice were obtained from Charles River (Maastricht, the Netherlands). Ccr2–/– mice were kindly provided by Dr Maeda (University of North Carolina, Chapel Hill). Mice that underwent bone marrow transplantation were housed in sterile filter-top cages and fed a sterile regular chow diet (RM3; Special Diet Services, Witham, United Kingdom). Drinking water was infused with antibiotics (83 mg/L ciprofloxacin and 67 mg/L polymyxin B sulfate) and 6.5 g/L sugar and was provided ad libitum.
In vitro transduction protocol for bone marrow cells
Bone marrow cell suspensions were isolated from C57Bl/6 mice by flushing the femurs and tibias with PBS. Single-cell suspensions were prepared by passing the cells through a cell strainer and 105 cells/well were plated in a 12-well plate. Viral transductions were performed by incubating the bone marrow cells with different titers of pRRl-cPPt-PGK-GFP-PreSIN in complete DMEM supplemented with 10 μg/mL diethylaminoethyl (DEAE)–dextran at 37°C. After 24 hours, cells were washed with PBS and fixed in 4% paraformaldehyde for FACS analysis to determine GFP expression levels. Viral copy number inserted in the bone marrow cells after 24 hours was determined by isolation of total genomic DNA (from 106 bone marrow cells) and PCR analysis.
Irradiation and bone marrow transplantation
To induce bone marrow aplasia, female C57Bl/6 mice (12 to 14 weeks of age) were exposed to a single dose of 9 Gy (0.19 Gy/min, 200 kV, 4 mA) total body irradiation, using an Andrex Smart 225 Röntgen source (YXLON International, Copenhagen, Denmark) with a 6-mm aluminium filter, one day before transplantation. Bone marrow cell suspensions were isolated from C57Bl/6 mice as described in the previous section. Subsequently, bone marrow cells (1.0 × 107) were transduced with either H1.Empty or H1.shCCR2 lentivirus in the presence of 10 μg/mL DEAE-dextran (multiplicity of infection [moi] = 15). After 24 hours, the cells were injected into the tail vein of the irradiated recipients (n = 4 per group). As a positive control, irradiated C57Bl/6 mice (n = 5) received 1.0 × 107 bone marrow cells isolated from male Ccr2–/– mice. In a similar set-up, C57Bl/6 mice (n = 2) were injected with bone marrow cells transduced with GFP lentivirus. In these mice, GFP expression levels in blood cells were determined 6 weeks after transplantation by preparing cytospins of white blood cells (9 g, 5 minutes) after lysis of the erythrocytes. The images were taken with a Leica DM-RE microscope and LeicaQwin software (Leica Imaging Systems).
Macrophage recruitment
Six weeks after bone marrow transplantation, the mice receiving H1.Empty or H1.shCCR2 lentivirus–transduced bone marrow and the mice reconstituted with Ccr2–/– bone marrow were injected intraperitoneally with sterile 3% (wt/vol) Brewer thioglycollate solution (Difco, Detroit, MI). Peritoneal macrophages were isolated 5 days later by lavage of the peritoneal cavity with PBS (10 mL) and counted after lysis of erythrocytes. Also, blood smears were prepared, stained with Wright Giemsa (Sigma), and scored manually. Total RNA was isolated from peritoneal macrophages and gene expression was determined by reverse transcription (RT)–PCR compared with murine HPRT (Table 1). Also, bone marrow was isolated from each individual mouse and, after lysis of the bone marrow cells, genotyped for the presence of the Ccr2 gene in a single reaction (primers: CCR2 forward, 5′-GATGATGGTGAGCCTTGTCA-3′; CCR2 reverse, 5′-CACAGCATGAACAATAGCCA-3′; and a pgk-neo specific primer, Neo 5′-TTAAGGGCC-AGCTCATTCCT-3′). In wild-type mice, a 360 base pair sequence is amplified corresponding with intact Ccr2, whereas in the Ccr2–/– mice a 290 base pair sequence is generated. Also, viral copy number in the bone marrow was determined. In addition, the percentage repopulation of male bone marrow in the female recipients at that time point was determined by means of Taqman PCR analysis on the Sry gene, located on the Y-chromosome. To quantify the percentage of male bone marrow, a calibration curve was used containing increasing amounts of male bone marrow supplemented with female bone marrow to standardize the amount of genomic DNA per well.
Statistical analysis
Data are expressed as mean plus or minus the standard error of the mean (SEM). A 2-tailed Student t test was used to compare individual groups. To determine the significance of the relative mRNA expression levels, statistical analysis was performed on ΔCt values.
Results
Silencing CCR2 expression by shRNA vectors
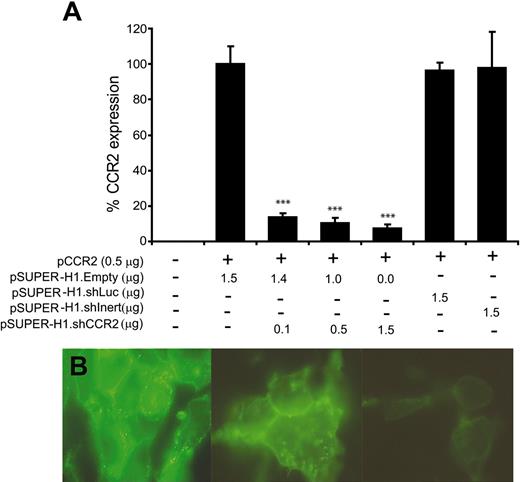
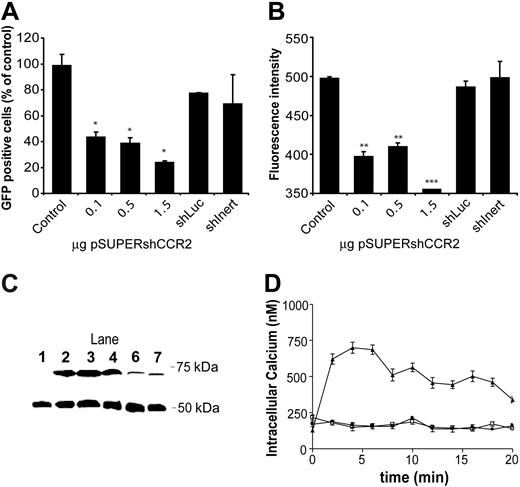
To determine the silencing capacity of an shRNA against CCR2 (pSUPER-H1.shCCR2) on murine CCR2 expression, HEK 293 cells were cotransfected with pEGFPN-1/CCR2 fusion vector and increasing amounts of pSUPER-H1.shCCR2. As controls, the effects of an shRNA sequence against luciferase and a nonsense shRNA sequence on CCR2 expression were also determined. CCR2 mRNA levels were already found to be significantly reduced after cotransfection with only 0.1 μg of pSUPER-H1.shCCR2 (Figure 1A, P < .001 compared with the shRNA controls at all concentrations) with a maximum reduction of 94% at 1.5 μg pSUPER-H1.shCCR2, whereas the shRNA controls and pSUPER-H1.Empty had no effect. In agreement, expression analysis by fluorescence microscopy revealed an almost complete quenching of GFP-tagged murine CCR2 protein expression in HEK 293 cells cotransfected with pSUPER-H1.shCCR2 compared with that of the pSUPER-H1.Empty cotransfected control (Figure 1B). Subsequent FACS analysis (Figure 2A) confirmed that CCR2/GFP fusion protein expression was dose-dependently blunted after cotransfection with increasing amounts of pSUPER-H1.shCCR2, whereas the shRNA controls (ie, pSUPER-H1.Empty, pSUPER-H1.shLuc, and the nonsense shRNA sequence) were ineffective. Also, the fluorescence intensity of the GFP-positive cells was reduced to the level of untransfected HEK 293–cell controls (on average 380) after cotransfection with pSUPER-H1.shCCR2 (Figure 2B). These FACS data were confirmed by Western blot analysis (Figure 2C).
Intracellular calcium levels were measured in HEK 293 cells expressing CCR2 and the H1.shCCR2 or H1.Empty sequence in response to the murine CCR2 ligand JE. Upon incubation with JE (1 nM), transient calcium influx was observed after 2 minutes, and peaked at 4 minutes, after which intracellular calcium levels gradually declined to base values. Transfection of these cells with pSUPER-H1.shCCR2 completely abolished the JE-induced calcium influx to levels comparable with that of nonstimulated cells (Figure 2D), indicating that the observed reduction in CCR2 mRNA and protein levels establishes a complete loss of CCR2 function in response to JE.
Silencing of CCR2 expression by pSUPER-H1.shCCR2. (A) Expression of murine CCR2 in HEK 293 cells after cotransfection with pEGFPN-1/CCR2 and pSUPER-H1.Empty (1.5 μg), relative to that of HPRT, was not affected by pSUPER-H1.shLuc or pSUPER-H1.shInert, whereas CCR2 mRNA levels were up to 94% reduced after cotransfection with increasing amounts of pSUPER-H1.shCCR2 (***P < .001). Error bars represent SEM. (B) HEK 293 cells cotransfected with either pEGFPN-1/CCR2 and pSUPER-H1.Empty (1.5 μg; left panel) or pEGFPN-1/CCR2 and pSUPER-H1.shCCR2 (0.5 μg or 1.5 μg; middle and right panels, respectively), which show increased quenching of GFP protein expression on the cell surface, indicating almost complete loss of CCR2 surface protein after transfection with pSUPER-H1.shCCR2. Cells were viewed with a Biorad 2-photon confocal laser scanning microscope (1000× magnification), and images were digitized with Adobe Photoshop 7.0.
Silencing of CCR2 expression by pSUPER-H1.shCCR2. (A) Expression of murine CCR2 in HEK 293 cells after cotransfection with pEGFPN-1/CCR2 and pSUPER-H1.Empty (1.5 μg), relative to that of HPRT, was not affected by pSUPER-H1.shLuc or pSUPER-H1.shInert, whereas CCR2 mRNA levels were up to 94% reduced after cotransfection with increasing amounts of pSUPER-H1.shCCR2 (***P < .001). Error bars represent SEM. (B) HEK 293 cells cotransfected with either pEGFPN-1/CCR2 and pSUPER-H1.Empty (1.5 μg; left panel) or pEGFPN-1/CCR2 and pSUPER-H1.shCCR2 (0.5 μg or 1.5 μg; middle and right panels, respectively), which show increased quenching of GFP protein expression on the cell surface, indicating almost complete loss of CCR2 surface protein after transfection with pSUPER-H1.shCCR2. Cells were viewed with a Biorad 2-photon confocal laser scanning microscope (1000× magnification), and images were digitized with Adobe Photoshop 7.0.
Transduction of CCR2-transfected HEK 293 cells with H1.shCCR2 lentivirus resulted in a significantly reduced expression of CCR2 relative to hHPRT (5.95 ± 0.67 versus 14.4 ± 3.6 in cells transduced with H1.Empty virus; P = .02).
Transduction of bone marrow cells
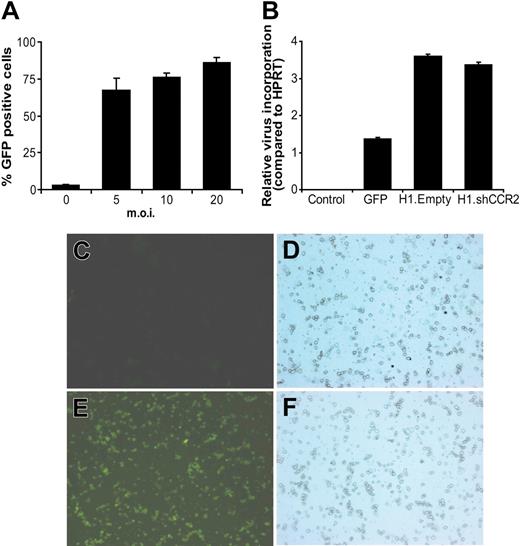
In the next stage, the transduction efficiency of lentivirus in bone marrow cells was optimized. FACS analysis of bone marrow cells showed that transduction was not satisfactory at low multiplicities of infection (moi 6.5% ± 0.6% and 9.8% ± 3.3% at moi's of 1 and 2, respectively). The use of higher titers and DEAE-dextran (10 μg/mL) led to a strong increase in infection efficiency from 3.1% ± 0.5% in control cells via 67.1% ± 6.0% (moi = 5) and 76.2% ± 2.0% (moi = 10) to 86.1% ± 2.3% (moi = 20) 24 hours after infection (Figure 3A). Viral incorporation analysis revealed that bone marrow cells contained on average 2 copies of virus per Hprt gene 24 hours after transduction of the cells with GFP, H1.shCCR2, or H1.Empty control lentivirus (Figure 3B). The high transduction, obtained at moi's of 10 to 20 in the presence of 10 μg/mL DEAE-dextran, suffices for in vivo transplantation purposes and circumvents the need of a laborious preselection step. Six weeks after injection of GFP lentivirus–transduced bone marrow into irradiated C57Bl/6 mice, cytospin analysis showed that approximately 80% to 90% of all white blood cells were GFP positive (Figure 3C-F).
Transplantation of H1.shCCR2-transduced bone marrow
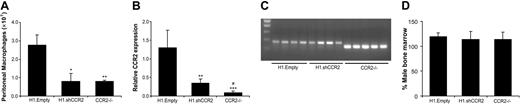
To generate chimeras with silenced CCR2 expression, bone marrow from C57Bl/6 mice was transduced with either H1.shCCR2 or H1.Empty control lentivirus in the presence of 10 μg/mL DEAE-dextran and intravenously injected into irradiated C57Bl/6 recipient mice, while a third group received Ccr2–/– bone marrow. No differences were observed in either weight gain during the experiment or in mononuclear-cell and neutrophil content of the total white blood cell population at 7 weeks after transplantation (data not shown). At that point, macrophages were recruited to the peritoneal cavity by an intraperitoneal thioglycollate challenge. Assessment of the number of peritoneal macrophages, 5 days after challenge, revealed a significant 70% reduction in macrophage recruitment in mice that had received H1.shCCR2 lentivirus–transduced bone marrow compared with the H1.Empty lentivirus controls (8 × 106 ± 4 × 106 cells versus 27 × 106 ± 6 × 106 cells, respectively; P = .03, Figure 4A). Interestingly, macrophage recruitment was comparable to that of mice reconstituted with Ccr2–/– bone marrow (8 × 106 ± 1 × 106 cells; P = .9 versus the H1.shCCR2 lentivirus group). Macrophage recruitment levels of mice reconstituted with H1.Empty-transduced bone marrow matched with those found in control mice that underwent transplantation (female C57Bl6 recipient mice that had received untransduced male C57Bl6 bone marrow, data not shown), which indicates that lentiviral transduction of bone marrow per se does not elicit an inflammatory response.
Silencing of CCR2 protein by pSUPER-H1.shCCR2. (A) FACS analysis of HEK 293 cells after cotransfection with pEGFPN-1/CCR2 and pSUPER-H1.Empty (1.5 μg, control), which was not significantly affected by cotransfection with pSUPER-H1.shLuc or pSUPER-H1.shInert. Cotransfection with increasing amounts of pSUPER-H1.shCCR2 led to a significant and dose-dependent silencing of CCR2 protein level up to 75%. *P ≤ .05. (B) The average fluorescence intensity of the GFP-positive cells was even more markedly reduced after cotransfection with pSUPER-H1.shCCR2 to the untransfected HEK 293 control level (approximately 390). The nonspecific controls had no effect on GFP fluorescence intensity compared with pSUPER-H1.Empty control levels. **P ≤ .01; ***P ≤ .001. (C) Western blot analysis of HEK 293–cell lysates for GFP protein expression using β-tubulin (50 kDa) as internal standard, showing a dose-dependent and specific reduction in GFP protein expression in cells cotransfected with increasing amounts of pSUPER-H1.shCCR2 (lanes D, E, and F) compared with only pEGFPN-1/CCR2-transfected (lane B) or pSUPER-H1.Empty cotransfected cells (lane C). Lane 1 represents a sample of untransfected HEK 293 cells. (D) Measurements of JE-induced calcium influx in HEK 293 cells expressing murine CCR2 and the H1.Empty (▴) or the H1.shCCR2 sequence (□). As a control, unstimulated HEK 293 cells expressing CCR2 were measured (•). In contrast to pSUPER-H1.Empty–transfected cells, no calcium influx was observed in pSUPER-H1.shCCR2–transfected cells after stimulation with JE, indicating that CCR2 signal transduction is silenced by shCCR2 treatment. Error bars represent SEM.
Silencing of CCR2 protein by pSUPER-H1.shCCR2. (A) FACS analysis of HEK 293 cells after cotransfection with pEGFPN-1/CCR2 and pSUPER-H1.Empty (1.5 μg, control), which was not significantly affected by cotransfection with pSUPER-H1.shLuc or pSUPER-H1.shInert. Cotransfection with increasing amounts of pSUPER-H1.shCCR2 led to a significant and dose-dependent silencing of CCR2 protein level up to 75%. *P ≤ .05. (B) The average fluorescence intensity of the GFP-positive cells was even more markedly reduced after cotransfection with pSUPER-H1.shCCR2 to the untransfected HEK 293 control level (approximately 390). The nonspecific controls had no effect on GFP fluorescence intensity compared with pSUPER-H1.Empty control levels. **P ≤ .01; ***P ≤ .001. (C) Western blot analysis of HEK 293–cell lysates for GFP protein expression using β-tubulin (50 kDa) as internal standard, showing a dose-dependent and specific reduction in GFP protein expression in cells cotransfected with increasing amounts of pSUPER-H1.shCCR2 (lanes D, E, and F) compared with only pEGFPN-1/CCR2-transfected (lane B) or pSUPER-H1.Empty cotransfected cells (lane C). Lane 1 represents a sample of untransfected HEK 293 cells. (D) Measurements of JE-induced calcium influx in HEK 293 cells expressing murine CCR2 and the H1.Empty (▴) or the H1.shCCR2 sequence (□). As a control, unstimulated HEK 293 cells expressing CCR2 were measured (•). In contrast to pSUPER-H1.Empty–transfected cells, no calcium influx was observed in pSUPER-H1.shCCR2–transfected cells after stimulation with JE, indicating that CCR2 signal transduction is silenced by shCCR2 treatment. Error bars represent SEM.
Efficient transduction of bone marrow cells using GFP lentivirus. (A) Transduction of whole bone marrow with GFP lentivirus led to a titer-dependent increase in percentage of GFP-expressing cells. (B) Virus particle incorporation in bone marrow cells compared with HPRT of untransduced control cells and 24 hours after transduction with GFP-, H1.Empty-, or H1.shCCR2-lentivirus. Error bars in panels A and B represent SEM. (C,D) Fluorescence and light microscopic high-power field, respectively, of a white blood cell cytospin from control mice 6 weeks after transplantation of nontransduced bone marrow (original magnification, ×200). (E,F) White blood cells from mice that underwent transplantation with lentiviral GFP-transduced bone marrow show approximately 90% GFP-positive cells (panel E; original magnification, ×200), indicating that transplantation of lentivirus-transduced bone marrow results in sustained transgene expression by blood cells. Images were captured with a Leica DR-ME microscope equipped with a 20× objective lens and a 10× ocular lens, and Leica QWin software (Leica Imaging Systems) was used.
Efficient transduction of bone marrow cells using GFP lentivirus. (A) Transduction of whole bone marrow with GFP lentivirus led to a titer-dependent increase in percentage of GFP-expressing cells. (B) Virus particle incorporation in bone marrow cells compared with HPRT of untransduced control cells and 24 hours after transduction with GFP-, H1.Empty-, or H1.shCCR2-lentivirus. Error bars in panels A and B represent SEM. (C,D) Fluorescence and light microscopic high-power field, respectively, of a white blood cell cytospin from control mice 6 weeks after transplantation of nontransduced bone marrow (original magnification, ×200). (E,F) White blood cells from mice that underwent transplantation with lentiviral GFP-transduced bone marrow show approximately 90% GFP-positive cells (panel E; original magnification, ×200), indicating that transplantation of lentivirus-transduced bone marrow results in sustained transgene expression by blood cells. Images were captured with a Leica DR-ME microscope equipped with a 20× objective lens and a 10× ocular lens, and Leica QWin software (Leica Imaging Systems) was used.
Analysis of CCR2 expression by peritoneal macrophages revealed a highly significant reduction in CCR2 mRNA levels in mice that received a transplant of H1.shCCR2-versus H1.Empty lentivirus–transduced bone marrow (expression relative to murine HPRT: 0.4 ± 0.1 and 1.3 ± 0.2, respectively; P = .002). For comparison, mice that had received Ccr2–/– bone marrow showed a relative CCR2 expression of 0.09 ± 0.02 (Figure 4B).
Lentiviral shCCR2 results in reduced macrophage influx due to reduced CCR2 expression. (A) Macrophage recruitment to the peritoneum in mice that underwent transplantation with H1.shCCR2 lentivirus–transduced bone marrow is significantly reduced compared with those that received H1.Empty lentivirus–transduced bone marrow. Recruitment levels were found to be similar to control mice that underwent transplantation with Ccr2–/– bone marrow (*P < .05, **P < .01). (B) CCR2 mRNA expression level in peritoneal macrophages of mice that underwent transplantation with H1.shCCR2-transduced bone marrow was reduced 7 weeks after transplantation (**P < .01, ***P < .001 compared with H1.Empty; #P < .01 compared with H1.shCCR2). (C) Ccr2 genotyping of transplanted C57Bl/6 mice, revealing a 360–base pair amplicon of wild-type Ccr2 in mice that underwent transplantation with H1.Empty- or H1.shCCR2 lentivirus–transduced bone marrow and the 290–base pair amplicon was observed in mice that underwent transplantation with Ccr2–/– bone marrow. (D) Percent male bone marrow in bone marrow cell lysates from recipient mice, determined by PCR analysis on the Sry gene, located on the Y-chromosome. Error bars in panels A, B, and C represent SEM.
Lentiviral shCCR2 results in reduced macrophage influx due to reduced CCR2 expression. (A) Macrophage recruitment to the peritoneum in mice that underwent transplantation with H1.shCCR2 lentivirus–transduced bone marrow is significantly reduced compared with those that received H1.Empty lentivirus–transduced bone marrow. Recruitment levels were found to be similar to control mice that underwent transplantation with Ccr2–/– bone marrow (*P < .05, **P < .01). (B) CCR2 mRNA expression level in peritoneal macrophages of mice that underwent transplantation with H1.shCCR2-transduced bone marrow was reduced 7 weeks after transplantation (**P < .01, ***P < .001 compared with H1.Empty; #P < .01 compared with H1.shCCR2). (C) Ccr2 genotyping of transplanted C57Bl/6 mice, revealing a 360–base pair amplicon of wild-type Ccr2 in mice that underwent transplantation with H1.Empty- or H1.shCCR2 lentivirus–transduced bone marrow and the 290–base pair amplicon was observed in mice that underwent transplantation with Ccr2–/– bone marrow. (D) Percent male bone marrow in bone marrow cell lysates from recipient mice, determined by PCR analysis on the Sry gene, located on the Y-chromosome. Error bars in panels A, B, and C represent SEM.
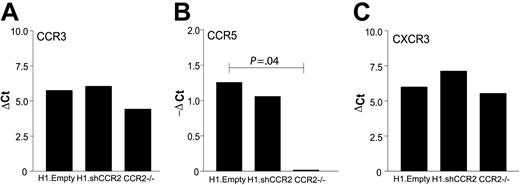
As expected, genotyping showed that C57Bl/6 mice that received a transplant of H1.Empty- or H1.shCCR2 lentivirus–transduced bone marrow displayed the 360 base pair amplicon, indicative of wild-type Ccr2 (Figure 4C). C57Bl/6 mice that had received Ccr2–/– bone marrow showed the Ccr2–/– phenotype (290 base pair sequence). Reconstitution with male bone marrow in the female recipient mice was quantitative and persistent for at least 7 weeks in all 3 groups, as judged from PCR analysis of the Sry gene (Figure 4D). PCR analysis of genomic DNA from bone marrow revealed that unpurified bone marrow from mice that received a transplant of H1.shCCR2 contained on average 2 virus integrants per Sry copy in both the H1.shCCR2 (2.30) and the H1.Empty groups (1.98), while as expected in bone marrow from mice that received a transplant of Ccr2–/– no virus particles were detected. Furthermore, no differences were observed in peritoneal macrophage expression levels of other chemokine receptors, such as CCR3, CCR5, and CXCR3 (Figure 5A-C, P = NS), or of inflammatory mediators like interferon β (IFNβ) and IFNγ (data not shown), indicating that the shRNA against CCR2 was highly specific and did not display significant off-target effects on the expression levels of other relevant genes. Intriguingly, macrophages of mice that received a transplant of Ccr2–/– showed an almost 50% reduction in relative CCR5 mRNA expression levels (P = .04).
Discussion
Transplantation of transgenic or knockout bone marrow to wild-type recipients is a widely used technique to determine the role of a particular leukocyte gene in several inflammatory disorders, such as arthritis1 and atherosclerosis.2-4 Unfortunately, the time-consuming generation of transgenic or knockout mice and the subsequent backcrossing to an appropriate disease background for analysis of gene function considerably delays progression in gene function research. In this study, we demonstrate the high potential of a novel lentiviral gene silencing approach on bone marrow cells for the in situ generation of leukocyte knockdowns of the aimed target gene. Targeted gene silencing by means of shRNA has, since its discovery, proven to be a powerful tool in the analysis of gene function in vitro as well as in vivo as it can result in prolonged selective inhibition of target gene expression even in eukaryotic cells.21-25 To deliver shRNA and warrant prolonged shRNA expression, we made use of third-generation self-inactivating lentiviruses,12,13 which have been demonstrated to efficiently infect and integrate their genome into a range of mitotic and nonmitotic cells.14,15,32 Although rather efficient retroviral transduction of stem cells of up to 64% has been reported,8-11 this required extensive manipulation of bone marrow stem cells to increase their mitotic capacity, possibly compromising the hematopoietic potential.40 Efficient lentiviral infection requires only a short in vitro incubation period and preventing undesired differentiation of the hematopoietic stem cells.
Absence of nonspecific effects on related chemokine receptors. No nonspecific effects were observed on peritoneal macrophage mRNA levels of other chemokine receptors, as CCR3 (A), CCR5 (B), and CXCR3 (C) in mice that underwent transplantation with H1.shCCR2 lentivirus–transduced bone marrow compared with the H1.Empty controls. Ccr2–/– bone marrow transplantation led to a significant decrease in macrophage CCR5 expression (P = .04). ▵Ct indicates delta cycle threshold.
Absence of nonspecific effects on related chemokine receptors. No nonspecific effects were observed on peritoneal macrophage mRNA levels of other chemokine receptors, as CCR3 (A), CCR5 (B), and CXCR3 (C) in mice that underwent transplantation with H1.shCCR2 lentivirus–transduced bone marrow compared with the H1.Empty controls. Ccr2–/– bone marrow transplantation led to a significant decrease in macrophage CCR5 expression (P = .04). ▵Ct indicates delta cycle threshold.
In this study, the transduction of bone marrow cells with GFP lentivirus was almost quantitative (80%-90%) when using titers of 10 moi to 20 moi. Transduction levels in our study exceeded reported transduction efficiencies for human bone marrow–derived CD34+ stem cells41-43 and even were sufficiently high to perform bone marrow transplantation without prior selection of infected cells. Generally, 2 or more consecutive transduction steps were required to achieve similarly high levels, although Banerjea et al, for example, communicated on efficient gene transfer in a single-step protocol.44 Our protocol is a compromise of a limited culture period of bone marrow cells in growth factor–free medium (24 hours) on the one hand, preventing undesired differentiation or maturation of the bone marrow cells, and lowest possible virus titer on the other. Furthermore, a single transduction protocol will also minimize the risk of multiple, potentially harmful viral integrants per cell due to, among others, insertional mutagenesis.45 However, it should be noted that in the case of shRNAs, multiple integrations per host cell may even be beneficial, as multiple integrations will be accompanied by higher transcript expression, leading to more efficient silencing of the target gene. In our study, the initial infection levels of the bone marrow cells by the H1.Empty and the H1.shCCR2 lentivirus were shown to be similar to that of GFP lentivirus–transduced cells.
As a model gene to illustrate the potential of lentiviral gene silencing in vivo, we have chosen the CC-chemokine receptor 2 (Ccr2). CCR2 is critically involved in monocyte recruitment to sites of inflammation in response to an inflammatory stimulus46 during atherosclerosis, renal fibrosis,47 or pulmonary inflammation.48 An shRNA sequence, targeting nucleotides 867 to 885 of the murine Ccr2 gene, reduced CCR2 mRNA levels by 94% and protein levels in HEK 293 cells to an almost similar extent. Furthermore, the shCCR2 construct completely prevented CCR2 signal transduction in response to its natural ligand JE, as revealed by intracellular calcium release measurements.
Interestingly, transplantation of bone marrow cells transduced with a lentivirus carrying the shCCR2 construct strongly attenuated thioglycollate-elicited, CCR2-dependent migration of macrophages into the peritoneal cavity. The amount of recruited peritoneal macrophages in mice that underwent transplantation with H1.shCCR2 lentivirus–transduced bone marrow was comparable to that of mice that received Ccr2–/– bone marrow transplants. Macrophage recruitment levels of the H1.Empty and H1.shCCR2 lentivirus group were essentially similar to values described by Kurihara et al46 for wild-type and Ccr2–/– mice, respectively, indicating that local macrophage recruitment and proliferation was not affected by lentiviral transduction of the bone marrow per se. Peritoneal macrophages derived from the H1.shCCR2-transduced bone marrow cells displayed a 70% reduction in CCR2 mRNA content compared with the H1.Empty group, which apparently translated in the observed Ccr2 knockout phenotype. In the mice that underwent transplantation with Ccr2–/– bone marrow, only approximately 7% of circulating blood cells originated from the recipient mice, which accounted for the residual CCR2 mRNA expression in blood cells.
The bone marrow transplantation led to a quantitative and persistent repopulation of donor bone marrow as determined by genomic DNA analysis of the Y-chromosomal Sry gene. Moreover, insertion of the target gene apparently did not result in a shifted differentiation pattern of hematopoietic stem cells. At the endpoint of the study, viral copy number in the bone marrow from recipient mice was essentially similar to that of the initial transduction level, implying that the lentivirus-infected cells are not outcompeted by residual or nontransduced cells and that the expansive capacity of bone marrow cells is not affected by lentiviral transduction. We realize that these data were all acquired at 7 weeks after transplantation and that more long-term studies are required on the effects and persistence of lentivirus-transduced bone marrow; however, we are confident that the results will not be very different.
In line with the in vitro data, the obtained reduction in CCR2 expression in vivo was sufficient for a complete and specific knockdown of CCR2 function. Importantly, IFNβ and IFNγ mRNA levels did not differ, indicating that the shRNA lentivirus–transduced bone marrow did not elicit an inflammatory response after transplantation.49 While CCR2 silencing by shCCR2 was not accompanied by an altered expression pattern of other chemokine receptors, the peritoneal macrophages isolated from mice that underwent transplantation with Ccr2–/– displayed a significant 50% reduction in CCR5 mRNA levels, pointing toward transcriptional modulation of the flanking Ccr5 gene by insertion of the PGK-neo cassette into the Ccr2 gene on mouse chromosome 9. Down-regulation of genes downstream of the site of PGK-neo cassette insertion has been reported previously50,51 and can be circumvented when using the shRNA approach.
In conclusion, here we show effective delivery of lentiviral shRNA to bone marrow cells and subsequent transplantation into irradiated recipient mice as a strategy to generate hematopoietic knockdowns with silenced CCR2 expression. shRNA-mediated silencing of gene expression persisted for at least 7 weeks after transplantation and led to a complete loss of CCR2 function without any noticeable side effects. Speed and efficiency render this strategy very valuable for addressing the role of other hematopoietic, and in particular leukocyte, genes in inflammatory disorders.
Prepublished online as Blood First Edition Paper, May 10, 2005; DOI 10.1182/blood-2004-12-4839.
Supported by grant 016.026.019 from the Netherlands Organization for Scientific Research (I.B.) and grants 2003T201 (E.A.L.B.), 2001T041 (M.V.E.), and M93.001 (P.J.V.S.) from the Netherlands Heart Foundation, and GlaxoSmithKline Pharmaceuticals, United Kingdom (J.G.).
I.B. and J.G. contributed equally to this study.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors would like to thank Miranda Stitzinger and Saskia de Jager from the Division of Biopharmaceutics and Hans de Bont from the Division of Toxicology (Leiden Amsterdam Center for Drug Research, Leiden University) for technical assistance.