Abstract
The SH2 domain–containing inositol 5′-phosphatase-1 (SHIP) has the potential to modulate multiple signaling pathways downstream of receptors that impact hematopoietic stem cell (HSC) biology. Therefore, we postulated that SHIP might play an important role in HSC homeostasis and function. Consistent with this hypothesis, HSC proliferation and numbers are increased in SHIP–/– mice. Despite expansion of the compartment, SHIP–/– HSCs exhibit reduced capacity for long-term repopulation. Interestingly, we observe that SHIP–/– stem/progenitor cells home inefficiently to bone marrow (BM), and consistent with this finding, have reduced surface levels of both CXCR4 and vascular cell adhesion marker-1 (VCAM-1). These studies demonstrate that SHIP is critical for normal HSC function, homeostasis, and homing.
Introduction
The survival of an organism is dependent upon the ability of hematopoietic stem cells (HSCs) to replenish the blood compartment on a daily basis. To accomplish this task, HSCs must maintain a fine balance between 3 possible fates: self-renewal, differentiation, or senescence. Although the decision process that determines the fate of an HSC clone remains to be completely defined, several molecules are already known to play a role in this process, including components of the HSC microenvironment or niche. This niche consists of both extracellular matrix molecules and cells, such as osteoblasts and stromal cells, which produce cytokines and chemokines important for the maintenance of the HSC pool.1-5
These microenvironmental or external cues engage receptors on HSCs, leading to the activation of signaling pathways governing cell proliferation, self-renewal, differentiation, mobilization, and bone marrow (BM) retention.5 Some of these pathways, such as those initiated by stem cell factor (SCF/c-kitL),6 stromal cell–derived factor 1 (SDF-1/CXCL12),7 and thrombopoietin (TPO),8 result in the activation of phosphatidylinositol 3′ kinase (PI3K) and the formation of phosphatidyl inositol 3,4,5-trisphosphate (PIP3). Therefore, the SH2 domain–containing inositol 5′-phosphatase 1 (SHIP) may influence these pathways in HSCs.9,10 SHIP is a 145-kDa protein primarily expressed by cells of the hematopoietic system,11 including HSCs,12 that can associate with various adapter proteins, scaffold proteins, or receptors following activation of hematopoietic cells.9,13 Formation of these complexes enables SHIP to hydrolyze the 5′phosphate on PIP3,11,14 thus preventing membrane recruitment and activation of pleckstrin homology domain–containing kinases that serve as effectors of PI3K signaling. This activity permits SHIP to limit the survival, activation, differentiation, and/or proliferation of hematopoietic cells.10 Thus, we hypothesized that SHIP might also influence these processes in the HSC compartment. Previous studies reported that SHIP–/– whole bone marrow (WBM) cells do not reconstitute lethally irradiated mice as well as wild-type (WT) WBM in a noncompetitive setting.15 Furthermore it was reported that SHIP–/– WBM has comparable numbers of competitive repopulating units (CRUs) relative to WT littermates in a limiting-dilution assay, which uses compromised competitor cells.15 However, because these analyses were performed with WBM rather than purified HSCs, they did not assess whether SHIP–/– HSCs are defective for repopulation in a WT environment. Thus, it is not possible from these previous studies to conclude that SHIP plays a direct role in signaling pathways active in the HSC compartment.
To study the effects of SHIP on HSCs we used SHIP–/– mice generated by a Cre-lox mutation strategy.16 We found that SHIP–/– mice contain significantly more HSCs in their bone marrow and spleen as measured by flow cytometry. We also observe that a greater proportion of SHIP–/– HSCs enter the cell cycle compared with WT HSCs. Since it was shown in previous studies that SHIP–/– BM contains reduced HSC activity relative to WT BM,15 it became important to assess the function of SHIP–/– HSCs with different assays. We find that when purified SHIP–/– HSCs or WBM are transplanted into lethally irradiated mice, they fail to compete effectively with WT HSCs or BM cells for long-term multilineage repopulation of the hematopoietic compartment. These results might have suggested that the absence of SHIP causes accelerated senescence of the HSC compartment; however, we observe fewer apoptotic HSCs in SHIP–/– BM compared with WT. We then assessed the ability of SHIP–/– HSCs to reach the BM niche, where they encounter the proper environment to support their function. In vivo homing studies performed using purified stem/progenitor cells suggest that SHIP–/– cells home to the BM with a decreased efficiency compared with WT cells. Most interestingly, we also observe that SHIP–/– HSCs have significantly lower surface expression of CXCR4 and vascular cell adhesion marker-1 (VCAM-1), key receptors for homing and retention of hematopoietic cells in the BM.17-19 Therefore, SHIP plays an important role in regulating HSC proliferation, survival, and self-renewal, as well as BM homing and retention.
Materials and methods
Mice
SHIP–/– mice were generated by deletion of the promoter and first exon of SHIP via a Cre-LoxP strategy and then backcrossed to the C57BL6/J background.16 SHIPΔIP/ΔIP mice were created using a Cre-LoxP strategy targeting the inositol phosphatase encoding region. SHIPΔIP/ΔIP mice are on a 129SvJ background and were kindly provided by Dr Jeffrey Ravetch (Rockefeller University, New York, NY).20 For all experiments, germline SHIP-deficient and WT mice were 6 to 8 weeks of age. We used 8- to 12-week-old B6.SJL-Ptprca Pep3b/BoyJ (CD45.1; Jackson Laboratory, Bar Harbor, ME) and a CD45.1 × CD45.2 strain generated by intercrossing C57BL6/J (CD45.2) and B6.SJL-Ptprca Pep3b/BoyJ (CD45.1) mice as recipients for transplantation experiments. Animal experiments were conducted in compliance with institutional guidelines at the University of South Florida.
Cell isolation
BM cells were flushed from intact femurs and tibia, and spleens were crushed with a syringe plunger. Collection of the cells was performed in tissue media (TM) consisting of RPMI 1640, 3% fetal bovine serum (FBS), and 10 mM HEPES (Invitrogen; Gibco BRL, Carlsbad, CA). Cells were then filtered through a 70-μm strainer (BD Bioscience, San Jose, CA), and red blood cells (RBCs) were lysed at room temperature (RT) for 2 to 5 minutes in 1 × RBC lysis buffer (Ebioscience, San Diego, CA), centrifuged, and resuspended in staining media (SM): 1 × Dulbecco phosphate-buffered saline (D-PBS), 3% FBS, 10 mM HEPES (Invitrogen, Gibco BRL). Peripheral blood (PB) was obtained by retro-orbital (RO) or submandibular bleeding, collected in microtainers with K2 EDTA (BD, Franklin Lakes, NJ), and RBC-lysed twice to obtain PB mononuclear cells (PBMCs), which were resuspended in SM.
HSC phenotype
All antibodies were from BD Biosciences (San Diego, CA) except where noted. All flow cytometry data were analyzed with FlowJo Software (Tree Star, Ashland, OR). BM cells, splenocytes, or PBMCs were treated with Fc block (2.4G2) on ice, then stained on ice for Lineage–/lowc-Kit+Sca1+Thy1+ (KTLS) phenotype.21 The stain included Sca1-PE (E13-161.7), c-Kit–APC (2B8), Thy1.2-Cychrome (53-1.2), and a lineage (Lin) panel-FITC: CD2(RM2-5), CD3ϵ (145-2C11), CD4(GK1.5), CD5 (53-7.3), CD8α (53-6.7), B220 (RA3-6B2), Gr-1 (RB6-8C5), Mac-1 (M1/70), NK1.1 (PK136), and Ter119 (TER-119; eBioscience). For Lin–c-kit+Sca1+CD48–CD150+ (KLSCD48–CD150+),22 the same Lin panel was used with the addition of CD34-FITC (RAM34) along with Sca1-biotin (E13-161.7), c-Kit-APCCy7 (2B8; eBioscience), CD48-PE (HM48-1), and CD150-Alexa647 (9D1; Serotec, Raleigh, NC), followed with streptavidin-PeCy7. The cells were then resuspended in SM containing 75 ng/mL DAPI for dead cell exclusion (Sigma, St Louis, MO). Analysis was done on a fluorescence-activated cell-sorting (FACS) Vantage or FACS Aria using DiVAII (BD Biosciences). For the c-Kit+Flk2–Lin–/lowSca1+ (KFLS) phenotype,23 we used Sca1-FITC, c-Kit–APC, and the previously mentioned Lin panel–PE with addition of the Flk2 antibody (A2F.10-1). After staining, the cells were washed and resuspended in SM containing 7-amino-actinomycin D (7-AAD; BD Biosciences) for dead-cell exclusion. Acquisition was done on a FACS Calibur. For the side population (SP) analysis, BM cells were stained following standard procedure.24 Analysis was done on a FACS Vantage using DiVaII. For homing marker analysis, Lin-depletion of BM cells was done using the Lin-PE panel as mentioned, with the exclusion of CD2, anti-PE magnetic beads, and the AutoMACS system (Miltenyi Biotech, Auburn, CA). The Lin-depleted fraction was stained for KTLS and CXCR4 (2B11/CXCR4), VCAM-1 (429), or very late antigen-4 (VLA-4; 9C10) on biotin, followed by staining with streptavidin-PeCy7. DAPI was used for dead-cell exclusion. Analysis was done on a FACS Aria using DiVaII.
Cell-cycle analysis
BM cells were enriched either for Sca1+ or c-Kit+ cells using AutoMACS (Miltenyi Biotech). Sca1+ or c-Kit+ cells were then stained for the KTLS or KFLS phenotypes, then fixed in Cytofix/Cytoperm (BD Biosciences) for 30 minutes on ice, washed in Perm/Wash buffer, and incubated with 4 to 10 ng/mL Hoechst dye 33342 (Sigma) in Perm/Wash buffer overnight at 4°C. The DNA content was measured 24 hours later on the FACS Vantage and analyzed using FlowJo software.
CRU assay
CD45.1-recipient mice were given antibiotic water (40 mg/mL sulfamethaxazole and trimethorprim oral suspension [Hi-Tech Pharmacal, Amityville, NY]) prior to receiving a single dose of 9.5 Gy 3 hours before transplantation. WBM cells (106) from CD45.2 SHIP–/– or WT mice along with 1 × 106 CD45.1 WBM cells were injected retro-orbitally into recipients, as per Harrison.25 Mice that had received transplants were later assessed for PB multilineage reconstitution. Repopulating units (RUs) were calculated as per Harrison:25 RU = (10 × % CD45.2 repopulation)/(100–% CD45.2 repopulation).25
Direct competition assay
The direct competition (DC) assay was modified from Domen et al.26 Donor BM cells were magnetically enriched for Sca1+ cells on AutoMACS. The Sca1+ fraction was stained for Lin–/lowc-Kit+Sca1+Thy1low or Lin–/lowFlk2–c-Kit+Sca1+ and sorted twice in the presence of DAPI using the FACS Vantage DiVAII. For the KTLS transplantation, 200 CD45.1 WT and 200 CD45.2 SHIP–/– KTLS cells were injected with 4 × 104 Sca1–CD45.1/45.2 WBM cells. For the KFLS transplantation, 100 CD45.1 WT and 100 CD45.2 SHIP–/– KFLS cells were injected with 4 × 105 CD45.1/45.2 Sca1–cells. CD45.1/45.2 recipients had been previously irradiated with a split dose of 10 Gy at a 3-hour interval and given antibiotic water, as mentioned.
Assessment of PB multilineage reconstitution
PBMCs were treated with Fc block, then stained with CD45.1-PE, CD45.2-FITC, and APC-conjugated Lin markers: B220, CD3, or Mac1/Gr1. The cells were then washed and resuspended in SM containing 7-AAD to exclude dead cells. Cells were analyzed on a FACSCalibur.
Annexin V assay and TUNEL assay
BM cells were isolated by flushing femurs and tibiae and then stained with c-Kit–APCCy7, Thy1.2-APC, Lin-FITC, Sca1-biotin, and Flk2-PE on ice; biotin was revealed by streptavidin-PeCy7. The cells were then incubated with Annexin V (BD Biosciences) according to the manufacturer's protocol. Analysis was done on a FACS Aria using DAPI for dead-cell exclusion. Transferase-mediated dUTP nick-end labeling (TUNEL) assay was performed using the in situ cell death detection kit following manufacturer's instruction (Roche Applied Sciences, Indianapolis, IN). First, BM cells were depleted of Lin+ cells using Lin-PE, anti-PE beads, and the AutoMacs (Miltenyi Biotech), Lin– cells were then stained for KTLS and TUNEL.
In vivo homing assay
The in vivo homing assay was established on the basis of protocols published by two different groups.27,28 BM cells from 6- to 8-week-old SHIP–/– and WT mice were isolated from hind/forelimbs by flushing, and from the vertebral column by crushing with mortar and pestle. BM cells were then RBC-lysed and Fc-blocked for 15 minutes, then depleted of Lin+ cells using Lin– PE, followed by anti-PE beads, using AutoMacs (Miltenyi Biotech). Lin– cells were then stained for Lin-PE, Sca1-FITC, and DAPI, and Sca1+Lin– cells were sorted on the FACS Aria. After sorting, the cells were stained with 1 μM DDAO-SE (Molecular Probes/Invitrogen, Carlsbad, CA) and/or 0.5 μM CFSE (Molecular Probes/Invitrogen) for 15 minutes at 37°C, washed, and centrifuged at 300g for 7 minutes at 4°C. Cells were then resuspended in PBS with 1 mM HEPES and an equal number of live Sca1+Lin– SHIP–/– and WT cells (2 × 105 cells of each genotype) were then injected into irradiated recipient mice (10 Gy [1000 rad] 3 hours prior to transplantation). For some experiments, both CFSE-stained SHIP–/– and WT Sca1+Lin– cells were injected in different recipients. In others, CFSE-stained SHIP–/– and DDAO-stained WT Sca1+Lin– were injected in the same recipients. After injection (12-14 hours), BM cells and splenocytes were isolated from recipients and directly analyzed on a FACS Aria in the presence of DAPI. Events (5 × 106) were collected for each recipient.
Results
SHIP–/– mice have an expanded HSC compartment
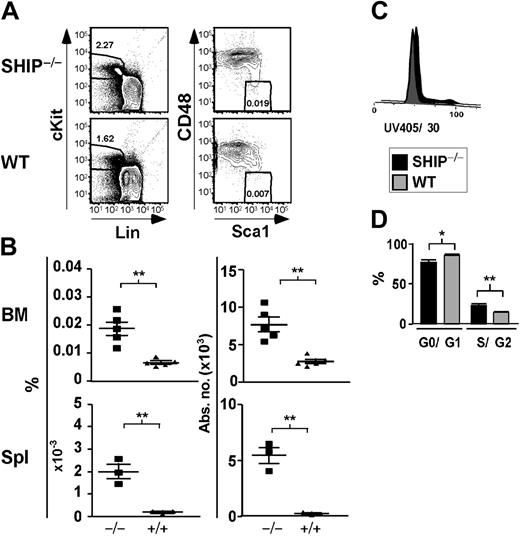
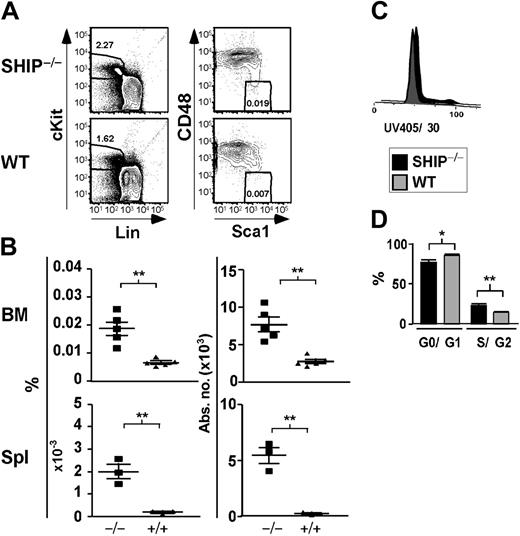
We initially quantitated HSC numbers based on a phenotype established by Morrison and Weissman, Lin–/lowThy1+c-Kit+Sca1+ (KTLS),21 which is highly enriched for long-term repopulating HSC (LT-HSC) activity.21 We found that SHIP–/– mice had an increased percentage and absolute number of KTLS cells in their BM, spleen (Table 1), and PB (data not shown). BM of SHIP–/– mice contained 6-fold more KTLS cells than WT controls (Table 1). In the spleen of SHIP–/– mice, we observed a greater than 6-fold increase in the percentage and absolute numbers of KTLS cells (Table 1). A comparable scenario was seen in the peripheral blood, where KTLS cells were increased by 2.5-fold compared with WT littermates (data not shown). A similar expansion of the KTLS compartment was seen in SHIPΔIP/ΔIP mice, another germline SHIP mutant model with a different genetic background (data not shown).20 Although Thy1.2 expression can be detected on HSCs from C57BL6/J mice, surface expression is lower, making negative and positive distinctions more difficult than for the Thy1.1 allele.23,29 Thus, we also quantified HSC numbers using different phenotypes, including the immunophenotype defined by Christensen and Weissman:23 Lin–/lowFlk2–c-Kit+Sca1+ (KFLS). KFLS cells, as opposed to KTLS, can be used regardless of genetic background.23 We found that SHIP–/– mice also exhibited an increased percentage and absolute number of KFLS cells in their BM and spleen (Table 1). KFLS cell numbers are expanded by 2-fold in the BM of SHIP–/– mice relative to WT mice. We also observed that both the percentage and absolute numbers of KFLS cells in SHIP–/– spleens were increased by more than 3-fold as compared to WT spleens (Table 1). In addition, Kiel et al22 recently published a new phenotype to identify HSCs that relies on the differential expression of the SLAM family receptors, CD48 and CD150, by HSCs and multipotential progenitor cells. We observe a 2.8-fold increase in the number of KLSCD48– cells in SHIP–/– BM compared with WT BM (Table 1; Figure 1A-B). Moreover, the percentage and absolute numbers of KLSCD48– cells are increased by 8.7- and 19-fold, respectively, in SHIP–/– relative to WT spleen (Table 1; Figure 1A-B). When using the KLSCD48–CD150+ immunophenotype we detect a 2-fold increase in the number of HSCs in the BM of SHIP–/– BM compared with WT (Table 1). In order to confirm our observations made using immunophenotypes relying on surface-marker expression, we assessed the level of HSCs using the SP phenotype. Identification of SP cells relies on the function of a transporter protein that exclude the dye Hoechst 33342:24,30 these cells are enriched approximately 1000-fold for LT-HSC activity compared with WBM cells.24 When comparing SHIP–/– versus WT BM, we observe a 2-fold increase in the percentage and absolute number of SP cells in SHIP–/– BM (Table 1). Thus, analysis of 5 different HSC phenotypes21-24 demonstrates a preferential expansion of the LT-HSC compartment in SHIP–/– mice. These observations implicate SHIP in the control of HSC compartment homeostasis.
Significant increase in the percentage and absolute number of KLSCD48– cells in SHIP–/– BM. (A) Representative FACS plots showing detection of KLSCD48– HSCs in SHIP–/– and WT BM. (B) Percentage and absolute number of KLSCD48– cells in the BM (per femur and tibia pair) and spleen (Spl) of SHIP–/– (▪) and WT (▴) mice. (C) Histogram of DNA content in SHIP–/– and WT KTLS cells. (D) Bar graph showing the percentage of SHIP–/– (▪) or WT (▦) KTLS/KFLS cells in each stage of cell cycle as calculated using the Watson Pragmatic model in the FlowJo cell-cycle platform. Statistical analysis was performed using Prism 4. Significance was established using the unpaired student t test (Prism 4; GraphPad Software, San Diego, CA). **P < .001 and *P < .01 (mean ± SEM; n ≥ 3).
Significant increase in the percentage and absolute number of KLSCD48– cells in SHIP–/– BM. (A) Representative FACS plots showing detection of KLSCD48– HSCs in SHIP–/– and WT BM. (B) Percentage and absolute number of KLSCD48– cells in the BM (per femur and tibia pair) and spleen (Spl) of SHIP–/– (▪) and WT (▴) mice. (C) Histogram of DNA content in SHIP–/– and WT KTLS cells. (D) Bar graph showing the percentage of SHIP–/– (▪) or WT (▦) KTLS/KFLS cells in each stage of cell cycle as calculated using the Watson Pragmatic model in the FlowJo cell-cycle platform. Statistical analysis was performed using Prism 4. Significance was established using the unpaired student t test (Prism 4; GraphPad Software, San Diego, CA). **P < .001 and *P < .01 (mean ± SEM; n ≥ 3).
Since SHIP has been shown to negatively regulate proliferation of different cell types,13 SHIP deficiency could lead to an increase in HSC cell-cycle activity. In agreement with this, we observe that SHIP–/– BM contains a greater proportion of KTLS in the S/G2 phase (23.2% ± 1.5% compared with 14.1% ± 0.1% for WT BM; Figure 1C). This study directly demonstrates that SHIP–/– HSCs themselves have increased cycling activity relative to WT. This is consistent with findings of Helgason et al15 that CRUs in SHIP–/– BM are more sensitive to 5-fluorouracil treatment. In SHIP–/– BM, we also observe a significant decrease in the proportion of HSCs in the quiescent (G0/G1) stage. Quiescent HSCs are enriched for long-term multilineage engraftment relative to those that have entered the cell cycle.31
SHIP–/– BM cells show decreased ability to reconstitute the hematopoietic compartment of lethally irradiated recipients
It was previously reported that SHIP–/– BM cells have a lower ability to reconstitute irradiated mice after transplantation, but contains similar CRU activity compared with WT BM when measured by limiting-dilution assay.15 Since these results are inconsistent with our finding that SHIP–/– BM contains increased number of HSCs (Figure 1), we addressed this discrepancy by measuring the ability of SHIP–/– HSCs to accomplish long-term multilineage repopulating activity. We first performed a well-defined CRU assay as described by Harrison25 to assess the level of repopulation activity in WBM. WBM is used for the CRU assay and therefore is not dependent on isolation of HSCs based on cell-surface markers, whose expression can be altered by genetic mutation of unrelated loci.32 Using this assay we observe that SHIP–/– BM cells do not reconstitute recipients as efficiently as WT littermates, with a significant reduction of 4.4-fold in CRU numbers in primary transplantation recipients (Figure 2A-B). SHIP–/– BM cells show a significant decrease in long-term reconstitution of both the lymphoid and myeloid cell lineages. For example, 16 weeks after transplantation only 13.2% ± 2.7% of B cells are SHIP–/– derived (CD45.2+) compared with 60.1% ± 2.8% for WT (CD45.1+) WBM, T-cell repopulation is 21.2% ± 4.7% SHIP–/– derived versus 39.7% ± 4.1% for WT and 34.7% ± 5.2% of myeloid cells are SHIP–/– BM derived versus 57.4% ± 2.8% from WT (Figure 2C). The reduction in the SHIP–/– B-cell representation observed in the CRU assay agrees with other studies showing that SHIP deficiency negatively impacts early B-lineage development.33 We do not observe that SHIP–/– BM cells have enhanced myeloid repopulation relative to WT BM cells (Figure 2C). However, we do observe a bias toward myeloid differentiation within cells derived from SHIP–/– BM, where 59.7% ± 2.7% of the CD45.2+ in the SHIP–/– WBM recipients are Mac1+/Gr1+ compared with 24.1% ± 2% for WT BM recipients. This occurred at the expense of the B-cell lineage, where only 25.3% ± 1.5% of CD45.2+ cells were B220+ compared with 61.8% ± 3.1% for WT (data not shown). The overall reduction in long-term multilineage repopulation by SHIP–/– BM cells demonstrates that despite the increase in HSC numbers observed (Table 1 and Figure 1A-B), HSC activity is significantly compromised in these mice (Figure 2A,C).
SHIP–/– WBM and purified HSCs have compromised reconstituting activity. (A) CRU activity calculated based on the percentage of global repopulation in the PB by donor WBM cells (SHIP–/– [▪] and WT [▦]). (B) Percentage of global repopulation of PB by SHIP–/– (▪) and WT (▦) donor in a CRU assay. (C) Percentage of lymphoid and myeloid PB cells derived from SHIP–/– (▪) or WT (▦) WBM 16 weeks after transplantation in a CRU assay. (D) FACS plots show the level of PB reconstitution 16 weeks after KTLS transplantation in a representative DC assay mouse using sorted SHIP–/– and WT KTLS cells. (E) Percentage of global reconstitution 16 weeks after transplantation of sorted KTLS (SHIP–/– [▪] and WT [▦]) (n = 11 over 2 different experiments). (F) Proportion of lymphoid and myeloid PB cells derived from SHIP–/– (▪) or WT (▦) WBM 16 weeks after KTLS transplantation. (G) Percentage of global reconstitution 12 weeks after sorted KFLS transplantation (SHIP–/– [▪] and WT [▦]) (n = 7). Significance was established using the unpaired student t test (Prism 4): **P < .001; *P < .01; ++P < .005; and +P < .05 (mean ± SEM).
SHIP–/– WBM and purified HSCs have compromised reconstituting activity. (A) CRU activity calculated based on the percentage of global repopulation in the PB by donor WBM cells (SHIP–/– [▪] and WT [▦]). (B) Percentage of global repopulation of PB by SHIP–/– (▪) and WT (▦) donor in a CRU assay. (C) Percentage of lymphoid and myeloid PB cells derived from SHIP–/– (▪) or WT (▦) WBM 16 weeks after transplantation in a CRU assay. (D) FACS plots show the level of PB reconstitution 16 weeks after KTLS transplantation in a representative DC assay mouse using sorted SHIP–/– and WT KTLS cells. (E) Percentage of global reconstitution 16 weeks after transplantation of sorted KTLS (SHIP–/– [▪] and WT [▦]) (n = 11 over 2 different experiments). (F) Proportion of lymphoid and myeloid PB cells derived from SHIP–/– (▪) or WT (▦) WBM 16 weeks after KTLS transplantation. (G) Percentage of global reconstitution 12 weeks after sorted KFLS transplantation (SHIP–/– [▪] and WT [▦]) (n = 7). Significance was established using the unpaired student t test (Prism 4): **P < .001; *P < .01; ++P < .005; and +P < .05 (mean ± SEM).
Measuring HSC activity by transplantation of WBM from mutant mice can be confounded by the fact that the mutation can endow lineage-committed progenitor cells with properties that may obscure the observation of compromised HSC function.34 This is a concern when working with SHIP–/– BM as the SHIP mutation enhances myelopoiesis, leading to an increase in myeloid progenitors with enhanced survival.35 Therefore, analysis of HSC activity in SHIP–/– mice must also be assessed with purified HSCs to rule out these potential experimental artifacts. To directly assess whether SHIP expression by HSCs is required for long-term multi-lineage repopulation and self-renewal, we directly compared the repopulating potential of purified HSCs from SHIP–/– and WT mice. Thus, CD45.2 SHIP–/– KTLS or KFLS cells were transplanted into lethally irradiated CD45.1/CD45.2 WT recipients with an equal number of CD45.1 WT KTLS or KFLS and Sca1–CD45.1/CD45.2 support cells. This assay, the DC assay,26 directly compares the capacity of genetically modified HSCs to compete with WT HSCs for engraftment and long-term multilineage repopulation. The level of repopulation in recipient PB was assessed at regular intervals (Figure 2D), and at 16 to 20 weeks after transplantation, we observe that global repopulation by SHIP–/– KTLS is reduced by 15-fold relative to WT KTLS (Figure 2E). Different lineage reconstitution by SHIP–/– KTLS is also significantly reduced (Figure 2F). Thus, SHIP–/– KTLS cells are capable of multilineage reconstitution, but are unable to sustain this activity for extended periods when in direct competition with WT KTLS (Figure 2F). Furthermore, SHIP–/– cells do not dominate the myeloid compartment as might have been expected from previous studies,15,35 with only 1% of Mac1+/Gr1+ cells being derived from SHIP–/– KTLS versus 21% from WT KTLS (Figure 2F). This indicates that although SHIP–/– myeloid lineage progenitors have enhanced survival and can prevail over serially transplanted competitors,15,35 they are, nonetheless, unable to outcompete normal WT myeloid progenitors when derived from purified HSCs in a chimeric transplant setting. As with KTLS cells, long-term global repopulation by SHIP–/– KFLS is significantly reduced (Figure 2G). These results demonstrate that SHIP expression is required by HSCs in order to sustain long-term multilineage repopulation.
SHIP–/– HSCs have a lower rate of spontaneous apoptosis
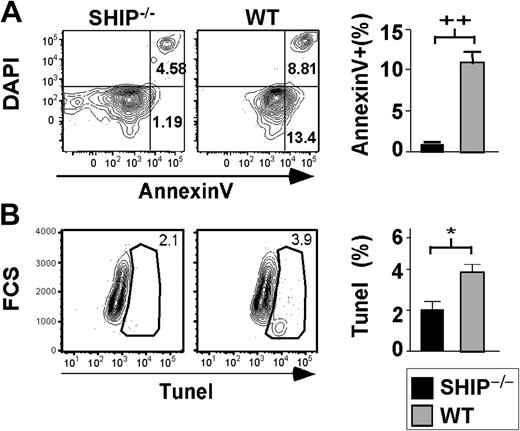
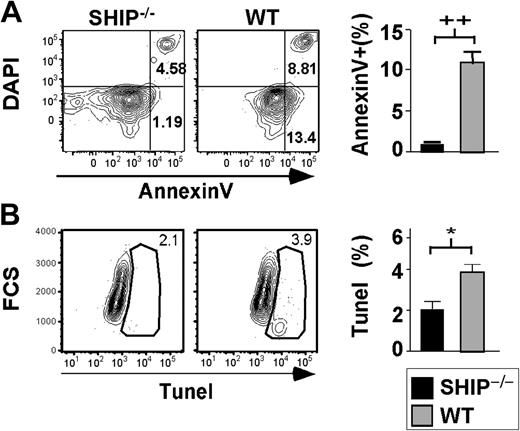
It was reported that SHIP–/– mice have increased osteoporosis caused by an increase in osteoclast activity.36 The HSC niche is dependent on the presence of osteoblasts, the counterpart of osteoclasts, to properly sustain the hematopoietic compartment.2 Thus, any disruption of the balance between osteoblasts and osteoclasts could destroy the niche necessary for HSC survival, resulting in anoikis-associated apoptosis of the HSC compartment. If SHIP–/– HSCs exhibited increased apoptosis, this could contribute to the decreased ability of SHIP–/– HSCs to accomplish long-term repopulation. Therefore, we assessed the level of apoptosis in SHIP-deficient HSCs. We find that a significantly lower proportion of SHIP–/– HSCs are apoptotic relative to WT HSCs as measured by Annexin V staining (Figure 3A). In fact, 11.1% of WT HSCs (KTLSFlk2–) stained with Annexin V, while only 1.1% of SHIP–/– HSCs did. TUNEL analysis done on SHIPΔIP/ΔIP HSCs also revealed that SHIP-deficient HSCs are undergoing apoptosis at half the rate of WT HSCs in the BM (Figure 3B). Thus, the lack of long-term repopulating activity of SHIP–/– HSCs activity appears not to be associated with anoikis-induced apoptosis of the compartment.
SHIP–/– HSCs exhibit decreased apoptotic rate. (A) Representative FACS plots of DAPI versus Annexin V after gating on KTLSFlk2– cells. Bar graph shows the percentage of KTLSFlk2– that are apoptotic based on the Annexin V+ and DAPI– staining: SHIP–/– (▪); WT (▦). (B) Representative FACS plots for TUNEL staining after gating on KTLS cells. Bar graph represents the percentage of KTLS cells positive for TUNEL staining: SHIP–/– (▪); WT (▦). Significance was established using the unpaired student t test (Prism 4): *P < .01; ++P < .005 (mean ± SEM; n ≥ 3).
SHIP–/– HSCs exhibit decreased apoptotic rate. (A) Representative FACS plots of DAPI versus Annexin V after gating on KTLSFlk2– cells. Bar graph shows the percentage of KTLSFlk2– that are apoptotic based on the Annexin V+ and DAPI– staining: SHIP–/– (▪); WT (▦). (B) Representative FACS plots for TUNEL staining after gating on KTLS cells. Bar graph represents the percentage of KTLS cells positive for TUNEL staining: SHIP–/– (▪); WT (▦). Significance was established using the unpaired student t test (Prism 4): *P < .01; ++P < .005 (mean ± SEM; n ≥ 3).
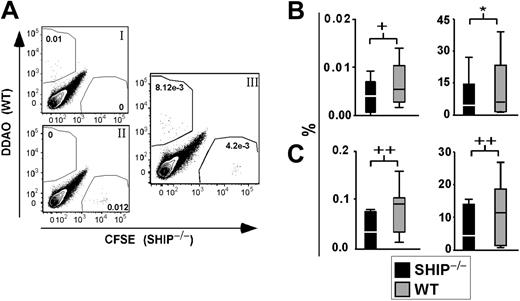
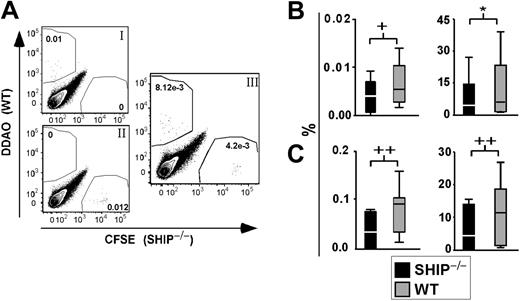
SHIP–/–Sca1+Lin– cells do not home to the BM as efficiently as WT Sca1+Lin– cells. (A) Representative FACS plot of BM from transplant recipients after gating on live cells. (Ai) BM from control recipient 12 to 14 hours after transplantation with 0.5 × 106 WBM stained with 1 μM DDAO. (Aii) BM from control recipient 12 to 14 hours after transplantation with 0.5 × 106 WBM stained with 0.5 μM CFSE. (Aiii) Representative FACS plot of BM from a mouse 12 to 14 hours after transplantation with 2 × 105 SHIP–/–Lin–Sca1+ cells stained with 0.5 μM CFSE and 2 × 105 WT Lin–Sca1+ cells stained with 1 μM DDAO. (B) Left panel shows percentage of dye+ SHIP–/– (▪) or WT(▦) Sca1+Lin– cells found in the recipient BM 12 to 14 hours after transplantation. Right panel shows percentage of SHIP–/– (▪) or WT (▦) Sca1+Lin– cells that trafficked to and were retained in BM of recipients over the total number of cells injected, 12 to 14 hours after transplantation. (C) Left panel shows percentage of stained SHIP–/– (▪) or WT (▦) Sca1+Lin– cells found in the spleen of recipients 12 to 14 hours after transplantation. Right panel shows percentage of SHIP–/– (▪) or WT (▦) Sca1+Lin– cells that reached the spleen of recipients over the total number of cells injected, 12 to 14 hours after transplantation. Significance was established using the stratified Wilcoxon-Mann Whitney test using StatXact (Cytel Software, Cambridge, MA): +P < .05; *P < .01; and ++P < .005 (mean ± SEM; n = 12).
SHIP–/–Sca1+Lin– cells do not home to the BM as efficiently as WT Sca1+Lin– cells. (A) Representative FACS plot of BM from transplant recipients after gating on live cells. (Ai) BM from control recipient 12 to 14 hours after transplantation with 0.5 × 106 WBM stained with 1 μM DDAO. (Aii) BM from control recipient 12 to 14 hours after transplantation with 0.5 × 106 WBM stained with 0.5 μM CFSE. (Aiii) Representative FACS plot of BM from a mouse 12 to 14 hours after transplantation with 2 × 105 SHIP–/–Lin–Sca1+ cells stained with 0.5 μM CFSE and 2 × 105 WT Lin–Sca1+ cells stained with 1 μM DDAO. (B) Left panel shows percentage of dye+ SHIP–/– (▪) or WT(▦) Sca1+Lin– cells found in the recipient BM 12 to 14 hours after transplantation. Right panel shows percentage of SHIP–/– (▪) or WT (▦) Sca1+Lin– cells that trafficked to and were retained in BM of recipients over the total number of cells injected, 12 to 14 hours after transplantation. (C) Left panel shows percentage of stained SHIP–/– (▪) or WT (▦) Sca1+Lin– cells found in the spleen of recipients 12 to 14 hours after transplantation. Right panel shows percentage of SHIP–/– (▪) or WT (▦) Sca1+Lin– cells that reached the spleen of recipients over the total number of cells injected, 12 to 14 hours after transplantation. Significance was established using the stratified Wilcoxon-Mann Whitney test using StatXact (Cytel Software, Cambridge, MA): +P < .05; *P < .01; and ++P < .005 (mean ± SEM; n = 12).
In vivo homing of SHIP–/– stem/progenitor cells to the BM is significantly reduced compared with WT
We considered that the diminished repopulation observed for SHIP–/– HSCs could result from an inefficiency of SHIP–/– HSCs to home and be retained in the BM niche. Thus, we assessed the homing capacity of SHIP–/– stem/progenitor cells compared with WT cells in an in vivo homing assay.27,28 Sca1+Lin– cells from the BM of SHIP–/– and WT mice were isolated, stained with fluorescent dyes, and injected into irradiated recipient mice. The frequency of SHIP–/– and WT Sca1+Lin– cells present in the recipient BM and spleen was later assessed by flow cytometry (Figure 4A). Using a total of 12 recipients for each genotype, we found that SHIP–/– stem/progenitor cells traffic to the BM with significantly reduced efficiency relative to WT stem/progenitor cells (Figure 4B). Furthermore, we observed that SHIP–/– Sca1+Lin– cells do not home to the spleen as well as WT Sca1+Lin– cells (Figure 4C). These results suggest that SHIP–/– HSCs are impaired in their ability to home and be retained in the BM. This deficiency likely contributes to their inability to engraft and sustain multilineage hematopoiesis.
SHIP–/– KTLS cells have lower surface densities of CXCR4 and VCAM-1
CXCR4, VCAM-1, and VLA-4 are all known to play a prominent role in homing and trafficking of HSCs and other hematopoietic cells to BM.17,19,37 Thus, we decided to examine the level of expression of these markers on SHIP–/– HSC cells compared with WT HSCs. By flow cytometry, we observed a 2.5-fold reduction in the surface expression of the CXCR4 receptor on SHIP–/– KTLS relative to WT KTLS (Figure 5A-B). Flow cytometry analysis revealed a reduction in surface expression of VCAM-1 on SHIP–/– HSC cells compared with WT HSCs (Figure 5A). We found that 22.1% ± 2.5% of SHIP–/– KTLS cells were positive for VCAM-1, while 50.8% ± 6.9% were positive for WT KTLS cells (Figure 5A-B). The VCAM-1 mean fluorescence intensity (MFI) values for SHIP–/– KTLS were also reduced by 3-fold compared with WT KTLS (Figure 5B). However, not all homing molecules showed a reduced level of expression. For example, the percentage of KTLS cells expressing VLA-4 as well as the MFI value in SHIP–/– BM cells was unchanged compared with WT BM. Furthermore, contrary to SHIP–/– KTLS, SHIP–/– Lin+cKit– cells did not exhibit a significant difference in the level of VCAM-1 and CXCR4 expression compared with WT Lin+cKit– cells (data not shown). Reduction of VCAM-1 and CXCR4 expression on the SHIP–/– HSCs suggests these cells may be hampered in their ability to traffic and be retained in the BM, consistent with the decreased BM homing and retention we observe (Figure 4).
SHIP–/–HSCs express lower levels of CXCR4 and VCAM-1 molecules as assessed by flow cytometry. (A) A representative histogram of CXCR4 (left) and VCAM-1 (right) expression on live (DAPI-) KTLS cells (isotype control [□]), SHIP–/– [▪], and WT [▦]). (B) The plots on the left represent the mean fluorescence intensity (MFI) of KTLS cells for CXCR4, VCAM-1, and VLA-4, respectively, and the plots on the right show the percentage of KTLS cells positive for VCAM-1 and VLA-4, respectively (SHIP–/– [▪], WT [▴]). Significance was established using the unpaired student t test (Prism 4): **P < .001 and +P < .05 (mean ± SEM; n ≥ 3).
SHIP–/–HSCs express lower levels of CXCR4 and VCAM-1 molecules as assessed by flow cytometry. (A) A representative histogram of CXCR4 (left) and VCAM-1 (right) expression on live (DAPI-) KTLS cells (isotype control [□]), SHIP–/– [▪], and WT [▦]). (B) The plots on the left represent the mean fluorescence intensity (MFI) of KTLS cells for CXCR4, VCAM-1, and VLA-4, respectively, and the plots on the right show the percentage of KTLS cells positive for VCAM-1 and VLA-4, respectively (SHIP–/– [▪], WT [▴]). Significance was established using the unpaired student t test (Prism 4): **P < .001 and +P < .05 (mean ± SEM; n ≥ 3).
Discussion
In this study, we show that SHIP–/– mice have more HSCs in their BM, spleen, and PB than WT littermates based on analysis using multiple HSC phenotypes. An expansion of the HSC compartment is also observed in a second SHIP-deficient model, where the inositol phosphatase region was targeted, the SHIPΔIP/ΔIP.20 This latter model is deficient for SHIP expression and, presumably stem-SHIP (sSHIP) expression, contrary to the SHIP–/– model. sSHIP is an isoform expressed in embryonic stem (ES) cells and HSCs but not in differentiated cells.12 Our studies show that SHIPΔIP/ΔIP HSCs have a similar phenotype to the SHIP–/– HSCs compared with WT: expansion of the HSC compartment, decreased apoptosis rate, and decreased level of CXCR4 expression. According to our data, the ablation of both isoforms does not lead to a more severe phenotype; however, additional studies should be done to further assess the role of sSHIP in HSCs.
The increase in HSC numbers observed in SHIP-deficient mice may result partly from the increased proliferation and decreased apoptotic frequency observed in the SHIP–/– HSC compartment. These results are consistent with SHIP being a negative regulator of several signaling pathways downstream of receptors that impact HSC proliferation and survival, such as c-mpl, interleukin-3R (IL-3R), CXCR-4, Flt-3, and c-Kit, which induce cell proliferation and/or survival through activation of the mitogen-activated protein kinase (MAPK) and PI3K/Akt pathways.10,38-46 Moreover, SHIP–/– myeloid and hematopoietic progenitor cells are hyperresponsive to several of these factors.13 The enhanced survival and proliferation of SHIP–/– HSCs could partially explain the decreased repopulating ability of these cells, as it was shown that cycling HSCs engraft less efficiently than resting HSCs.31,47,48
Homing and eventual engraftment of HSCs following transplantation has been shown to rely on the expression and activity of several surface molecules, including VLA-4, VLA-5, and CXCR4.17,49,50 The CXCR4 receptor induces cell migration toward an increasing gradient of SDF-1/CXCL12.51 Treatment of nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice with SDF-1 leads to the mobilization of HSCs to PB,52 and treatment of human HSCs with anti-CXCR4 antibody prior to transplantation in NOD/SCID mice results in HSC engraftment failure.17 In addition, one of the ligands for VLA-4, VCAM-1, which is usually found on stromal/endothelial cells, has been shown to be expressed in hematopoietic cells such as Mac-1+, B220+, and, more interestingly, cKit+ cells.19 Even though the role of VCAM-1 on hematopoietic stem/progenitor cells remains to be fully elucidated, there is evidence that it is involved in their retention in the BM, as shown for B-cell progenitors.19 With that information in mind, the reduction in the percentage of HSCs expressing CXCR4 and VCAM-1 receptors in SHIP–/– BM can provide a basis for the decrease in SHIP–/– HSCs ability to home to the BM, as shown by our in vivo homing assays (Figure 4B). This inefficiency of SHIP–/– HSCs to reach and be retained in the HSC niche could contribute to their impaired self-renewal and long-term repopulating capacity. In previous studies, in vitro stimulation of murine BM stem/progenitor cells with different cytokines was shown to induce cell cycle, decrease VLA-4 expression, and compromise engraftment.47 In our study, the increase in HSC cell cycle observed in vivo did not necessarily correlate with a decrease in VLA-4 expression, but caused a decline in long-term engraftment capacity. We observed that decreased CXCR4 and VCAM-1 expression on KTLS cells correlates with a reduction in the homing process. Moreover, CXCR4 has been shown to participate in the activation of VLA-4 and VLA-5 to promote human HSC attachment and extravasation, resulting in homing of HSCs to the BM niche.50 The reduction in the CXCR4 receptor seen on SHIP–/– HSCs might also dampen this process and, thus, HSC homing. Furthermore, it has been shown that CXCR4 and VCAM-1 are important for the retention of HSCs or hematopoietic progenitors in the BM.18,19 Interestingly, we find that SHIP–/– mice have an increased number of HSC cells in their spleen (Table 1; Figure 1A-B) and peripheral blood (data not shown), consistent with the down modulation of CXCR4 and VCAM-1 on SHIP–/– HSCs.
Even though we observed a reduction in engraftment by both the DC and CRU assay, the defect seems greater in the DC assay, where equal numbers of purified SHIP–/– and WT HSCs are cotransplanted. This suggests that in the CRU assay, where WBM cells are used, the increased number of HSCs in SHIP–/– BM might partially compensate for the reduced levels of CXCR4 and VCAM-1 on SHIP–/– HSCs, resulting in better engraftment by SHIP–/– HSCs in the CRU assay compared with the DC assay.
The CRU assay included in this study showed a decrease in the repopulating ability of SHIP–/– WBM, while previous studies revealed that SHIP–/– WBM had comparable repopulating activity in a limiting-dilution CRU assay.15 The different results observed between the 2 CRUs might stem from the variation in the assays themselves. In the Harrison CRU assay, healthy competitor cells are used, while the limiting-dilution CRU assay requires the use of competitor WBM weakened by 2 serial transplantations.53 Thus, the compromised WBM competitors in the latter CRU assay may be unable to compete for the niche as efficiently as normal WT WBM against SHIP–/– WBM. Furthermore, the calculation method for the Harrison CRU assay differs from the one used for the limiting-dilution assay, which might result in a different assessment of repopulating activity by the 2 assays.
In summary, our findings demonstrate a role for SHIP in the maintenance of the HSC compartment. SHIP appears to be critical for the negative control of HSC proliferation and survival as well as impacting the ability of HSCs to home and be retained in the BM niche.
Prepublished online as Blood First Edition Paper, February 7, 2006; DOI 10.1182/blood-2005-12-5021.
Supported in part by grants from the National Institutes of Health (RO1 DK54767, PO1 NS27405, and RO1 HL72523), and the academic development funds from Moffitt Cancer Center and the University of South Florida. W.G.K. is the Newman Scholar of the Leukemia and Lymphoma Society of America.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We appreciate discussions with John Ninos, MD. We thank Steven L. Highfill, Amy Costello, Joe A. Wahle, Daniela S. Wood, and Meghan Rieger for technical assistance; and Davina Mulchan and Sarah May for genotyping. We thank Scott N. Freeman for revision of the manuscript. We thank the staff of the Flow Core and Alan Cantor, PhD, from the Biostatistic Core at the H. Lee Moffitt Cancer Center and Research Institute.

![Figure 2. SHIP–/– WBM and purified HSCs have compromised reconstituting activity. (A) CRU activity calculated based on the percentage of global repopulation in the PB by donor WBM cells (SHIP–/– [▪] and WT [▦]). (B) Percentage of global repopulation of PB by SHIP–/– (▪) and WT (▦) donor in a CRU assay. (C) Percentage of lymphoid and myeloid PB cells derived from SHIP–/– (▪) or WT (▦) WBM 16 weeks after transplantation in a CRU assay. (D) FACS plots show the level of PB reconstitution 16 weeks after KTLS transplantation in a representative DC assay mouse using sorted SHIP–/– and WT KTLS cells. (E) Percentage of global reconstitution 16 weeks after transplantation of sorted KTLS (SHIP–/– [▪] and WT [▦]) (n = 11 over 2 different experiments). (F) Proportion of lymphoid and myeloid PB cells derived from SHIP–/– (▪) or WT (▦) WBM 16 weeks after KTLS transplantation. (G) Percentage of global reconstitution 12 weeks after sorted KFLS transplantation (SHIP–/– [▪] and WT [▦]) (n = 7). Significance was established using the unpaired student t test (Prism 4): **P < .001; *P < .01; ++P < .005; and +P < .05 (mean ± SEM).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/11/10.1182_blood-2005-12-5021/4/m_zh80110696400002.jpeg?Expires=1765057965&Signature=ZSAG-gkuxFr4CsYx~ljIOuYtTe9XMA72vzxCcQEKJsba78zDCWZQfRtH~AN49fHqvqj6MNFM61~gtu6WA2MmMzBifvREyRHG4kRqJmPjaOqkSZ8gR9WaCh4MzwQEl-sNUDUHSLzoQSavg62VJGsZPNbWaSGq5YP-hl27FJ3u0yw6SX2wbjTq7ZnRIsSAtS9p1rwl1I4S-W5JvcEvsmFJ4MK~JoCKkey-XvUkXnkuhjRnOxMUXJbG7hYNGm01f1s8vA5SspBzxeNmHeaeh5cU3Y4AYNlIksEbVbG1qA5R2dzUzDgbmjGljqtYSBujpXngWSMN584-sn1tU5F6YXF-dg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. SHIP–/–HSCs express lower levels of CXCR4 and VCAM-1 molecules as assessed by flow cytometry. (A) A representative histogram of CXCR4 (left) and VCAM-1 (right) expression on live (DAPI-) KTLS cells (isotype control [□]), SHIP–/– [▪], and WT [▦]). (B) The plots on the left represent the mean fluorescence intensity (MFI) of KTLS cells for CXCR4, VCAM-1, and VLA-4, respectively, and the plots on the right show the percentage of KTLS cells positive for VCAM-1 and VLA-4, respectively (SHIP–/– [▪], WT [▴]). Significance was established using the unpaired student t test (Prism 4): **P < .001 and +P < .05 (mean ± SEM; n ≥ 3).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/11/10.1182_blood-2005-12-5021/4/m_zh80110696400005.jpeg?Expires=1765057965&Signature=2z2FO1WVy~mReuJTF217BVu9Etzr7yRepbfs8A~LuG6kB5VcMvgGrTkYnRR0hTTKHyEnzQB3paymb-zCM9CorN-qYG-6P0leKVY~mQ-yEkxrYWPZrnFO3jwM9-FuN9APk4~jA7myKXc4AB11GCSD1yHvx3aRcLUhd-fWzMea-~CXkQNpZko3bRqFAPtsRu94V363k3p6tSKFZfWm~2KngDT7WBloBL1BPdDTicvC11TaQmgUM8I7LotejiewQqVQOjgBePPeoxi98MwxBOMEeAdez22-H02NOJcttb33omPuah-lxcGQxZys8AZzY602AUvIRmQtJtqfQxxIg0tZLA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 2. SHIP–/– WBM and purified HSCs have compromised reconstituting activity. (A) CRU activity calculated based on the percentage of global repopulation in the PB by donor WBM cells (SHIP–/– [▪] and WT [▦]). (B) Percentage of global repopulation of PB by SHIP–/– (▪) and WT (▦) donor in a CRU assay. (C) Percentage of lymphoid and myeloid PB cells derived from SHIP–/– (▪) or WT (▦) WBM 16 weeks after transplantation in a CRU assay. (D) FACS plots show the level of PB reconstitution 16 weeks after KTLS transplantation in a representative DC assay mouse using sorted SHIP–/– and WT KTLS cells. (E) Percentage of global reconstitution 16 weeks after transplantation of sorted KTLS (SHIP–/– [▪] and WT [▦]) (n = 11 over 2 different experiments). (F) Proportion of lymphoid and myeloid PB cells derived from SHIP–/– (▪) or WT (▦) WBM 16 weeks after KTLS transplantation. (G) Percentage of global reconstitution 12 weeks after sorted KFLS transplantation (SHIP–/– [▪] and WT [▦]) (n = 7). Significance was established using the unpaired student t test (Prism 4): **P < .001; *P < .01; ++P < .005; and +P < .05 (mean ± SEM).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/11/10.1182_blood-2005-12-5021/4/m_zh80110696400002.jpeg?Expires=1765258802&Signature=LqEmumYYzZSfX6hr64ROstWTxM2BfAUD6irZnEtG3dcbu7Te4qWUeLSf9etzgJS~By7Qcd8qtoUbUBUxvSlpILOGx3Tp1JdxUhJ4DZ7t4Fqg-f6DvmGcfhaNthLxjSCiSyAbprd-04B9OaXuwc~rzk4--g6rAhWKlCQAo56melQtdLsfXrYsGKJNbswgB5fyUZOImP89DdWQYlQD2KeZl1oBe8LOkMMa8UUBgqTibf2rQxIECQuThURg~BI3nFmjedv6No5Of7xxDUpKdvfjlA~FK2xGojWRYiVwuQ6-TI0Oq4WP0mkOH8niT039k905Aip6BR3EONhlwxUxnViJMA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. SHIP–/–HSCs express lower levels of CXCR4 and VCAM-1 molecules as assessed by flow cytometry. (A) A representative histogram of CXCR4 (left) and VCAM-1 (right) expression on live (DAPI-) KTLS cells (isotype control [□]), SHIP–/– [▪], and WT [▦]). (B) The plots on the left represent the mean fluorescence intensity (MFI) of KTLS cells for CXCR4, VCAM-1, and VLA-4, respectively, and the plots on the right show the percentage of KTLS cells positive for VCAM-1 and VLA-4, respectively (SHIP–/– [▪], WT [▴]). Significance was established using the unpaired student t test (Prism 4): **P < .001 and +P < .05 (mean ± SEM; n ≥ 3).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/11/10.1182_blood-2005-12-5021/4/m_zh80110696400005.jpeg?Expires=1765258802&Signature=nYMyvz94AG7K6Ks7-6ZGeeQsc37kJXhztIwuMw~6Hvp9w0bBw44TH2KUsO0~TNX1Dqp0lxFSQYiDIxeqzUbtTKDzEIYVE4Dh1Z16HvPxJtB~cT7Ie7ilIScQfbEznCgO5UukE07tHeC05fUxEBL1WPwNo5syEe6d6Uw48uFjKsplK-yOg4AE~n5RMXn3Re9Em538ulyvchDD8EG46mHJTvX3TSK2y58tdA6Lw~amQcKlxZkg~dCsfaw5MyAMvbM1ZEtffkkwUnjgzjmE5nplyx7hWN2EHVHRCMa72JxcThyxZcvzPg2lBeO-H1ginwsnYRkjEjGe06L2SLeeczs1lA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)