Abstract
Apoptosis plays a central role in the regulation of the size of the hematopoietic stem cell pool as well as in the processes of cell differentiation along the various hematopoietic lineages. TRAIL is a member of the TNF family of cytokines with a known apoptogenic role against a variety of malignant cells and an emerging role in the modulation of normal hematopoiesis. Here we worked on the hypothesis that PKCϵ could act as a switch of the cellular response to TRAIL during erythropoiesis. We demonstrate that EPO-induced erythroid CD34 cells are insensitive to the apoptogenic effect of TRAIL at day 0 due to the lack of specific receptor expression. From day 3 onward, erythroid cells express surface death receptors and become sensitive to TRAIL up to day 7/8 when, notwithstanding death-receptor expression, the EPO-driven up-regulation of PKCϵ intracellular levels renders differentiating erythroid cells resistant to TRAIL likely via Bcl-2 up-regulation. Our conclusion is that in human CD34 cells, EPO promotes a series of events that, being finely regulated in their kinetics, restricts the sensitivity of these cells to TRAIL to a specific period of time, which therefore represents the “TRAIL window” for the negative regulation of erythroid-cell numbers.
Introduction
It is now well established that apoptosis plays a central role in the regulation of the size of the hematopoietic stem cell pool1 as well as in the processes of cell differentiation along the various hematopoietic lineages. Fetal erythropoiesis can be negatively regulated by Fas ligand,2,3 although bone marrow hematopoiesis does not appear to be affected by Fas deficiency.3 However, Fas expression on hematopoietic stem cells or hematopoietic progenitors can be induced by certain cytokines such as IFNγ or tumor necrosis factor α (TNFα),3-5 reducing hematopoietic repopulating potential. Relatively little is known on the effects of other members of the TNF family on hematopoietic progenitors. We have demonstrated that TNF-related apoptosis-inducing ligand (TRAIL) acts as a negative regulator of adult erythropoiesis, selectively reducing the number of erythroblasts in liquid culture, as well as reducing the number and size of erythroid colonies in semisolid assays.6 Recently, Secchiero et al demonstrated that TRAIL inhibited the generation of mature erythroblasts in liquid culture through the activation of an ERK 1/2–mediated signaling pathway.7
TRAIL is a member of the TNF family of cytokines, which are structurally related proteins playing important roles in regulating cell death, immune response, and inflammation.8,9 The unique feature of TRAIL, compared with other members of the TNF family, is its ability to induce apoptosis in a variety of malignant cells both in vitro and in vivo, displaying minimal toxicity on normal cells and tissues.10,11 TRAIL interacts with 4 high-affinity transmembrane receptors belonging to the apoptosis-inducing TNF-receptor (R) family. TRAIL-R1 (DR4) and TRAIL-R2 (DR5) transduce apoptotic signals on binding of TRAIL, whereas TRAIL-R3 (DcR1) and TRAIL-R4 (DcR2) are homologs to DR4 and DR5, but they lack the intracellular death domain and apoptosis-inducing capability. It has been proposed that TRAIL-R3 and TRAIL-R4 function as decoy receptors protecting normal cells from apoptosis.12,13
Most normal human cell types tested to date, including bone, epithelial, endothelial, fibroblastic, and smooth muscle cells, are refractory to TRAIL. Nevertheless, TRAIL can induce hepatocyte apoptosis,14 as well as cell damage in the human prostate15 and brain.16 The earliest biochemical event following engagement of TRAIL death receptors by their ligand is the recruitment of proteins to the intracellular death domain of the receptor to form a structure known as the death-inducing signaling complex (DISC).17
Of the several known isoforms of protein kinase C, we have demonstrated that PKCϵ is selectively posttranscriptionally down-modulated in the EPO-dependent murine 32D-Epo.1 cells, while it is expressed in the parental cell line 32D as well as in the 32D-GM1 and 32D-G1 cells with granulomacrophagic and granulocytic phenotype, respectively. The subsequent observation that the pharmacologic inhibition of PKCϵ increased the number of erythroid colonies in vitro strongly suggested a relevant role for this isoform of PKC in erythropoiesis.18 Previous observations19 had already established a link between PKCϵ and apoptosis in different model systems. On the basis of the observation that PKCϵ up-regulation increased the formation and growth rate of tumors in nude mice,20 Gubina et al21 demonstrated that PKCϵ prevents apoptosis of the factor-dependent TF-1 cells cultured in the absence of cytokines via Bcl-2 up-regulation.
Given this complex background, we decided to investigate the potential role of PKCϵ in the protection against TRAIL activity during human erythropoiesis.
Materials and methods
CD34+ cell purification
Primary CD34+ cells were isolated from peripheral blood of healthy donors by immunomagnetic positive selection using the CD34+ cell isolation Kit (Miltenyi Biotech, Gladbach, Germany) in the magnetic field of a Vario-MACS apparatus (Miltenyi Biotech), according to the manufacturer's protocol. Purity of CD34+ cells was immediately checked by anti–CD34-PE mAb (Beckman Coulter, Miami, FL) and flow cytometry. Only samples exceeding 95% purity were used for subsequent experiments.
Cell cultures and treatment
Purified human CD34+ cells were cultured up to 18 days, at an optimal cell density of 1 × 106 cell/mL, in serum-free X-vivo medium supplemented with 3 ng/mL recombinant human interleukin-3 (rIL-3) and 40 ng/mL stem cell factor (SCF), with or without 5 U/mL erythropoietin (EPO). Cytokines were readded every 3 days, up to 18 days.
HeL, K562, and TF-1 cell lines were grown in 10% FBS-enriched RPMI medium at the optimal density of 0.5 × 106 cells/mL. TF-1 cells were maintained in the presence of 3 ng/mL IL-3.
IL-3, recombinant human SCF, and recombinant human EPO were all from PeproTech (London, United Kingdom). As a PKCϵ inhibitor, we used the translocation inhibitor H-Glu-Ala-Val-Ser-Leu-Lys-Pro-Thr-OH at 250 μg/mL for 48 hours (Calbiochem, San Diego, CA).
Flow cytometric analysis
Aliquots of 0.3 × 106 cells/experimental point were labeled by a panel of anti–TRAIL-R MoAbs (Alexis Biochemical, San Diego, CA). Expression of TRAIL-R1, TRAIL-R2, TRAIL-R3, and TRAIL-R4 was analyzed by indirect staining using 1 μg HS101 anti–human TRAIL-R1, HS201 anti–human TRAIL-R2, HS301 anti–human TRAIL-R3, and HS401 anti–human TRAIL-R4 monoclonal antibodies, followed by PE-labeled goat anti–mouse IgG (Beckman Coulter) as a second reagent. Analysis was performed by an Epics XL flow cytometer (Beckman Coulter) and Expo ADC software (Beckman Coulter).
siRNA design and transfection
Double-strand siRNAs (dsRNA) were designed to target sequences corresponding to nt's 223 to 244, 429 to 450, 942 to 963, and 1158 to 1179 on human PKCϵ mRNA (NM005400). The target sequences were as follows: 5′-AAGAT CAAAA TCTGC GAGGCC-3′, 5′-AAGAT CGAGC TGGCTG TCTTT-3′, 5′-AACTA CAAGG TCCCT ACCTTC-3′, and 5′-AAAAA GCTCA TTGCT GGTGCC-3′.
The respective sense and antisense RNA sequences were synthesized by Silencer siRNA Construction Kit (Ambion, Austin, TX).22 Nonspecific siRNA duplexes containing the same nucleotides, but in irregular sequence (ie, scrambled PKCϵ siRNA), were prepared according to the manufacturer's protocol and used as controls.
The GFP-PKCϵ expression and control plasmid were kindly provided by Professor Peter Parker (Cancer Research UK, London Research Institute).
To maximize transfection efficiency, siRNAs (100 nM each) and GFP-PKCϵ plasmids (1 μg/transfection) were delivered using the Amaxa nucleofection technology (Amaxa, Koeln, Germany) according to the manufacturer's protocols.
Semiquantitative reverse-transcriptase–polymerase chain reaction (RT-PCR) analysis
Total RNA was isolated using the RNeasy mini kit (Qiagen, Hilden, Germany). Total RNA (1 μg) was reverse transcribed with Malone murine leukemia virus (MMV) reverse transcriptase, and progressive dilutions (1/10, 1/100, 1/1000, 1/10 000) were subjected to PCR amplification to detect β-actin and PKCϵ cDNA.
PCR was performed under the following reaction conditions: 95°C for 30 seconds, 56°C for 30 seconds, 72°C for 30 seconds, and a final extension at 72°C for 5 minutes. We used 35 cycles of amplification. The sequence of primers used for PCR was as follows: β-actin, 5′-TGACG GGGTC ACCCA CACTG TGCCC ATCTA-3′ (sense) and 5′-CTAGA AGCAT TTGCG GTGGA CGATG GAGGG-3′ (antisense); PKCϵ, 5′-CAATGGC CTTCTTAAG ATCAAAA-3′ (sense) and 5′-CCTGA GAGATC GATGATC ACATAC-3′ (antisense).
Western blot
Cultured cells were counted and 2 × 106 cells were collected at specific time points, washed in PBS, and centrifuged at 200g for 10 minutes. Pellets were resuspended in a cell-lysis buffer (50 mM Tris-HCl, pH 7.4; 1% NP-40; 0.25% sodium deoxycholate; 150 mM NaCl; 1 mM EDTA; 1 mM PMSF; 1 mM Na3VO4; 1 mM NaF) supplemented with fresh protease inhibitors, and protein concentration was determined using BCA protein assay kit (Pierce, Rockford, IL). Proteins from each sample (14 μg) were then migrated in 5% SDS–acrylamide gels and blotted onto nitrocellulose filters.
Blotted filters were blocked and incubated with specific primary antibodies diluted as described in the manufacturers' protocols. Specifically, rabbit polyclonal anti-PKCϵ and anti–phospho-PKCϵ antibodies (Upstate, Lake Placid, NY) were used at the concentration of 1 μg/mL. MoAbs anti-PKCδ (Becton Dickinson, Heidelberg, Germany), anti–Bcl-2 (Santa Cruz Biotechnology, Santa Cruz, CA), anti–β-actin, and anti–β-tubulin (Sigma, St Louis, MO) were diluted 1:500, 1:50, 1:5000, and 1:20 000, respectively. Anti–Bax rabbit polyclonal antibody (Cell Signaling Technology, Beverly, MA) was diluted 1:1000 before use.
Filters were washed and further incubated for 1.5 hours at room temperature with 1:5000 peroxidase-conjugated anti–rabbit or with 1:2000 peroxidase-conjugated anti–mouse IgG (Pierce) in the primary antibody working solution at room temperature. Specific reactions were revealed with the ECL Supersignal West Pico Chemiluminescent Substrate detection system (Pierce).
Assessment of apoptosis
Cell-culture viability was assessed by trypan blue exclusion. Apoptotic cells were identified by flow cytometry as subdiploid peaks generated either by DNA fragmentation or by annexin V/PI staining. Briefly, cells were permeabilized by ethanol in the presence of RNAse H buffer and stained with 50 μg/mL propidium iodide, or phosphatidylserine was stained by FITC conjugate annexin V (ACTIPLATE; Valter Occhiena, Torino, Italy) in Ca2+ and PI staining buffer, following the manufacturer's protocol.
Results
TRAIL induces apoptosis in human erythromyeloid cell lines
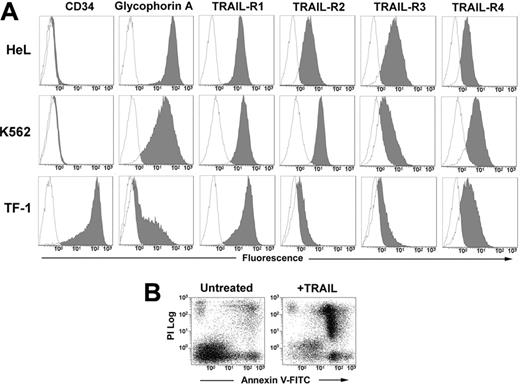
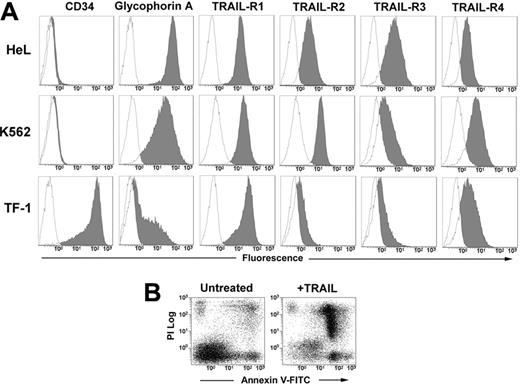
The human erythromyeloid cell lines HeL, K562, and TF-1 all expressed at different levels surface glycophorin A. The phenotypic analysis with anti–TRAIL-receptor (TRAIL-R) moAbs revealed that the 3 cell lines express all the receptors, with a very neat expression of death receptors R1 and R2 in HeL and K562 cells, and a lower expression of R2 in TF-1 cells (Figure 1A). After 48 hours of treatment with 50 ng/mL TRAIL, apoptosis was induced in all 3 cell lines (HeL: 52.3% ± 9.1; K562: 49.6% ± 5.0; and TF-1: 50.2% ± 9.1; data are expressed as mean percentages of annexin V+ cells ± SD of 3 independent experiments) as detected by annexin V–FITC/PI staining and flow cytometry (Figure 1B).
PKCϵ reduces TRAIL-mediated apoptosis in human erythromyeloid cell lines
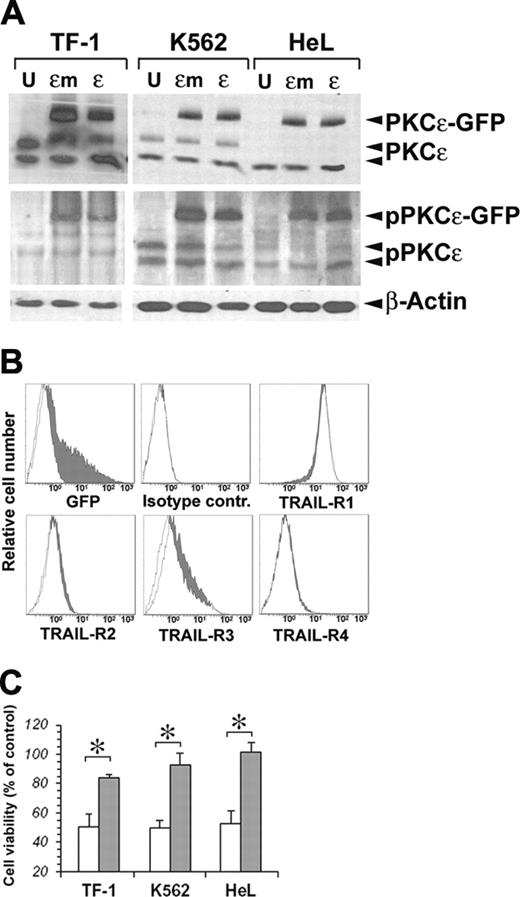
On the basis of the known prosurvival effects of PKCϵ on TF-1 cells21 and its protection against TRAIL-mediated apoptosis in glioma cells,23 we performed a series of experiments to investigate whether PKCϵ could modulate the sensitivity of erythromyeloid cell lines to the apoptogenic effects of TRAIL. For this purpose, we first overexpressed PKCϵ in our cell lines, using as negative control an inactive PKCϵ K522M mutated (PKCϵm) kinase.24 As shown in Figure 2A, both PKCϵ and PKCϵm could be very well overexpressed in the 3 cell lines, and both the wild-type and mutated forms of the enzyme could be phosphorylated, as revealed by immunoblotting with anti–phospho-PKCϵ antibody. As PKCϵ overexpression did not modify the surface expression of TRAIL-Rs in either cell line (Figure 2B), we treated transfected and mock K562, HeL, and TF-1 cell lines with 50 ng/mL TRAIL for 48 hours. Figure 2C shows a significant reduction of TRAIL-mediated apoptosis in all 3 cell lines overexpressing PKCϵ, but not PKCϵm.
Phenotype of erythroleukemic cell lines and their sensitivity to TRAIL. (A) HeL, K562, and TF-1 cells were stained with anti-CD34, anti–glycophorin A, and with specific MoAbs to TRAIL-R1, TRAIL-R2, TRAIL-R3, and TRAIL-R4 as described in “Materials and methods.” Specific fluorescence histograms (gray) are superimposed to negative controls (empty histograms). (B) TRAIL-induced apoptosis of TF-1 cells treated for 48 hours with 50 ng/mL TRAIL. Apoptosis was detected by staining cells with annexin V–FITC and PI.
Phenotype of erythroleukemic cell lines and their sensitivity to TRAIL. (A) HeL, K562, and TF-1 cells were stained with anti-CD34, anti–glycophorin A, and with specific MoAbs to TRAIL-R1, TRAIL-R2, TRAIL-R3, and TRAIL-R4 as described in “Materials and methods.” Specific fluorescence histograms (gray) are superimposed to negative controls (empty histograms). (B) TRAIL-induced apoptosis of TF-1 cells treated for 48 hours with 50 ng/mL TRAIL. Apoptosis was detected by staining cells with annexin V–FITC and PI.
TRAIL induces apoptosis of CD34-derived erythroblasts
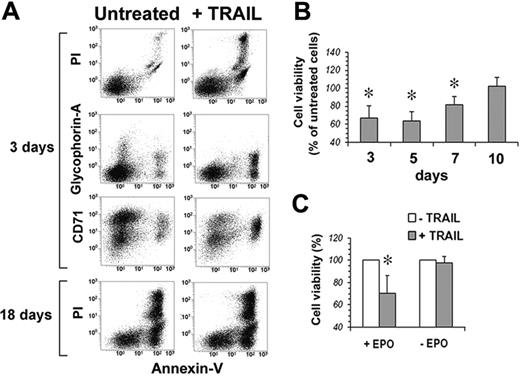
Human peripheral-blood–purified CD34 cells were first analyzed for the surface expression of TRAIL-Rs. Figure 3 shows that none of the 4 TRAIL-Rs is expressed on freshly purified CD34 cells. However, when cultured in serum-free medium in the presence of IL-3, SCF, and EPO, starting from day 3 CD34 cells showed a progressive increase of the surface expression of TRAIL-R1 and TRAIL-R2. On the contrary, TRAIL decoy receptors were hardly expressed at all, with the exception of a transient expression of TRAIL-R4 around day 13. Accordingly, when TRAIL was added to EPO-cultured CD34 cells, they were resistant to apoptosis at day 0. Starting at day 3, cells became sensitive to the apoptogenic effect of TRAIL (Figure 4A-B). Surprisingly, however, notwithstanding the stable surface expression of TRAIL death receptors (and in the virtual absence of TRAIL-decoy receptor expression), the sensitivity to TRAIL of EPO-differentiating CD34 cells decreased progressively from day 7 onward, and from day 10 erythroblasts became resistant to TRAIL (Figure 4B). Control cultures of purified CD34 cells in the absence of EPO were always resistant to the apoptogenic effect of TRAIL (Figure 4C), confirming that the sensitivity to TRAIL of normal CD34 cells between day 3 and day 7 was EPO dependent.
Transfection of PKCϵ and PKCϵm in erythroleukemic cell lines. (A) Detection of exogenous PKCϵ-GFP and endogenous wild-type PKCϵ protein by Western blot. Phosphorylated PKCϵ (pPKCϵ) was detected by specific C-terminal anti–phospho-Ser729 antibody. β-Actin was monitored for protein loading. (B) Cell-surface expression of TRAIL-Rs after PKCϵ transfection. TF-1 cells were transfected with PKCϵ-GFP (filled histograms) or PKCϵm-GFP (open histograms), and TRAIL-R expression was monitored by flow cytometry 48 hours later. (C) PKCϵ reduces the sensitivity of erythroleukemic cell lines to TRAIL-induced apoptosis. K562, HeL, and TF-1 cell lines were transfected with PKCϵ (▪) or with PKCϵm (□) and 48 hours later were treated with 50 ng/mL TRAIL. Residual cell viability was analyzed by flow cytometry, staining cells with annexin V–FITC and PI. The mean of 3 independent experiments is reported as percentage of control. Controls were represented by mock-transfected cell lines treated with TRAIL. *P < .05.
Transfection of PKCϵ and PKCϵm in erythroleukemic cell lines. (A) Detection of exogenous PKCϵ-GFP and endogenous wild-type PKCϵ protein by Western blot. Phosphorylated PKCϵ (pPKCϵ) was detected by specific C-terminal anti–phospho-Ser729 antibody. β-Actin was monitored for protein loading. (B) Cell-surface expression of TRAIL-Rs after PKCϵ transfection. TF-1 cells were transfected with PKCϵ-GFP (filled histograms) or PKCϵm-GFP (open histograms), and TRAIL-R expression was monitored by flow cytometry 48 hours later. (C) PKCϵ reduces the sensitivity of erythroleukemic cell lines to TRAIL-induced apoptosis. K562, HeL, and TF-1 cell lines were transfected with PKCϵ (▪) or with PKCϵm (□) and 48 hours later were treated with 50 ng/mL TRAIL. Residual cell viability was analyzed by flow cytometry, staining cells with annexin V–FITC and PI. The mean of 3 independent experiments is reported as percentage of control. Controls were represented by mock-transfected cell lines treated with TRAIL. *P < .05.
TRAIL-R expression of CD34-derived erythroblasts. CD34 purified cells were cultured for 18 days in serum-free medium with EPO, SCF, and IL-3, showing a progressive expression of glycophorin A. At the indicated time intervals (T), TRAIL-R expression was evaluated by specific MoAb staining. Filled histograms indicate specific fluorescence; open histograms, isotype-matched irrelevant Ab, negative control.
TRAIL-R expression of CD34-derived erythroblasts. CD34 purified cells were cultured for 18 days in serum-free medium with EPO, SCF, and IL-3, showing a progressive expression of glycophorin A. At the indicated time intervals (T), TRAIL-R expression was evaluated by specific MoAb staining. Filled histograms indicate specific fluorescence; open histograms, isotype-matched irrelevant Ab, negative control.
TRAIL kills EPO-responsive CD34 cells. (A) Flow cytometric analysis of CD34 cells at day 3 and day 18 of culture (EPO, SCF, and IL-3) treated with 50 ng/mL TRAIL for 48 hours; residual cell viability was evaluated by staining cells with annexin V–FITC and PI, while cell differentiation was monitored by glycophorin A–PE and CD71-CY5 expression. (B) CD34-derived erythroblast sensitivity to TRAIL-induced apoptosis. At the indicated time intervals, cells were treated for 48 hours with 50 ng/mL TRAIL, and the percentage of residual cell viability was monitored by staining with annexin V–FITC and PI. Each histogram is the mean of 3 independent experiments expressed as percentages of control (CD34 cells cultured without TRAIL), *P < .05. (C) Sensitivity to TRAIL of CD34 cells cultured 3 days with IL-3 and SCF in serum-free medium with (+EPO) or without (–EPO) EPO. Data are expressed as percentages of TRAIL-untreated cells (3 independent experiments, *P < .05).
TRAIL kills EPO-responsive CD34 cells. (A) Flow cytometric analysis of CD34 cells at day 3 and day 18 of culture (EPO, SCF, and IL-3) treated with 50 ng/mL TRAIL for 48 hours; residual cell viability was evaluated by staining cells with annexin V–FITC and PI, while cell differentiation was monitored by glycophorin A–PE and CD71-CY5 expression. (B) CD34-derived erythroblast sensitivity to TRAIL-induced apoptosis. At the indicated time intervals, cells were treated for 48 hours with 50 ng/mL TRAIL, and the percentage of residual cell viability was monitored by staining with annexin V–FITC and PI. Each histogram is the mean of 3 independent experiments expressed as percentages of control (CD34 cells cultured without TRAIL), *P < .05. (C) Sensitivity to TRAIL of CD34 cells cultured 3 days with IL-3 and SCF in serum-free medium with (+EPO) or without (–EPO) EPO. Data are expressed as percentages of TRAIL-untreated cells (3 independent experiments, *P < .05).
EPO-mediated up-regulation of PKCϵ is responsible for TRAIL resistance of human CD34-derived erythroblasts
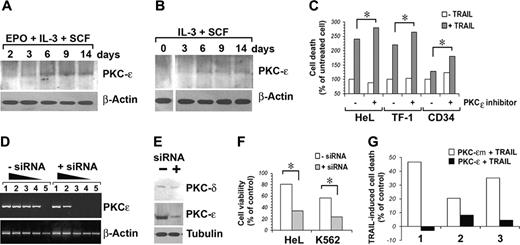
Our next question was therefore if PKCϵ could be implicated in the observed resistance to TRAIL of erythroblasts from day 10 onward. Figure 5A-B shows the kinetic of PKCϵ induction in our primary CD34 cell cultures in the presence or absence of EPO.
Induction of PKCϵ and its effects on TRAIL-induced apoptosis in human erythroblasts. (A-B) Western blot detection of total PKCϵ protein expression in CD34 cells cultured in serum-free medium with the indicated cytokines. β-Actin was monitored for protein loading. (C) TRAIL-induced apoptosis in cells pretreated for 48 hours with 250 μg/mL PKCϵ inhibitor (+) or control peptide (–). Cell death is expressed as percentage of controls. (D) Semiquantitative RT-PCR analysis of residual PKCϵ mRNA expression after PKCϵ-siRNA transfection. RNA was recovered 48 hours after siRNA transfection. Lanes 1 to 5: PCRs of 10–1 to 10–5 dilutions of total cDNA obtained from 1 μg reverse-transcribed total RNA. (E) Western blot analysis of residual PKCϵ protein expression after PKCϵ-siRNA transfection. PKC-δ was used as control for PKCϵ-siRNA specificity. (F) PKCϵ-siRNA transfection increases cell sensitivity to TRAIL. Residual cell viability of erythroid cell lines HeL and K562 treated with PKCϵ-siRNA and challenged with TRAIL is reported as percentage of controls (some samples, in absence of TRAIL). Mean of 3 independent experiments is reported. *P < .05. (G) PKCϵ overexpression reduces TRAIL sensitivity of CD34-derived erythroblast. CD34 cells, derived from 3 unrelated donors and cultured with IL-3, SCF, and EPO for 24 hours, were transfected with GFP-PKCϵ or GFP-PKCϵm. Forty-eight hours later, cells were treated with 50 ng/mL TRAIL. Apoptosis was monitored 48 hours after TRAIL treatment, staining cells with annexin V–FITC and PI. TRAIL-induced cell death is reported as percentage of controls (mock-transfected CD34 cells from the same patients).
Induction of PKCϵ and its effects on TRAIL-induced apoptosis in human erythroblasts. (A-B) Western blot detection of total PKCϵ protein expression in CD34 cells cultured in serum-free medium with the indicated cytokines. β-Actin was monitored for protein loading. (C) TRAIL-induced apoptosis in cells pretreated for 48 hours with 250 μg/mL PKCϵ inhibitor (+) or control peptide (–). Cell death is expressed as percentage of controls. (D) Semiquantitative RT-PCR analysis of residual PKCϵ mRNA expression after PKCϵ-siRNA transfection. RNA was recovered 48 hours after siRNA transfection. Lanes 1 to 5: PCRs of 10–1 to 10–5 dilutions of total cDNA obtained from 1 μg reverse-transcribed total RNA. (E) Western blot analysis of residual PKCϵ protein expression after PKCϵ-siRNA transfection. PKC-δ was used as control for PKCϵ-siRNA specificity. (F) PKCϵ-siRNA transfection increases cell sensitivity to TRAIL. Residual cell viability of erythroid cell lines HeL and K562 treated with PKCϵ-siRNA and challenged with TRAIL is reported as percentage of controls (some samples, in absence of TRAIL). Mean of 3 independent experiments is reported. *P < .05. (G) PKCϵ overexpression reduces TRAIL sensitivity of CD34-derived erythroblast. CD34 cells, derived from 3 unrelated donors and cultured with IL-3, SCF, and EPO for 24 hours, were transfected with GFP-PKCϵ or GFP-PKCϵm. Forty-eight hours later, cells were treated with 50 ng/mL TRAIL. Apoptosis was monitored 48 hours after TRAIL treatment, staining cells with annexin V–FITC and PI. TRAIL-induced cell death is reported as percentage of controls (mock-transfected CD34 cells from the same patients).
To formally prove that the induction of PKCϵ in EPO-cultured erythroblasts was responsible for their resistance to TRAIL, we first pharmacologically inhibited PKCϵ by the selective inhibitor of PKCϵ translocation.6,18 Figure 5C shows that PKCϵ inhibition increases TRAIL-induced cell death both in cell lines and in day-10 CD34 cells. Subsequently, we designed and successfully transfected in HeL and K562 cells siRNAs targeting PKCϵ mRNA. Figure 5D shows a semiquantitative RT-PCR analysis of HeL cells treated with anti-PKCϵ siRNA, showing a more than 100-fold decrease of specific cDNA amplification. Specific targeting of PKCϵ mRNA with consequent protein synthesis inhibition (Figure 5E) sensitized both HeL and K562 cells to TRAIL-induced apoptosis (Figure 5F). Finally, we overexpressed PKCϵ in CD34 cells differentiated with EPO for 3 days, observing acquired resistance to the apoptogenic effect of TRAIL, while PKCϵm-transfected CD34 cells did not (Figure 5G).
PKCϵ modulates Bcl-2 levels in erythroid progenitors
Given that the general activation of PKC by PMA does not affect TRAIL-R aggregation at the cell surface,25 and that overexpression of PKCϵ does not modulate TRAIL-R surface density, we looked at Bcl-2 levels as one possible antiapoptotic factor involved in the protection of erythroid progenitor cells.
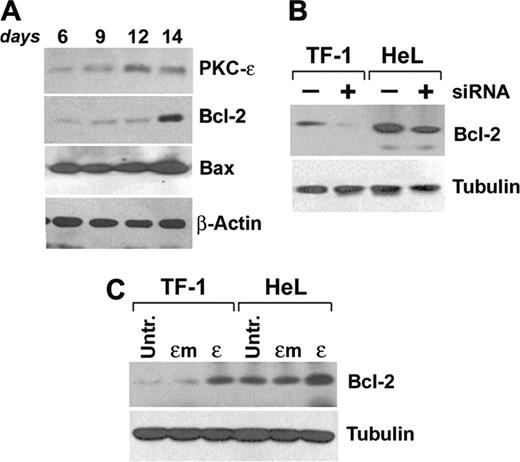
Starting experiments on primary CD34-derived erythroblasts are shown in Figure 6A. Kinetic analysis of Bcl-2 expression during EPO-driven erythroid maturation showed an up-regulation of the protein expression at day 14, while Bax levels remained constant.
Since it had been previously demonstrated that Bcl-2 levels could be modulated by PKCϵ,21 we either inhibited or overexpressed PKCϵ in our cell lines and subsequently immunoblotted for Bcl-2. Results show that siRNA-mediated downmodulation of PKCϵ induces a reduction of Bcl-2 protein expression (Figure 6B), while PKCϵ, but not PKCϵm, overexpression increases Bcl-2 protein levels (Figure 6C).
Discussion
Early erythroid progenitor cells are largely quiescent and require EPO to enter cell cycle, while late erythroid progenitors essentially require EPO as a survival factor that allows them to reach terminal differentiation. In vivo, physiologic EPO concentrations are suboptimal, causing apoptosis of about 20% of the bone marrow erythroid cells.26 It is now well established that erythroid differentiation can be further negatively regulated by death-receptor ligands present in the marrow microenvironment, such as Fas and TRAIL.
We demonstrate that, in differentiating erythroid progenitors, PKCϵ levels are regulated by EPO and control the protection against the apoptogenic effect of TRAIL. PKCϵ is important for cytokine and growth factor receptor–mediated signaling. This has been demonstrated by the observations that loss of PKCϵ results in a severely attenuated response of macrophages to LPS and IFNγ in PKCϵ–/– animals, as well as in impaired PDGF-receptor signaling.27,28 It has also been demonstrated that PKCϵ acts as a link between the integrin and IFNγ signaling pathways.29 We had previously demonstrated, indeed, that PKCϵ was one of the PKC isoforms modulated during human and mouse erythroid-cell differentiation18 and that TRAIL, as one of the TNF family members, could negatively regulate erythropoiesis.6 The data that we describe in this paper demonstrate that EPO promotes in human CD34 cells a series of events that, being finely regulated in their kinetics, determine the sensitivity of these cells to TRAIL. In fact at day 0, CD34 cells are insensitive to TRAIL because they do not express death receptors on the cell surface. PKCϵ is virtually absent at this early stage of differentiation. From day 3 onward, differentiating erythroid progenitors express surface death receptors TRAIL-R1 and TRAIL-R2, while PKCϵ levels are still undetectable. Cells become sensitive to TRAIL, and overexpression of PKCϵ at this stage abrogates TRAIL-induced apoptosis. Around day 7/8, PKCϵ levels increase rapidly, while surface death-receptor expression remains stable. PKCϵ induction is EPO dependent, since control CD34 cells cultured in the presence of SCF + IL-3 do not up-regulate PKCϵ synthesis. The induction of PKCϵ levels confers to erythroid cells resistance to TRAIL, notwithstanding the surface expression of death receptors and the virtual absence of surface decoy receptors. The resistance to apoptosis is dependent on PKCϵ induction, since both silencing of PKCϵ mRNA with consequent downmodulation of the protein synthesis and pharmacologic inhibition of PKCϵ activation are able to restore cell sensitivity to TRAIL. Very recently, Gillespie et al30 have shown that PKCϵ levels are correlated with the sensitivity of melanoma cell lines to TRAIL, suggesting that PKCϵ might have a general role in the fine tuning of the signaling emanating from death-receptor triggering.
PKCϵ modulates Bcl-2 levels. (A) CD34-derived erythroblasts were cultured with IL-3, SCF, and EPO up to 14 days and total Bcl-2, PKCϵ, and Bax protein expression levels were monitored at the indicated times by Western blot. (B) Western blot of Bcl-2 in TF-1 and HeL cell lines in the presence (+) or absence (–) of PKCϵ-specific siRNA. (C) Western blot of Bcl-2 in TF-1 and HeL cell lines overexpressing wild-type (ϵ) or mutated (ϵm) PKCϵ.
PKCϵ modulates Bcl-2 levels. (A) CD34-derived erythroblasts were cultured with IL-3, SCF, and EPO up to 14 days and total Bcl-2, PKCϵ, and Bax protein expression levels were monitored at the indicated times by Western blot. (B) Western blot of Bcl-2 in TF-1 and HeL cell lines in the presence (+) or absence (–) of PKCϵ-specific siRNA. (C) Western blot of Bcl-2 in TF-1 and HeL cell lines overexpressing wild-type (ϵ) or mutated (ϵm) PKCϵ.
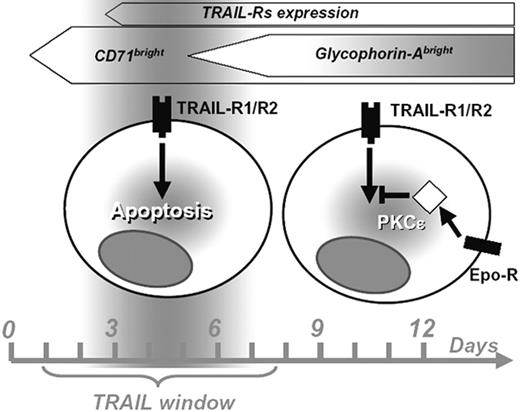
Proposed scheme of the period of sensitivity to TRAIL (“TRAIL window”) along human erythroid differentiation and the role of PKCϵ in the downstream intracellular signaling pathway.
Proposed scheme of the period of sensitivity to TRAIL (“TRAIL window”) along human erythroid differentiation and the role of PKCϵ in the downstream intracellular signaling pathway.
Our data show that the antiapoptotic molecule Bcl-2 is a candidate mediator of erythroid-cell resistance to TRAIL. Bcl-2 levels are in fact up-regulated in erythroid progenitors with a kinetic that is compatible with that of EPO-driven PKCϵ induction. The observation of the reciprocal effects of PKCϵ downmodulation or overexpression on Bcl-2 levels in our model system demonstrates that Bcl-2 is modulated by PKCϵ levels. Indeed, these data parallel those from Gubina et al21 that demonstrated that the overexpression of PKCϵ in the TF-1 cell line was able to induce Bcl-2 expression. However, we cannot exclude that other antiapoptotic intermediates could counteract the effects of TRAIL. For example, although in our experiments Bax levels remained constant, McJilton et al31 demonstrated that in the human prostate cancer cell line LNCaP, PKCϵ could interact with Bax blocking its conformational changes required for the mitochondrial death-signaling pathway.
Altogether, our data prompt us to hypothesize a progression of the differentiating erythroid progenitor through an initial phase of resistance to TRAIL (due to the lack of specific surface-receptor expression), followed by a period of sensitivity that ends around day 7, due to the EPO-driven up-regulation of PKCϵ with downstream positive effects on Bcl-2 (Figure 7).
PKCϵ therefore is a novel quantitative regulator of EPO-dependent cell expansion and survival in erythropoiesis, similarly to what was recently suggested by Schmidt et al32 for Btk.
It will be therefore important to re-examine the pathophysiology of diseases such as myelodysplastic syndrome (MDS), characterized by an enhanced response to death ligands, in the light of PKCϵ levels as key regulators of the physiologic cellular response to TRAIL.
Prepublished online as Blood First Edition Paper, September 15, 2005; DOI 10.1182/blood-2005-07-2676.
Supported by Ministero dell'Istruzione, dell'Università e della Ricerca–Fondo per gli Investimenti della Ricerca di Base (MIUR-FIRB) (RBNE0189JJ), Programmi di Ricerca Cofinanziati (COFIN), and Fondazione Cassa di Risparmio di Parma (CARIPARMA) grants.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Instrumentation Laboratory, Italy, for technical support and to Vincenzo Palermo, Domenico Manfredi, and Davide Dallatana for technical support.