The ability of glucocorticoids (GCs) to kill lymphoid cells led to their inclusion in essentially all chemotherapy protocols for lymphoid malignancies, particularly childhood acute lymphoblastic leukemia (ALL). GCs mediate apoptosis via their cognate receptor and subsequent alterations in gene expression. Previous investigations, including expression profiling studies with subgenome microarrays in model systems, have led to a number of attractive, but conflicting, hypotheses that have never been tested in a clinical setting. Here, we present a comparative whole-genome expression profiling approach using lymphoblasts (purified at 3 time points) from 13 GC-sensitive children undergoing therapy for ALL. For comparisons, expression profiles were generated from an adult patient with ALL, peripheral blood lymphocytes from GC-exposed healthy donors, GC-sensitive and -resistant ALL cell lines, and mouse thymocytes treated with GCs in vivo and in vitro. This generated an essentially complete list of GC-regulated candidate genes in clinical settings and experimental systems, allowing immediate analysis of any gene for its potential significance to GC-induced apoptosis. Our analysis argued against most of the model-based hypotheses and instead identified a small number of novel candidate genes, including PFKFB2, a key regulator of glucose metabolism; ZBTB16, a putative transcription factor; and SNF1LK, a protein kinase implicated in cell-cycle regulation.
Introduction
Glucocorticoid (GC)–induced apoptosis is a phenomenon of considerable physiologic and therapeutic significance. Physiologically, it has been implicated in the shaping of the immune repertoire and in controlling immune responses,1 and therapeutically it has been exploited in the treatment of lymphoid malignancies, most notably childhood acute lymphoblastic leukemia (ALL),2 where good response to introductory hormone treatment predicts favorable overall outcome.3 Thus, defining the molecular basis of GC-induced cell death4-7 and the clinically relevant phenomenon of GC resistance8-12 has obvious bearing on understanding immune system regulation and developing improved therapy protocols for lymphoid malignancies.
GCs mediate most of their effects via their cognate receptor (GC receptor [GR]), a ligand-activated transcription factor of the large nuclear receptor family.13 GC-induced apoptosis critically depends on sufficient levels of GRs and subsequent alterations in gene expression, but the precise nature of the GC-regulated genes responsible for the antileukemic GC effects remains elusive.4-7 To address this issue, we and others performed expression profiling with subgenome microarrays (up to ∼ 10 000 genes) and various model systems of GC-induced cell death (mouse and human leukemia cell lines and mouse thymocytes). These studies identified a large number of GC-regulated genes and led to several hypotheses (reviewed in Schmidt et al4 ). Specifically, GCs may induce cell death by directly regulating the expression of components of the cell death machinery, such as components of the intrinsic pathway, including the Bcl-2 rheostat;14 the extrinsic pathway, comprising membrane death receptors and their signaling proteins;15,16 or the effector molecules of the death machinery (ie, the caspases).17,18 In support of this theory, transcriptional induction of the Bcl-2 homology 3 (BH3)–only molecule Bim,19 the caspase-activating granzyme A,20 or a potentially proapoptotic molecule, called GPR65/TDAG8,21 have been suggested to cause GC-induced apoptosis. Alternatively, GCs may deregulate cellular homeostasis, which, in turn, is interpreted by the cell as a death signal and subsequently triggers the apoptotic response. As one controversial example,22,23 GC repression of c-myc has been proposed to generate a “conflicting signal” that is not tolerated by proliferating leukemia cells and activates a cell death program. Other proposed examples include GC-mediated deregulation of metabolism24 and/or macromolecule neosynthesis.25 These GC effects may be critically enhanced by GR autoinduction, which is observed in several models of GC-induced apoptosis, but not in tissues that do not undergo cell death upon GC exposure (reviewed in Kofler26 ). Very recently, a weak, but significant, induction of mitogen-activated protein kinase (MAPK) kinase 3 was suggested to contribute to GC sensitivity by activating p38 MAPK which, in turn, phosphorylates the GR, thereby increasing its transactivation potential.27 However, whether any of these hypotheses can be extended to the clinical setting has not been investigated.
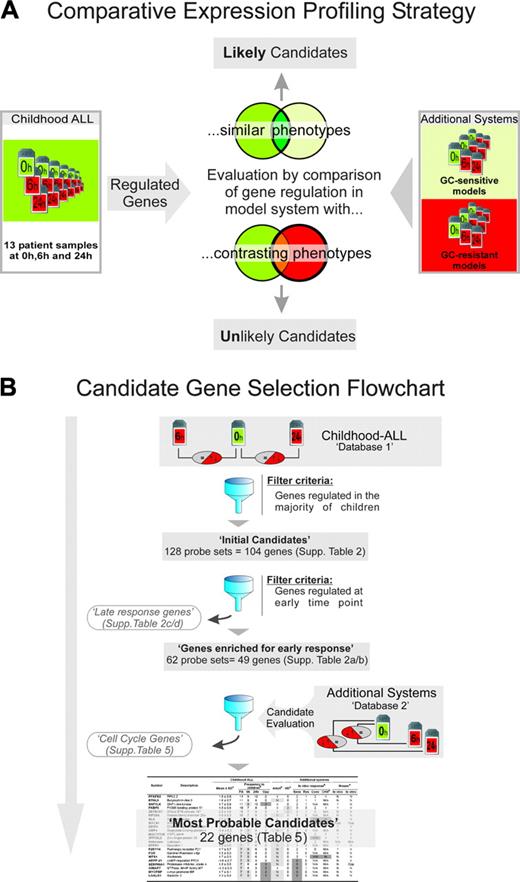
In this study, we addressed the molecular basis of GC-induced apoptosis by a novel comparative expression profiling strategy that used children with ALL and several other biological systems of GC sensitivity and resistance (Figure 1). First, we determined the genes regulated in malignant lymphoblasts from 13 GC-sensitive children with ALL during GC monotherapy using Affymetrix-based “whole-genome” expression profiling (Affymetrix, Santa Clara, CA). This database (Database 1 in Figure 1) can be used to query essentially any gene in the genome for in vivo regulation in ALL, a prerequisite for a potential upstream GC response gene in this death pathway. In a second step, we determined the genes that were coordinately regulated in most patients (“Initial candidates list”), followed by enrichment for early responding genes. The remaining 49 genes should include the upstream component(s) of a putative canonical pathway to GC-induced apoptosis mingled with genes not involved in the death response. To further address this issue, we determined the expression profiles of several additional biological systems in which GCs do or do not induce cell death (Database 2 in Figure 1). As explained in Table 1, this database provides evidence for or against a possible role in GC-induced apoptosis of response genes identified in childhood patients with ALL, and aided in both identifying a set of genes unlikely to be directly regulated by GC (cell-cycle genes; Figure 1) and in evaluating the significance of the final list of most probable candidates (Table 5). In conclusion, our study provides new and essential insight into the GC response genes and possible molecular mechanism of GC-induced apoptosis in essentially all relevant biological systems, most importantly, children with ALL.
Patients, materials, and methods
Patients and other biologic systems
Patients. Children with ALL admitted to the Department of Pediatrics, Innsbruck Medical University, and treated according to Berlin-Frankfurt-Münster (BFM) protocol 2000 were enrolled in this study. The study was approved by the Ethics Committee of the Innsbruck Medical University (EK1-1193-172/35) and written informed consent was obtained from parents or custodians. For comparison purposes, a 72-year-old white male with B-cell precursor (BCP)–ALL treated at the Department of Hematology and Oncology of the Innsbruck Medical University was included after giving written informed consent.
Blood sampling, GC treatment, and GC response. EDTA blood was taken by venous puncture prior to initiation of GC treatment, and at 6- to 8-hour intervals after initiation. To avoid tumor lysis syndrome, the daily GC dose was gradually increased over the first 3 to 4 days to reach 60 mg prednisolone/m2/day. Treatment was initiated with a single intravenous or oral application of 6% to 38% of the final dose, depending on peripheral blast counts, T- or B-cell phenotype, and clinical conditions. On the second day, the children received 30% to 60% of the final GC dose in 3 applications. To account for treatment differences, GC bioactivity was determined in the sera (Table 1). The adult patient received a single oral application of 20 mg dexamethasone on day 1 and another 12 mg on the morning of day 2. All patients responded to the treatment with a reduction of peripheral lymphoblasts within the first 24 to 48 hours. All children except BCP-ALL-24 scored as “prednisolone good responders” by day 8, as defined by the BFM protocol (< 1000 blasts/μL on day 8). Further details on in vivo and ex vivo treatment and purification of lymphoblasts for expression profiling are detailed below and in the Supplemental Materials' sections 1 and 2 on the Blood website; click on the Supplemental Materials link at the top of the online article.
Comparative expression profiling strategy and general work flow. (A) The principle of comparative expression profiling exemplified by evaluating candidate genes (ie, genes regulated in the majority of childhood ALL samples) in additional GC-sensitive and GC-resistant systems. Coregulation in the former supports, whereas coregulation in the latter argues against, a potential role of a candidate gene in the death response. Other relevant information, such as interspecies conservation, de novo protein biosynthesis-dependence, etc, can also be derived by comparisons with the additional systems as outlined in Table 1. (B) Summary of workflow described in “Introduction.” The complete databases are available at www.ncbi.nlm.nih.gov/geo/ (GEO accession numbers: GSE2677, GSE2842, GSE2843). Tables corresponding to various database subsets are shown in the Supplemental Materials, as indicated. The final 22 genes are presented in Table 5. The additional systems comprising database 2 and their use for evaluating the possible significance of the candidate genes in the death response are explained in Table 1.
Comparative expression profiling strategy and general work flow. (A) The principle of comparative expression profiling exemplified by evaluating candidate genes (ie, genes regulated in the majority of childhood ALL samples) in additional GC-sensitive and GC-resistant systems. Coregulation in the former supports, whereas coregulation in the latter argues against, a potential role of a candidate gene in the death response. Other relevant information, such as interspecies conservation, de novo protein biosynthesis-dependence, etc, can also be derived by comparisons with the additional systems as outlined in Table 1. (B) Summary of workflow described in “Introduction.” The complete databases are available at www.ncbi.nlm.nih.gov/geo/ (GEO accession numbers: GSE2677, GSE2842, GSE2843). Tables corresponding to various database subsets are shown in the Supplemental Materials, as indicated. The final 22 genes are presented in Table 5. The additional systems comprising database 2 and their use for evaluating the possible significance of the candidate genes in the death response are explained in Table 1.
In vitro models of GC sensitivity, resistance, and restored sensitivity. As in vitro models for GC-induced leukemia apoptosis we used CCRF-CEM-C7H2 T-ALL cells28 and preB697 BCP-ALL cells.29 Both cell lines undergo almost complete cell death after 48- to 72-hour incubation with 10-7 M dexamethasone. As GC resistance models, CEM-C1,30 CEM-C7R1,31 CEM-C7R1low, and PreB697-R4G4 (described in the supplement, section 1.2) were used. GC sensitivity was restored in resistant CEM-C1 cells by stable, constitutive expression of rat GRwt (CEM-C1ratGR clone C1-4G4),30 and in resistant CEM-C7R1 by high-level expression of human GRA458T (CEM-C7R1dim-high) (Supplemental Materials section 2.2).
Cycloheximide treatment. To assess whether gene regulations were dependent upon de novo protein biosynthesis, we used CCRF-CEM-C7H2 cells treated with dexamethasone in the presence of cycloheximide (CHX) for 6 hours.
Mouse models. For in vitro GC response, thymocytes from 4- to 6-week-old CD-1 mice were treated with 10-7 M dexamethasone or 0.1% ethanol as vehicle control for 4 hours. To determine the in vivo response to GCs, 4- to 6-week-old CD-1 mice were injected intraperitoneally with 0.2 mg dexamethasone per mouse or phosphate-buffered saline as control, and their thymocytes were used for RNA preparation.
Healthy donors. After giving written informed consent, 2 healthy adults were treated with dexamethasone according to a similar protocol as that used for the children. Subsequently, their peripheral blood mononuclear cells were purified by Lymphoprep separation (AXIS-Shield, Rodelokka, Norway) and used for RNA preparation.
Purification of peripheral lymphoblasts from patients
Mononuclear cells were purified from peripheral EDTA blood by centrifugation on Lymphoprep and the percentages of blasts determined by fluorescence-activated cell-sorting (FACS) analysis. If blast purity was less than 90%, the blasts were enriched to 90% or more by magnetic field separation as detailed in the supplement, section 2.1.
GC bioactivity assay
GC bioactivity (GBA) in the patients' plasma prior to and during GC therapy was measured from 20-μL samples (cell supernatants after Lymphoprep separation) using a recombinant cell bioassay in which COS-1 cells are transfected with expression vectors encoding human GR and a nuclear receptor coregulator, ARIP3, together with an appropriate reporter gene.32
Generation and characterization of transfected cell lines
The production of CEM-C1ratGR-4G4 has been described.30 CEM-C7R1dim-high and CEM-C7R1dim-low were generated by stable transduction of GC-resistant CEM-C7R1 cells (which contained 2 mutated GR alleles) with a retroviral vector expressing human GR containing the point mutation A458T. CEM-C7R1dim-high showed high-level GR expression and was sensitive to GC-induced apoptosis, whereas CEM-C7R1dim-low expressed much lower levels and remained resistant (Figure S1).
Microarray analysis and quality parameters
For microarray analysis, 1.5 μg high-quality total RNA (Supplemental Materials section 2) was processed into a biotinylated hybridization target using corresponding kits from Affymetrix, hybridized to U133 Plus 2.0 microarrays and analyzed in an Affymetrix scanner 3000. Image analysis was performed with the Affymatrix GCOS software (Santa Clara, CA). Data processing and analysis was performed in “R” (Bioconductor, http://www.bioconductor.org) using the robust multiarray analysis (RMA) method for normalization and Bioconductor's hgu133plus2 annotation package for annotation. Normalized expression values (E values) were inserted into a database using Bioconductor's developmental package maDB and used to calculate regulation values (M values). Section 4 in the Supplemental Materials summarizes quality parameters, including 3′ to 5′ signal ratios and percentages of “present calls” for each array, variance in technical replicates, regulation of methotrexate (MTX) response genes, and real time reverse transcriptase–polymerase chain reaction (RT-PCR) verification results for 25 genes.
Data verification by real time RT-PCR
Total RNA (500 ng) was reversely transcribed into cDNA using Superscript II (Invitrogen, Carlsbad, CA) according to manufacturer's protocol. cDNA (100 ng) was assayed on microfluidic cards containing 24 human genes in duplicate (Applied Biosystems, Foster City, CA). For MYC, SNF1LK, BTNL9, GZMA, and GPR65/TDAG-8, individual premanufactured ABI assays were used (Supplemental Materials section 4.3).
Results
To identify possible common upstream component(s) of the cell death pathway induced by GCs in childhood ALL, we exploited a comparative expression profiling strategy (Figure 1, Table 1) using “whole-genome” arrays (Affymetrix; U133 plus 2.0) and a number of additional biological systems in which GCs do or do not induce apoptosis (Tables 2, 3). To this end, we first determined the expression profiles of peripheral lymphoblasts from 13 children with ALL prior to and under treatment with GCs for 6 to 8 hours and 24 hours (for GC bioactivity levels in the sera and other clinical features, see Tables 2, 3). The expression profiles of these 39 arrays were used to generate 26 comparisons (0 hours versus 6 to 8 hours, and 0 hours versus 24 hours) that were entered into a database (GC response—childhood ALL database in Figure 1).
Regulation of previously identified candidates
First, we used this database to investigate whether genes implicated in GC-induced apoptosis in experimental systems (reviewed in Schmidt et al4 ) might be regulated in children with ALL. As shown in Table 4, of the current candidate genes, LDH-A/lactate dehydrogenase-A,24 GPR65/TDAG-8,21 MAP2K3/MAP kinase kinase 3,27 GZMA/granzyme A,20 MYC/c-myc,23 NR3C1/GR,24 and BCL2L11/Bim,8,19,36 none was regulated more than 2-fold in most children, as might be expected from key players in a conserved pathway. Two deserve further attention: the GR that was induced in all 3 T-ALLs, and the proapoptotic BH3-only molecule Bim where probe sets corresponding to this locus, but not necessarily to the known major bim transcripts, were induced in up to 6 of 13 children (“Discussion”). We further investigated the remaining 26 genes in a currently established list of experimental system–derived candidates4 but, with the exception of FKBP51, SOCS1, and DDIT4/Dig2, which will be discussed further, none scored in more than 4 children (Table S1).
Genes frequently regulated in childhood ALL
To directly define candidate genes relevant for induction of apoptosis by GC in childhood ALL, we first identified all probe sets that revealed M values of 1 or more (2-fold regulation) in at least 7 of 13 patients (128 probe sets, 104 genes; Table S2). Since we were mainly interested in primary response genes, we focused on the probe sets within this collection that were regulated with an M value of 0.7 or more at 6 to 8 hours in at least 6 of 13 patients. Twenty-five induced and 37 repressed probe sets (“top 62”), corresponding to 19 and 30 genes, respectively, met this requirement. Within the limitations of the assay system, this collection can be assumed to contain the critical upstream gene(s) responsible for GC-induced apoptosis, although hidden by genes unrelated to the death response. To distinguish the former from the latter, we performed comparative expression profiling using the additional biological systems shown in Tables 2, 3. The expression profiles from these systems prior to and after dexamethasone exposure were used to generate a second database (GC-response genes— additional systems database 2 in Figure 1). Subsequently, we determined the performance of the “top 62” probe sets derived from the children with ALL in this database (Table S4).
A cluster of regulated cell-cycle genes
Thirty-four of the 37 repressed probe sets resulted in a remarkably distinct pattern: they were highly expressed in cell lines in contrast to all other systems (Table S5). They remained unregulated in 6 of 7 cell lines, in peripheral lymphocytes from healthy controls and in mouse thymocytes, but were repressed in the adult patient, resembling the situation in children. Moreover, in patient BCP-ALL-40, the 34 probe sets were strongly regulated in vivo but much less so during ex vivo treatment, whereas the opposite behavior was shown by known GC response genes like FKBP51 (Tables S3 and S4 for regulation of these probe sets in children and additional systems, respectively). Combined with the fact that none of the 27 genes corresponding to these 34 probe sets has previously been reported to be GC regulated, the above results suggested that they are not direct transcriptional GC targets (“Discussion”). Since all of them are involved in late cell-cycle progression,37,38 we referred to this coordinately regulated group as “cell-cycle genes.” Because the aim of this study was to identify primary response genes in the GC-induced cell death pathway (which these genes are probably not), and since previous observations suggested that cell-cycle arrest is not required for cell death,39 we focused our further analyses on the remaining 28 probe sets.
Candidate genes for GC-induced apoptosis
After reduction of the 28 probes sets to their corresponding genes (by using the M values of the probe sets with the strongest regulation), the performance of the resulting 22 candidate genes in the children and additional systems was compiled in Table 5. Although not formally ruling out any gene, the combined information might prove useful for selection of candidates for future functional analyses. Thus, genes no longer regulated in the presence of the translation inhibitor cycloheximide (SOCS1, SLA, WFS1), and/or genes not regulated in any of the additional in vivo, ex vivo, and in vitro systems of GC-induced apoptosis (ARPP-21, SERPINA1, MYCBP, LGALS3) may not be direct transcriptional GC targets. In contrast, absent or reduced gene regulation in 4 instances of in vitro GC resistance and/or in mature peripheral blood lymphocytes (which are considered to be insensitive to GC-induced cell death when in a resting state)40,41 might argue in favor of functional relevance of the respective gene (eg, PFKFB2, BTNL9, SNF1LK). Finally, genes coregulated in childhood ALL and mouse thymocytes (eg, SNF1LK, FKBP5, DDIT4) would qualify as possible components of a canonical pathway conserved between species and systems (mouse thymocytes, human ALL cells).
In conclusion, we generated 2 databases encompassing comprehensive lists of GC-regulated candidate genes in children with ALL and a number of additional systems, permitting immediate analysis of any gene with respect to its regulation, and thus potential significance for GC-induced apoptosis. The study provided important evidence for some of the key questions in the field: several current model-based hypotheses could essentially be ruled out for childhood leukemia, and the number of potential candidates for a common upstream regulator in mouse thymocyte and human leukemia cells was dramatically reduced. Gene induction rather than gene repression might account for cell death in childhood ALL, although this conclusion must be viewed with caution since down-regulation may be more difficult to detect than gene induction, and only a handful of genes qualified for a critical upstream component of the GC-evoked death pathway in children with ALL, most notably 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 (PFKFB2), a key regulator of glucose metabolism; zinc finger and BTB domain–containing gene 16 (ZBTB16), a putative transcription factor, and SNF1-like kinase (SNF1LK), a protein kinase implicated in cell-cycle regulation.
Discussion
Despite its clinical relevance and decades of research, the molecular basis of GC-induced leukemia apoptosis has remained a mystery. Numerous hypotheses have been proposed, but whether the corresponding gene regulations occur in a clinical setting has not been investigated. Our study addressed, for the first time, the molecular basis of GC-induced leukemia apoptosis in a clinical setting by expression profiling using the currently most complete probe collection (U133 plus 2.0; 54 000 probe sets). The underlying hypothesis was that GCs induce apoptosis by altering gene expression at the mRNA level and that the basic mechanism is shared among different children with ALL. Within these premises and the limitations of the technology (presence of an appropriate probe set on the array, regulation 2-fold or more), the key component(s) of the respective pathway should become apparent given the number of children investigated. To provide further information regarding the possible significance of the identified genes, we also analyzed a number of additional systems.
Previously identified candidates and related hypotheses
Regarding candidates and related hypotheses derived from experimental systems, our data argued against a general role of lactate dehydrogenase A,24 granzyme A,20 TDAG-8,21 MAP kinase kinase 3,27 and c-myc23 in cell death induction in childhood ALL, although some of these genes may be relevant for cell death induction in experimental systems or in subgroups of children. In the case of c-myc, TDAG8, GZMA, bim, and GR, the findings were further reconfirmed by real-time RT-PCR for all patients where sufficient mRNA remained (section 4.3 in the Supplemental Materials). GR autoinduction, which we and others have proposed to be important for GC-induced apoptosis in the CCRF-CEM model for T-ALL,24,42,43 was observed in all 3 patients with T-ALL, but only in 2 of 10 patients with BCP-ALL. Although the number of patients is too small to draw firm conclusions for subgroups, GR autoinduction may be relevant for patients with T-ALL, an entity that shows a relatively high rate of tumor lysis syndrome.44 Three of the 8 probe sets for the BCL2L11/Bim locus on chromosome 2 were induced 2-fold or more in our children with ALL. Probe set 1 555 372_s_at, induced most frequently (6 of 13), matches the 3′ end of a multiple myeloma–derived cDNA referred to as Bam.45 The reported Bam mRNA starts 94 bp upstream from the BH3-containing Bim exon and encodes a predicted 73–amino acid protein with a BH3 domain and 40 amino acids not present in any known Bim protein. The second probe set, 225 606_at, maps about 1 kb downstream from the currently known 3′ end of Bim transcripts and might have resulted from alternative polyadenylation. It was regulated in 4 of the 6 children who showed induction of probe set 1 555 372_s_at. Probe set 1 558 143_s_at recognized the reported 3′ end of all major Bim isoforms (including BimEL, BimL, and BimS)46 and was induced in 3 of the 6 children after 24 hours, but not after 6 hours. Thus, although the complexity of the BCL2L11 locus precludes final conclusions, transcripts from this locus may contribute to cell death induction in at least a subgroup of children either as primary GC targets or as downstream effector molecules.
Three of the previously identified candidates (FKBP5/FKBP51, DDIT-4/Dig2, and SOCS-1) were reconfirmed in most patients. FKBP51 has recently been proposed as general indicator of GC sensitivity, and a corresponding assay was developed.47,48 It is a GR cochaperone that is transcriptionally induced by GC48,49 and competes with FKBP52 for dynein binding sites,50,51 thereby reducing nuclear transport of the ligand-bound receptor. Its induction reduces transcriptional GC effects, and cells overexpressing FKBP51 are more resistant to GC-induced apoptosis.50 DDIT4/Dig-2 has been suggested to mediate both prosurvival and proapoptotic functions52 and its overexpression, like that of FKBP51, reduced sensitivity to dexamethasone-induced apoptosis.53 Provided these findings in the cell lines can be extended to patients, these 2 genes, although regulated by GCs in many systems, may not be causally involved in cell death induction. SOCS1, a potentially antisurvival protein, as implicated by its name (“suppressor of cytokine signaling”), was induced in 9 of 13 patients, but its regulation in CCRF-CEM cells was sensitive to cycloheximide and its expression repressed in mouse thymocytes. However, should the data in the model systems be irrelevant for the clinical situation, SOCS1, by virtue of its function as an antisurvival protein, remains 1 of the most attractive candidates.
Cell cycle genes: an example of apparent “gene regulations” caused by population shift?
The significant decrease in expression levels of the cell-cycle genes (a set of genes known to be expressed in the G2 and/or M phases of the cell cycle) observed after GC exposure in 11 of 13 children and the adult subject deserves further discussion. Based on the arguments put forward in “Results,” we consider it unlikely that these genes are direct transcriptional GC targets. A possible explanation for the decline in expression levels after GC treatment might be that proliferating cells within the tumor were retained in the bone marrow, migrated out of the bloodstream, and/or were selectively killed by GCs. Thus, the observed changes in gene expression might reflect a treatment-induced shift from a more proliferative population of leukemia cells at 0 hours toward a less proliferative population after 6 and 24 hours rather than resulting from direct transcriptional regulation by GCs. Since in vitro migration does not occur and apoptotic cells are not effectively removed, this phenomenon might be more easily detectable in vivo than in tissue culture.
Candidates conserved between human leukemia cells and mouse thymocytes
Whether GC-induced apoptosis in mouse thymocytes and human patients with ALL is controlled by the same gene(s) is of considerable interest for various reasons, including functional testing of candidates in vivo. Our study uncovered only a small number of genes coordinately regulated in mice and most children with ALL (Table 4). With the possible exception of SNF1LK (which will be discussed in “New candidates”), these candidates do not appear very promising: two of them (FKBP51 and DDIT4) protected cells from GC-induced apoptosis,50,53 and none of the remaining coregulated genes has been implicated in apoptotic or survival pathways thus far, supporting the notion that GC-induced apoptosis in mouse thymocytes and human lymphoblastic leukemia cells might be controlled by different genes.
Gene induction versus gene repression
Another general suggestion from Table 4 concerns the question of whether GC-induced leukemia apoptosis results from gene induction or repression. After subtraction of the cell-cycle genes (which may not be direct GC targets nor responsible for cell death induction), only 3 down-regulated genes remained: GBP4 (guanylate-binding protein 4),54 ARPP-21 (cAMP-regulated phosphoprotein 21);55 and GIMAP7 (GTPase immune-associated protein 7, also known as human immune-associated nucleotide, hIAN7).56 Since these genes have not been implicated in any known death pathway and performed rather moderately in the various systems (Table 4), a prominent role as initiator of the death pathway seems unlikely. Thus, the data from the patients do not support the cell line–derived conclusion that GC-induced leukemia depends on gene repression.57
New candidates
A particularly interesting candidate in the upper part of Table 5 is SNF1LK, a member of the SNF/AMPK family of protein kinases. Although its role is currently not well understood, it has been implicated in regulation of the G2/M phase of the cell division cycle,58 and shows homology to genes controlling carbohydrate metabolism in plants.59 Thus, SNF1LK may be involved in the observed effects on cell-cycle genes or may lead to (potentially harmful) metabolic alterations. It was induced in 11 of 13 children, the adult patient, and in mouse thymocytes in vivo, making it a possible candidate for a critical upstream component in a pathway conserved between species. However, the reason for its repression in 2 children with T-ALL (reconfirmed by real-time RT-PCR; Supplemental Materials section 4.3) is unclear. The functions of KIF26A and BTNL-9 are currently unknown; hence, their potential role in apoptosis induction is difficult to assess. KIF26A belongs to the N11 kinesins, a subgroup of the large kinesin family that has been implicated in cellular transport processes.60 BTNL-9 shares structural similarity with butyrophilin, a structural component of the human milk fat globule.61 This gene was regulated in all 10 patients with BCP-ALL and none of the 3 patients with T-ALL, and may thus encode a protein regulating B-cell–specific GC actions. SLA (Src-like adaptor) encodes an adaptor protein that negatively regulates T-cell receptor (TCR) signalling.62 If it has a similar activity in B cells, its induction (like that of SOCS-1) might interfere with survival signals. However, regulation of this gene was sensitive to cycloheximide in CEM-C7H2 cells and it was not induced in C7R1dim4 cells, although they underwent GC-induced apoptosis (again resembling SOCS-1). One of the most frequently regulated probe sets was 228 854_at. It mapped about 4 kb downstream of the reported 3′ end of a putative transcription factor called ZBTB16/PLZF/ZFP-145, which is required for spermatogonial stem cell renewal63,64 and limb and axial skeletal patterning,65 and has been found to be rearranged in promyelocytic leukemia.66,67 As detailed in section 4.3 of the Supplemental Materials, there was a strong correlation (R2 = 0.8266) between the regulation data obtained with 228 854_at in the Affymetrix screen and the ZBTB16 real-time RT-PCR results, strongly suggesting that this probe set recognizes an undescribed variant ZBTB16 mRNA generated by alternative polyadenylation. Thus, even though ZBTB16 was regulated in peripheral blood lymphocytes from both healthy donors (who are supposedly relatively resistant to GC-induced apoptosis40,41 ), it remains a valid candidate for an upstream regulator of GC-induced apoptosis.
One of the most promising candidates is 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 (PFKFB2), a key enzyme in glucose metabolism.68 This gene was most frequently regulated at both the early and late time points. Its regulation was resistant to the translation inhibitor cycloheximide. Although not regulated in preB-697 cells, it was rapidly and strongly induced in CCRF-CEM-C7H2 but not, or much less so, in its GC-resistant derivatives, and showed clear induction in both “converted” models where GC sensitivity was restored by transgenesis. The gene was not regulated in peripheral blood lymphocytes from 2 healthy donors or in mouse thymocytes. Regarding possible functional consequences, recent data suggest that cellular metabolism and apoptosis might be intertwined with connections between regulation of cellular bioenergetics and apoptosis.69,70 Malignant cells, known for their altered glucose metabolism,71,72 might be particularly sensitive to disturbances in glycolytic pathways. In support of this concept, regulation of glucose metabolism in thymocytes has been reported many years ago73 and combination with 2-deoxy glucose (2-DG), a specific inhibitor of hexokinase (the enzyme phosphorylating glucose, thereby making it a substrate for further metabolic transformation), dramatically sensitized CCRF-CEM cells to cell death triggered by GCs, but not several other apoptosis inducers (K. Renner, C. Seger, and R. Kofler, manuscript submitted). Clearly, the possible functional role of PFKFB2 (and the remaining candidates in Table 4) needs to be directly assessed. Given the limitations of existing test systems, we are currently developing lentiviral transduction systems to allow functional testing in primary cells from patients.
Relation to previously defined resistance genes
Finally, we wondered whether genes previously implicated in resistance to GCs or other chemotherapeutics might be among the probe sets frequently regulated by GCs in children with ALL. Interestingly, none of 33 genes predictive for poor GC response74 was among the top 128 probe sets depicted in Table S2. On our microarray, we identified corresponding probe sets for 45 of 54 genes that predicted molecular treatment response in childhood ALL75 (the remaining 10 cDNAs could not be unambiguously annotated). Two of them (CDCA1 [cell division cycle–associated 1], probe set ID: 223 381_at; and TTK protein kinase, probe set ID: 204 822_at), were among our collection of regulated probe sets. Finally, we analyzed 45 genes associated with cross-resistance to 4 mechanistically distinct antileukemic agents and 139 genes related to discordant resistance to vincristine and asparaginase.76 MELK (204 825_at) was the only 1 of the 45 cross-resistance predictor genes, and HGFL/MGC17330 (221 756_at) the only member of the 139 discordant resistance predicting genes found in the top 128 probe sets (Table S2).
Prepublished online as Blood First Edition Paper, November 17, 2005; DOI 10.1182/blood-2005-07-2853.
Supported by the Austrian Science Fund (SFB-F021), the European community (QLG1-CT-2001-01 574), the Austrian Ministry for Education, Science and Culture (GENAU-CHILD, GENAU-BIN), the Tiroler Wissenschafts funds, and the Finnish Cultural Foundation. The Tyrolean Cancer Research Institute and this study were supported by donations from the “Tiroler Landeskrankenanstalten (TILAK),” the “Tyrolean Cancer Society,” various businesses, financial institutions, and the people of Tyrol.
S. Schmidt participated in most experiments, coordinated the data analyses, and contributed to the writing of the manuscript; J.R. performed the bioinformatic data analyses; S.R. generated the GRdim transfected cell lines; C.P. coordinated and performed the mouse work; S.J. performed the microarray analyses; C.A., E.P., S. Skvortsov, and R.C. were involved in patient sample preparation and analyses; M.F. coordinated the adult ALL data; T.R. and O.A.J. performed and analyzed the GBA; S.G. assisted in writing and data interpretation; B.M. coordinated the clinical work with children; and R.K. coordinated the entire study and contributed to the writing of the manuscript. The final version was seen and approved by all authors.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs R. Panzer, O. Haas, and H. Gadner (St. Anna Kinderspital, Vienna, Austria) for providing clinical data; all members of the EUGIA project for valuable discussions; Dr L. A. Huber for support in establishing the microarray facility; M. Amort, I. Jaklitsch, A. Kofler, and S. Lobenwein for technical assistance; and M. Kat Occhipinti-Bender for editing the manuscript.
Supplemental data
1The Table summarizes the M-values for the indicated genes in all children at both time points. Negative values indicate gene repression. The 3 columns at the right show the number of children with up- (U, M≥1.0) or down-regulation (D, M≤ -1.0) at either one or both time points. B (both) indicates up- and down-regulation in the same patient at different time points.2Unigene numbers and descriptions for each symbol are given in Table S2.3The column shows the same identifier as the one used in Table 1 of reference15 to facilitate comparisons.
Shown are logarithmic regulation values (M-values) for the indicated 62 probe sets (from Table 2 Part A) for all 13 children and both investigated time points. ID, Affymetrix ID. The description for each probe set is given in Table S2.
Shown are logarithmic regulation values (M-values) for the indicated 62 probe sets (from Table 2 Part A) for all additional biologic systems (see Table 1 in the main text). ID, Affymetrix ID for human probe sets, ID-Mouse, Affymetrix ID (Mouse Genome 430 2.0 microarray). The description for each probe set is given in Table S2.
Shown are expression (E) -values of the 34 probe sets corresponding to 27 cell cycle genes as determined at the onset of treatment (0h-values) in all 13 ALL children. In the last row, all E-values were added up as a tentative measure for proliferative activity of the respective patient (Total). ID, Affymetrix ID. The description for each probe set is given in Table S2.
Shown are expression (E) -values of the 34 probe sets corresponding to 27 cell cycle genes as determined at the onset of treatment (0h-values) in the adult (Adult) and the two healthy donors (HD-1 and 2). In cell lines and mice, the values of the untreated controls are depicted. In the last row, all E-values were added up as a tentative measure for proliferative activity of the respective biologic system (Total). ID, Affymetrix ID. The description for each probe set is given in Table S2.