Abstract
To characterize human hematopoietic stem cells (HSCs), xenotransplantation techniques such as the severe combined immunodeficiency (SCID) mouse repopulating cell (SRC) assay have proven the most reliable methods thus far. While SRC quantification by limiting dilution analysis (LDA) is the gold standard for measuring in vitro expansion of human HSCs, LDA is a statistical method and does not directly establish that a single HSC has self-renewed in vitro. This would require a direct clonal method and has not been done. By using lentiviral gene marking and direct intra-bone marrow injection of cultured CD34+ CB cells, we demonstrate here the first direct evidence for self-renewal of individual SRC clones in vitro. Of 74 clones analyzed, 20 clones (27%) divided and repopulated in more than 2 mice after serum-free and stroma-dependent culture. Some of the clones were secondary transplantable. This indicates symmetric self-renewal divisions in vitro. On the other hand, 54 clones (73%) present in only 1 mouse may result from asymmetric divisions in vitro. Our data demonstrate that current ex vivo expansion conditions result in reliable stem cell expansion and the clonal tracking we have employed is the only reliable method that can be used in the development of clinically appropriate expansion methods.
Introduction
Ex vivo expansion of hematopoietic stem cells (HSCs) is a major challenge for clinical and experimental transplantation protocols.1-3 Over the past 15 years many investigators have undertaken experiments to study ex vivo expansion of HSCs. These studies have led to a number of clinical trials to evaluate ex vivo-expanded cells in patients. However, no significant clinical benefit has been demonstrated to date. Clonal kinetics of ex vivo-expanded HSCs is one of the basic transplantation biology questions to be addressed before we can optimize ex vivo expansion approaches.
To characterize human HSCs, the severe combined immunodeficiency (SCID) mouse repopulating cell (SRC) assay has proven the most reliable method thus far.4,5 When this assay was used, the maximal expansion that was achieved was 2- to 4-fold expansion.6-9 While quantitative SRC assay using limiting dilution analysis (LDA) is the gold standard for measuring expansion, it is a statistical method and does not directly establish that a single HSC has self-renewed in vitro. This would require a direct clonal method and has not been done.
Retrovirus-mediated gene marking has been widely used for clonal analysis. Since these vectors essentially integrate at random, each genomic integration site serves as a distinct clonal marker that can be used to trace the progeny of individual stem cells after transplantation.10,11 Gene-marking studies in SRC assays have successfully elucidated the in vivo kinetics of human HSCs such as proliferation, self-renewal, and multilineage differentiation.12,13
We previously reported serum-free and stroma-dependent (SFSD) culture conditions for the ex vivo expansion of human CD34+ cells using the murine bone marrow stromal cell line HESS-5 and human cytokines Flk-2/Flt-3 ligand (FL), thrombopoietin (TPO), and stem cell factor (SCF).14,15 To obtain direct evidence that self-renewal division of HSCs in this culture system results in SRC “expansion,” we applied lentiviral gene marking and direct intra-bone marrow transplantation (iBMT) of cultured CD34+ cord blood (CB) cells. We demonstrate here the first direct evidence for self-renewal of individual SRC clones in vitro.
Materials and methods
Collection and purification of human CB CD34+ cells
Informed consent was provided according to the Declaration of Helsinki. CB was obtained from full-term deliveries according to procedures approved by the institutional review board of the Tokai University School of Medicine (Kanagawa, Japan). Mononuclear cells (MNCs) were isolated from CB by Ficoll-Hypaque (Lymphoprep, 1.077 ± 0.001 g/mL; Nycomed, Oslo, Norway) density gradient centrifugation. CD34+ cells were purified by positive selection using an immunomagnetic bead system (MACS; Miltenyi Biotec, Glodbach, Germany). More than 95% of the enriched cells were CD34+ cells, as shown by flow cytometric analysis (FACSCalibur; Becton Dickinson, San Jose, CA).
Serum-free and stroma-dependent (SFSD) culture system for ex vivo expansion
In this culture system, CB CD34+ cells were physically separated from the stromal layer by a polyethylene-terephthalate track-etched membrane in cell culture inserts (BD Labware, Franklin Lakes, NJ), as previously described.14 First, the murine hematopoietic-supportive stromal cells HESS-516 were cultured on the reverse side of the track-etched membrane of the insert in 12-well microplates in minimal essential medium-alpha (MEM-α; Nikken Bio Medical Laboratory, Kyoto, Japan) supplemented with 10% (vol/vol) horse serum (GibcoBRL) at 37°C under 5% CO2 in humidified air. Once cells grew to confluency, stromal cells were irradiated with 15 Gy using a 137Cs γ-irradiator, before being washed 5 times with StemPro, and the media changed for coculture. Transduced CB CD34+ cells were seeded on the upper side of the membrane in the insert where the cytoplasmic villi of HESS-5 cells passed through the etched 0.45-μm pores (pore density; 1.0 × 108/cm2) and were cultured in StemPro TM-34SFM (GibcoBRL) supplemented with StemPro TM-34 Nutrient Supplement (GibcoBRL), 2 mM l-glutamine (GibcoBRL), and penicillin/streptomycin with recombinant human TPO (50 ng/mL; a gift from Kirin Brewery, Tokyo, Japan), SCF (50 ng/mL; a gift from Kirin Brewery) and FL (50 ng/mL; R&D Systems, Minneapolis, MN).
5-and 6-carboxyfluorescein diacetate succinimidyl ester (CFSE) labeling
CFSE labeling of CB CD34+ cells was carried out as described previously.17 Cells were incubated with 10 μM CFSE (Molecular Probes, Eugene, OR) for 10 minutes at 37°C and then further dye uptake was prevented by washing with cold fetal calf serum (FCS). Labeled cells were cultured in the SFSD system and their kinetics were evaluated using a FACS Calibur (Becton Dickinson).
SRC assay
Male or female NOD/Shi-scid (NOD/SCID) mice were obtained from CLEA JAPAN (Tokyo, Japan). All experiments were approved by the animal care committee of Tokai University. Mice were housed in microisolated cages and given autoclaved food and water, acidified just before and after total body irradiation (300 cGy-350 cGy). Mice were anesthetized briefly with ether and injected with transduced cells through the tail vein. After 10 weeks, mice that had undergone transplantation were killed and bone marrow cells were flushed from the femura and tibias with PBS.
iBMT of human hematopoietic cells
iBMT was carried out as previously described.18 In brief, a 29-gauge needle was inserted into the joint surface of the right tibia of anesthetized mice, and human hematopoietic cells in a 10-μL suspension were injected into the BM cavity. For injections, a 1-mL Hamilton syringe with fixed 31-gauge needle was used.
Lentivirus production and infection of CD34+ cells
The vesicular stomatitis virus-G protein (VSV-G) pseudotyped lentiviral vector was generated by transient cotransfection of the self-inactivating vector construct pCS-CG19 with the VSV-G-expressing construct pMD.G, the rev-expressing construct pRSV, and the packaging construct pMDLg/p.RRE into 293T cells (ATCC, Manassas, VA) using calcium phosphate transfection, according to the manufacturer's recommendations (Invitrogen, Carlsbad, CA). 293T cells were grown on collagen I-coated 100-mm dishes (BD Labware, Bedford, MA) in Dulbecco modified Eagle medium (DMEM; Biofluids, Rockville, MD) containing 10% heat-inactivated FCS (JRH Biosciences, Lenexa, KS). The vector-containing supernatant was collected every 24 hours for 3 days, filtered through a 0.45-μm pore size filter, and superconcentrated twice by centrifugation at 50 000g for 90 minutes at 20°C. Viral supernatants were concentrated 100 to 200 times by ultracentrifugation, resuspended in serum-free medium StemPro TM-34SFM, and stored at -80°C until use.
CD34+ cells (100 000) in 200 μL StemPro TM-34SFM were seeded in each well of 96-well V-bottomed plates and cultured for 5 hours with superconcentrated lentiviral supernatant (200 μL/well) at a multiplicity of infection (MOI) of 50.20
FACS analysis and sorting
On day 4 after infection, the cells were washed once with PBS and stained with a monoclonal antibody against CD34 conjugated with PE (Becton Dickinson). Infection efficiency was evaluated using a FACS Calibur (Becton Dickinson). At 10 weeks after transplantation, the mice were killed and the bone marrow and spleen were collected. Cells were stained with monoclonal antibodies (mAbs) to human leukocyte differentiation antigens: PE-conjugated anti-human CD19 (SJ25C1) and CD33 (Leu-M9) (all Becton Dickinson); and APC-conjugated anti-human CD45 (J.33; Coulter/Immunotech, Hialeah, FL). A FACS analysis was conducted by 3- or 4-color flow cytometric analysis using a FACSCalibur. For isolation of CD19+EGFP+ and CD33+EGFP+ cells, cells were stained with PE-conjugated anti-human ECD-conjugated CD19 and PE-conjugated CD33, and APC-conjugated anti-CD45 mAbs and sorted using the FACSVantage flow cytometer (BD Biosciences).
3′-LTR integration site analysis using linear amplification-mediated-PCR
To identify the genomic-proviral junction sequence, linear amplification-mediated polymerase chain reaction (LAM-PCR) was performed on DNA isolated from total bone marrow as described by Schmidt et al.21 In brief, linear amplification of target DNA was performed by repeated primer extension using a vector-specific 5′-biotinylated primer LTR1 (5′ GAA CCC ACT GCT TAA GCC TCA 3′) with Taq polymerase (2.5 U; Qiagen, Valencia, CA) from 100 ng of each sample DNA. After selection with magnetic beads (Dynal, Oslo, Norway), the extension products were incubated with Klenow polymerase (2 U), dNTPs (300 mM; Pharmacia, Uppsala, Sweden), and random hexanucleotide mixture (Takara, Otsu, Japan). The samples were then washed on the magnetic particle concentrator (Dynal) and incubated with Sac I (5 U in 20 μL; Promega, Madison, WI) for 2 hours at 37°C, and then Tsp509 I (5 U in 20 μL; New England BioLabs, Ipswich, MA) for 2 hours at 55°C. After an additional wash step, a double-stranded asymmetric linker cassette and T4 DNA Ligase (6 U; New England BioLabs) were incubated with the beads at 16°C overnight. After denaturing with 0.1 N NaOH, each ligation product was amplified with Taq polymerase (5 U), 25 pmol of vector-specific primer LTR2 (5′ AGC TTG CCT TGA GTG CTT CA 3′), and linker cassette primer LC1 (5′ GTA CAT ATT GTC GTT AGA ACG CGT AAT ACG ACT CA 3′). Of each PCR product, 0.2% served as a template for a second, nested PCR with internal primers LTR3 (5′ AGT AGT GTG TGC CCG TCT GT 3′) and LC2 (5′ CGT TAG AAC GCG TAA TAC GAC TCA CTA TAG GGA GA 3′) under identical conditions. Amplified products were loaded on 2% agarose gels. To quantify the number of each band in cases where several clones of infected cells contributed to the graft, densitometric analysis was done using Densitograph software (ATTO, Tokyo, Japan). The final products were sequenced after cloning into the TOPO TA cloning vector (Invitrogen).
PCR tracing of individual clones
To trace individual clones by amplifying unique genomic-proviral junction, primer pairs located in the genome and vector were used. LTR3 was used as the 5′ primer. The 3′ primer was designed from the genomic-proviral junction sequence after determining its precise location in the human genome by comparison to known genomic sequences using a basic local alignment search tool (BLAST) search (Table S1 on the Blood website; see the Supplemental Table link at the top of the online article). For semiquantitative PCR, primers located in GFP cDNA (5′ primer: CCA GTT CAG CGT GTC CGG CG; 3′ primer: GGG GTC TTT GCT CAG GGC GG) were used. AmpliTaq and reagents were purchased from Takara Shuzo (Otsu, Japan). PCR conditions for 100 ng of each sample DNA were set as follows: 30 cycles consisting of denaturation at 94°C for 1 minute, annealing at 60°C for 1 minute, and elongation at 72°C for 2 minutes with a 2-second extension in each additional cycle, and final reaction at 71°C for 8 minutes followed by gradual cooling to 4°C. Amplified products were loaded on 2% agarose gels.
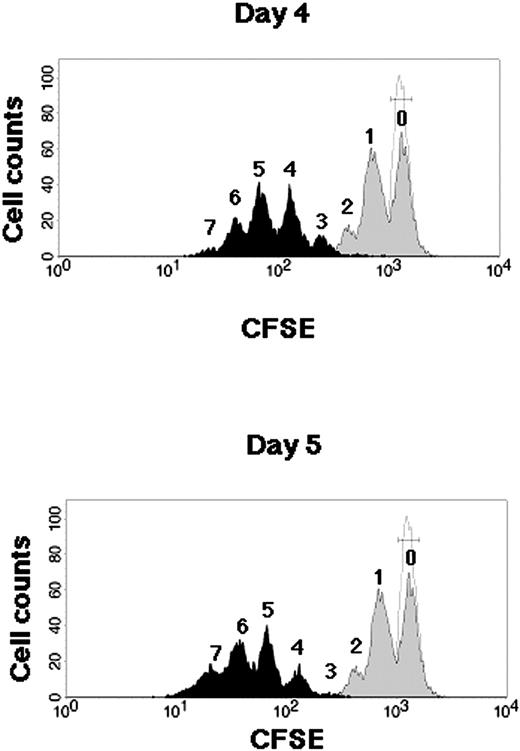
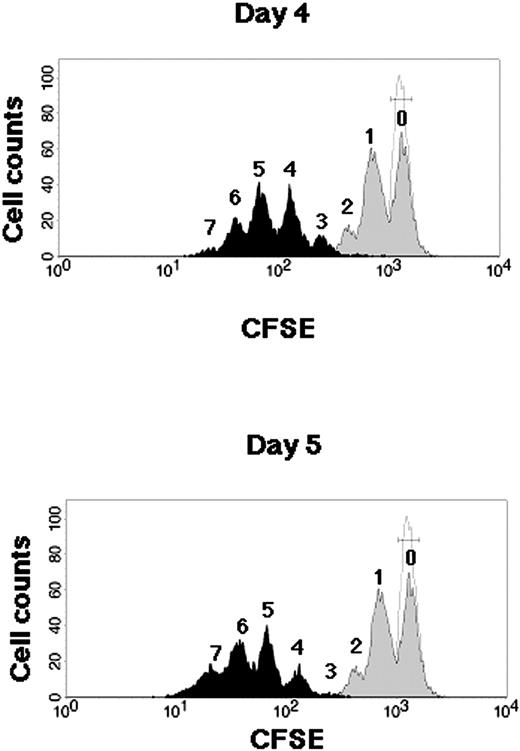
Kinetics of CB CD34+ cells during SFSD culture. Representative flow cytometric profiles of CFSE-labeled CB CD34+ cells. CB CD34+ cells were stained with CFSE and cultured in the SFSD system. The fluorescence intensity was analyzed at 2 days (gray histogram), 4 days (top, black histogram), and 5 days (bottom, black histogram) after culture. The number above each peak represents the number of divisions according to the fluorescence intensity. The relative frequencies of undivided cells were 51.24% ± 3.12% % at day 2, 0.09% ± 0.04% at day 4, and 0.04% ± 0.03% at day 5 (n = 3). The fluorescence intensity of the cells incubated with colcemid confirms the position of the cells that have not divided (dotted line).
Kinetics of CB CD34+ cells during SFSD culture. Representative flow cytometric profiles of CFSE-labeled CB CD34+ cells. CB CD34+ cells were stained with CFSE and cultured in the SFSD system. The fluorescence intensity was analyzed at 2 days (gray histogram), 4 days (top, black histogram), and 5 days (bottom, black histogram) after culture. The number above each peak represents the number of divisions according to the fluorescence intensity. The relative frequencies of undivided cells were 51.24% ± 3.12% % at day 2, 0.09% ± 0.04% at day 4, and 0.04% ± 0.03% at day 5 (n = 3). The fluorescence intensity of the cells incubated with colcemid confirms the position of the cells that have not divided (dotted line).
Statistical analysis
The data from several limiting dilution experiments were combined and used for analysis. The frequency of SRCs in the test BM sample was calculated by applying Poisson statistics to the proportion of negative recipients at different dilutions using L-Calc software (StemCell Technologies, Vancouver, BC, Canada). Data are represented as mean plus or minus standard error (SE). The 2-sided P value was determined, testing the null hypothesis that the 2 population medians are equal. P values less than .05 were considered to be significant.
Results
Cell division kinetics and SRC expansion during SFSD culture
Cell division kinetics during culture were assessed by a high-resolution procedure for tracking cells using their proliferation history, based on the loss of cellular fluorescence after staining with CFSE (Figure 1). Flow cytometric analysis of CD34+ cells harvested from 4- and 5-day cultures resolved more than 3 and 4 generations of progeny, respectively. Undivided cells were not detectable, which suggested the possibility of no dormant cells in this system after 4 days of culture.
A quantitative SRC assay was used to compare the frequency of SRCs between uncultured and cultured CD34+ cells in this system. When purified CD34+ cells were cultured for 5 days, the total number of cells had expanded 20.1-fold ± 2.7-fold (n = 10). Since 63.6% ± 4.2% (n = 10) of cells were CD34+ after culture, CD34+ cells had increased around 12.8-fold. A graded number of CD34+ cells were transplanted intravenously into irradiated NOD/SCID mice. Human cell engraftment in mice was determined 10 weeks after transplantation (Table 1). The frequencies of SRCs in uncultured and cultured CD34+ cells were 1 per 25 794 cells (95% CI; 14 408 to 46 177 cells) and 1 per 34 056 cells (95% CI; 19 072 to 60 810 cells), respectively. Thus, the calculated change in the total SRC numbers following culture was about a 9.8-fold increase. However, the result does not directly demonstrate that each HSC has multiplied 9.8 times through self-renewal divisions in vitro, since the changes of homing and proliferative abilities following culture may also affect results of the SRC assay. Clonal analysis is a more accurate method to demonstrate true SRC multiplication.
Quantitative SRC assay of gene-marked CD34+ CB cells before and after SFSD culture
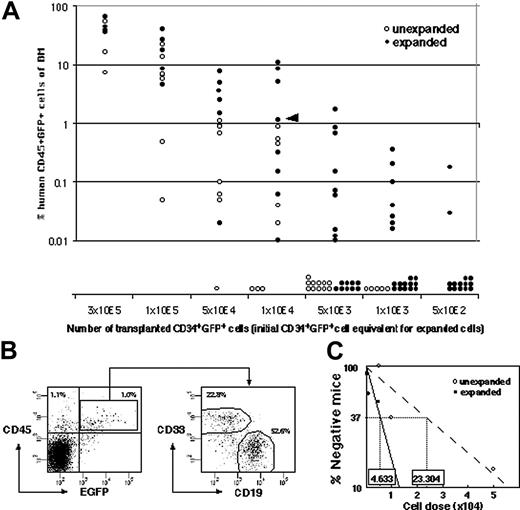
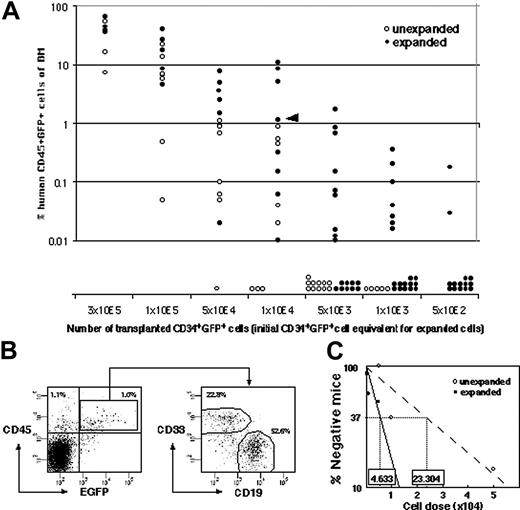
To set up clonal analysis, ex vivo expansion of gene-marked SRC was measured by quantitative SRC assay. CD34+ CB cells were infected with recombinant lentivirus containing cDNA encoding for enhanced green fluorescent protein (EGFP), and 42% ± 7% (n = 26) of the CD34+ cells subsequently expressed EGFP 4 days after infection. The infection conditions were optimized to minimize the probability of multiple integrations into target cells.22 This time, the SRC “expansion” was measured by transplanting the graded number of cells before culture and their whole progeny after 4 days of culture (Figure 2A). Both CD19+EGFP+ lymphoid and CD33+EGFP+ myeloid reconstitution were detected in each mouse when human CD45+EGFP+ cells were engrafted (Figure 2B). The calculated frequencies of EGFP+ SRCs before and after culture were 1 per 23 304 cells (95% CI; 17 147 to 31 672 cells) and 1 per 4633 cells (95% CI; 3674 to 5842 cells), respectively, making the fold expansion 5.03 (Figure 2C).
In vitro SRC multiplication at clonal level
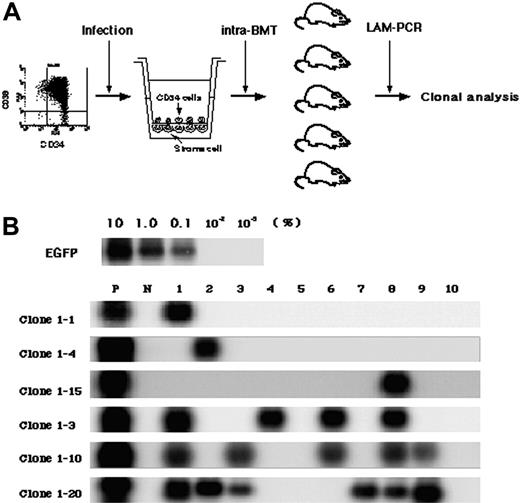
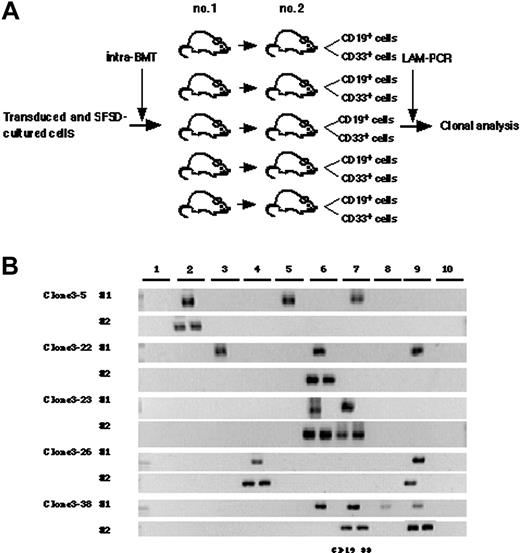
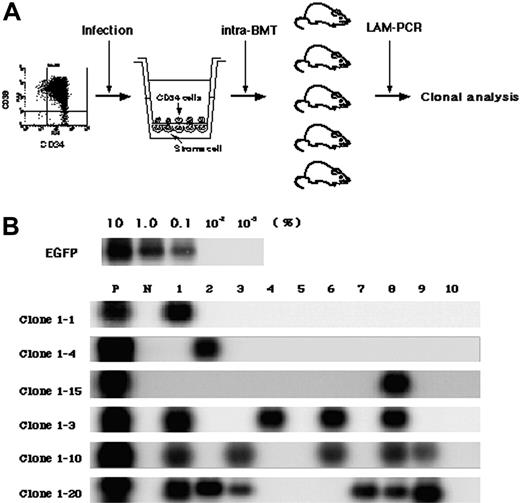
To detect multiplied clones, 5 × 104 gene-marked CD34+ cells were cultured for 4 days and then divided into 10 lots, each of which was transplanted directly into the bone marrow of a NOD/SCID mouse (Figure 3A). While the seeding efficiency of intravenously transplanted human cells into NOD/SCID mice is less than 10%, iBMT negates the effect of homing, allowing efficient detection of amplified SRCs.18 We used linear amplification-mediated (LAM)-PCR23 to detect unique genomic-proviral junctions as clonal markers. Detection of the same clones in different mice would provide direct evidence of ex vivo multiplication of an SRC clone. We identified 20 clone-specific genomic-proviral junction sequences by LAM-PCR on 10 mice (Table 2). Clone-specific primers were designed and PCR was performed to trace each clone within 10 mice. Since the detection sensitivity of a clone by PCR was assumed to be 0.1% by semiquantitative PCR using primers within the EGFP cDNA, clones comprising more than 104 cells would be detected by this method. Representative clones are shown in Figure 3B. Although 14 clones were detected in only 1 mouse (such as clones 1-1, 1-4, and 1-15), 6 clones were detected in more than 2 mice. Clones 1-2, 1-7, and 1-16 were detected in 2 mice (data not shown). Clone 1-3 was detected in 4 mice, clone 1-10 was detected in 5 mice, and clone 1-20 was detected in 6 mice. The data suggested that some of clones divided during culture and participated in hematopoietic reconstitution of 2, 4, 5, or 6 independent mice.
Comparison of the frequency of infected SRCs with and without SFSD culture. (A) The infected cells were divided into 2 groups consisting of the same number of cells (5 × 102, 1 × 103, 5 × 103, 1 × 104, 5 × 104, 1 × 105, 3 × 105 cells/mouse). One group was instantly transplanted (n = 41), and the other was cultured for 4 days in SFSD culture, and then transplanted into NOD/SCID mice (n = 75). At 10 weeks after transplantation, the mice were killed and the percentages of CD45+EGFP+ cells in the mouse bone marrow cells were determined by flow cytometry. The presence of cells was also confirmed by PCR using EGFP primers to rule out the false-positive results by flow cytometry (data not shown). (B) Representative FACS profiles. Data from mouse, indicated by the triangle in panel A, are shown. The numbers in the top left and right quadrants show the percentage of CD45+EGFP- and CD45+EGFP+ populations, respectively. Multilineage repopulation capacity of transduced SRCs was also examined. The numbers in the right panel show the percentage of CD33+ and CD19+ populations within CD45+EGFP+ cells from bone marrow. (C) The frequency of infected SRCs before and after culture. The number shown within each box indicates the calculated frequency of SRCs using L-Calc software (StemCell Technologies).
Comparison of the frequency of infected SRCs with and without SFSD culture. (A) The infected cells were divided into 2 groups consisting of the same number of cells (5 × 102, 1 × 103, 5 × 103, 1 × 104, 5 × 104, 1 × 105, 3 × 105 cells/mouse). One group was instantly transplanted (n = 41), and the other was cultured for 4 days in SFSD culture, and then transplanted into NOD/SCID mice (n = 75). At 10 weeks after transplantation, the mice were killed and the percentages of CD45+EGFP+ cells in the mouse bone marrow cells were determined by flow cytometry. The presence of cells was also confirmed by PCR using EGFP primers to rule out the false-positive results by flow cytometry (data not shown). (B) Representative FACS profiles. Data from mouse, indicated by the triangle in panel A, are shown. The numbers in the top left and right quadrants show the percentage of CD45+EGFP- and CD45+EGFP+ populations, respectively. Multilineage repopulation capacity of transduced SRCs was also examined. The numbers in the right panel show the percentage of CD33+ and CD19+ populations within CD45+EGFP+ cells from bone marrow. (C) The frequency of infected SRCs before and after culture. The number shown within each box indicates the calculated frequency of SRCs using L-Calc software (StemCell Technologies).
Multilineage differentiation of multiplied SRCs at the clonal level
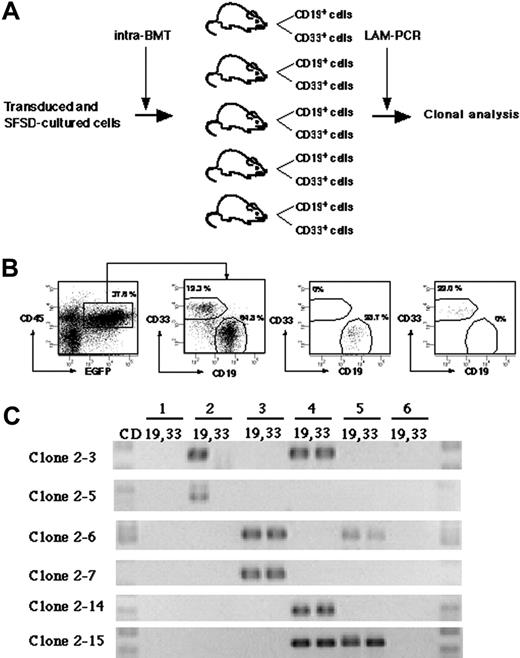
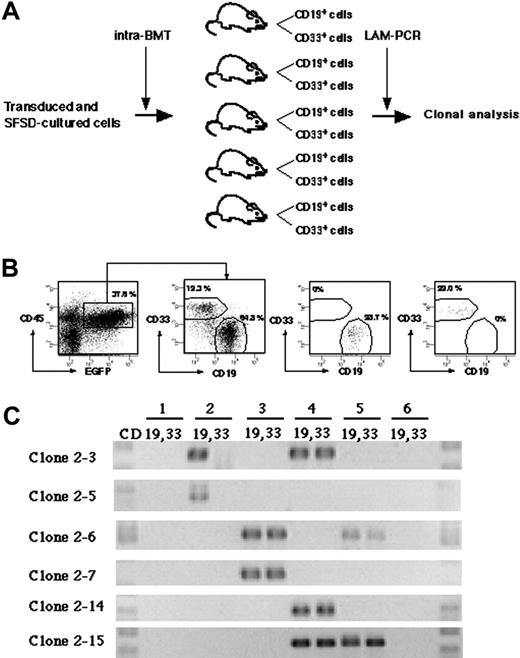
To detect multilineage differentiation of amplified SRCs, purified CD19+EGFP+ and CD33+EGFP+ cells from each mouse were analyzed for each clone (Figure 4A). Although gene-marked cultured cells were transplanted into 10 mice, 4 mice died before analysis. We identified 15 clonal markers from the remaining 6 mice by LAM-PCR (Table 2). Purified CD19+EGFP+ and CD33+EGFP+ cells from each mouse were examined for the presence of each clone (Figure 4B-C). Twelve clones were present in only 1 mouse. Eleven were present in both CD19+ and CD33+ cells (such as clone 2-7 and 2-14), whereas 1 clone (clone 2-5) repopulated only CD19+ cells. There were 3 clones present in 2 independent mice. Two of them reconstituted both CD19+ and CD33+ cells (clone 2-6 and 2-15), but 1 clone (clone 2-3) reconstituted only CD19+ cells in mouse no. 2 and both CD19+ and CD33+ cells in mouse no. 4. These results demonstrate the multilineage differentiation ability of the multiplied SRC clones in our system.
Self-renewal divisions of multiplied SRCs at the clonal level
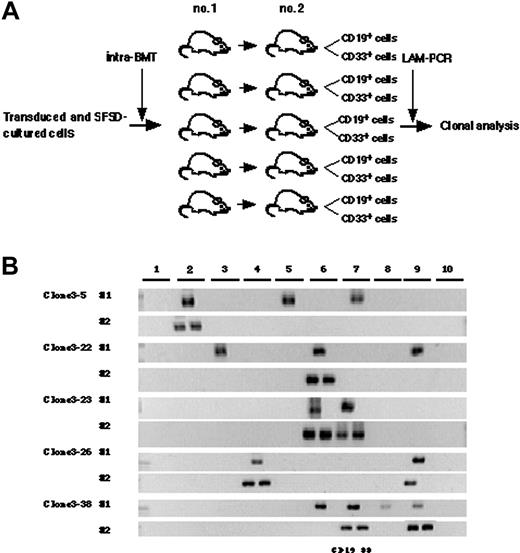
Finally, we designed a secondary transplantation experiment to confirm the self-renewal ability of each clone (Figure 5A). We identified 39 clonal markers from 10 primary and 10 secondary transplanted mice by LAM-PCR, 11 of which were detected in multiple mice (Table 2). Twenty-eight clones were present in a single primary recipient and 16 were detected in a secondary recipient. Eleven clones were detected in multiple primary recipients and 14 of 29 clones were detected in a secondary recipient. Representative clones are shown in Figure 5B. Clones 3-5, 3-22, and 3-24 were present in 3 primary mice but only in 1 secondary mouse with lymphomyeloid differentiation ability. Clone 3-13 was present in 4 primary mice but also only in 1 secondary mouse. Similarly, clones 3-7, 3-29, 3-32, and 3-35 were present in 2 primary mice but in only 1 secondary mouse. Clones 3-23, 3-26, and 3-38 were present in 2 to 4 primary mice and transmitted to 2 secondary mice with maintenance of both self-renewal and multilineage differentiation ability. This result demonstrated that some human HSC clones are able to multiply in vitro.
Discussion
Several protocols for ex vivo expansion of human HSCs demonstrated 2- to 4-fold expansion of SRCs by using quantitative SRC assay.6-9 However, it does not mean a 2- to 4-fold increase of SRC numbers because it is difficult to distinguish between a truly increased SRC number or increased SRC proliferative potential following culture (and therefore increased human engraftment detection) by this assay.22 On the other hand, recent data suggest that surface expression of CXCR4 and CD26 in CD34+ cells by ex vivo manipulation have a positive and negative effect, respectively, on their homing and engraftment in NOD/SCID mice.24,25 Thus, the true self-renewal division of SRCs in vitro could not be assessed by the quantitative SRC assay. By using lentiviral gene-marking and iBMT of cultured CD34+ CB cells, we have successfully dissected the relative contribution of clonal multiplication from homing efficiency and/or proliferative potential on SRC “expansion.”
Detection of multiplied SRCs in SFSD culture. (A) Experimental protocol for the detection of multiplied SRCs in SFSD culture. (B) PCR tracing of individual clones in mice that underwent transplantation with SFSD-cultured cells. Representative clones are shown. Each clone was detected by amplifying a unique genomic-proviral junction sequence with primer pairs located in the genome (clone number shown in Table S1) and vector (LTR3). For semiquantitative PCR, genomic DNA from the indicated ratio of mixture of Jurkat cells with and without transduction by recombinant retrovirus MFG-EGFP22 was used. The numbers at the top indicate each mouse. The numbers on the left indicate each SRC clone. P indicates positive control; N, negative control.
Detection of multiplied SRCs in SFSD culture. (A) Experimental protocol for the detection of multiplied SRCs in SFSD culture. (B) PCR tracing of individual clones in mice that underwent transplantation with SFSD-cultured cells. Representative clones are shown. Each clone was detected by amplifying a unique genomic-proviral junction sequence with primer pairs located in the genome (clone number shown in Table S1) and vector (LTR3). For semiquantitative PCR, genomic DNA from the indicated ratio of mixture of Jurkat cells with and without transduction by recombinant retrovirus MFG-EGFP22 was used. The numbers at the top indicate each mouse. The numbers on the left indicate each SRC clone. P indicates positive control; N, negative control.
We have previously demonstrated that iBMT is 15 times more sensitive than conventional intravenous injection for the detection of SRCs.18 In an attempt to improve the efficiency of homing, seeding, and repopulation, iBMT has been applied to detect clones used to repopulate different individuals.26 If the number of SRCs amplified 5-fold during culture, we should ideally find every clone in 5 different mice when cultured cells are divided and transplanted by this method into 10 mice. However, as shown in Table 2, 74 clones were detected in 110 mice that underwent transplantation. Therefore, the net amplification of SRCs was calculated to be at least 1.5-fold. The discrepancy between 1.5-fold amplification from clonal analysis and 5.03-fold expansion from the quantitative SRC assay may be due to the increased SRC proliferative potential and/or homing and engraftment efficiency following culture, which affects the quantitative SRC assay using intravenous injection.22,24,25 To further dissect these 2 factors, comparing cultured cells by limiting dilution using iBMT and clonal analysis would be useful. Further study is required to elucidate their relative contribution.
Detection of multilineage differentiation by multiplied SRCs. (A) Experimental protocol for the detection of multilineage differentiation by multiplied SRCs. (B) At 10 weeks after transplantation, the mice were killed and CD33+ and CD19+ populations within CD45+EGFP+ cells from bone marrow were sorted. The numbers in the panels show the percentage of the gated cells. Sorted CD19+EGFP+ and CD33+EGFP+ cells show more than 99% pure fractions. (C) PCR tracing of individual clones in CD19+EGFP+ and CD33+EGFP+ cells from mice that underwent transplantation with SFSD-cultured cells. Each clone was detected by amplifying a unique genomic-proviral junction sequence with primer pairs located in the genome (clone number shown in Table S1) and vector (LTR3). The numbers at the top indicate each mouse. The numbers on the left indicate each SRC clone.
Detection of multilineage differentiation by multiplied SRCs. (A) Experimental protocol for the detection of multilineage differentiation by multiplied SRCs. (B) At 10 weeks after transplantation, the mice were killed and CD33+ and CD19+ populations within CD45+EGFP+ cells from bone marrow were sorted. The numbers in the panels show the percentage of the gated cells. Sorted CD19+EGFP+ and CD33+EGFP+ cells show more than 99% pure fractions. (C) PCR tracing of individual clones in CD19+EGFP+ and CD33+EGFP+ cells from mice that underwent transplantation with SFSD-cultured cells. Each clone was detected by amplifying a unique genomic-proviral junction sequence with primer pairs located in the genome (clone number shown in Table S1) and vector (LTR3). The numbers at the top indicate each mouse. The numbers on the left indicate each SRC clone.
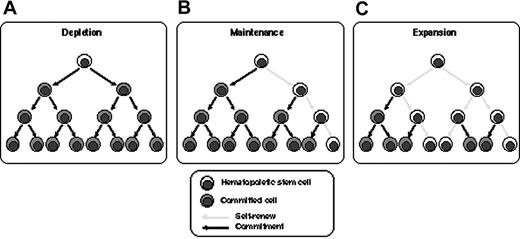
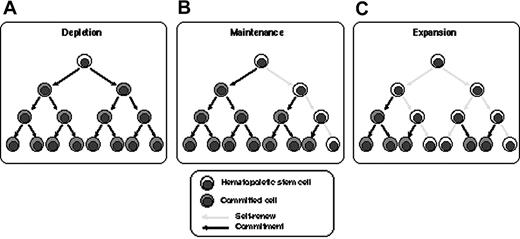
Dividing HSCs in vitro resulted in depletion, maintenance, or expansion (Figure 6). Of 74 clones analyzed in the 3 experiments presented here, 54 clones (73%) were present in 1 individual, which means that HSCs were dormant or maintained during culture. Contribution of dormant HSCs to this result is negligible, since more than 99.9% of CD34+ cells proliferated in the SFSD culture system (Figure 1) and the SRC frequency by iBMT was 1 in 44 CD34+CD38- cells and 1 in 1010 Lin-CD34+ cells.18,27 Glimm and Eaves17 also demonstrated that the vast majority of SRCs present in 5-day cultures of CB CD34+ cells stimulated with cytokines had executed at least 3 full cell cycles. Therefore, asymmetric self-renewal division would result in HSC maintenance in this population (Figure 6B). Twenty clones (27%) divided and resulted in multiplication up to 6 times after culture. Furthermore, 3 clones, clones 3-23, 3-26, and 3-38, multiplied and maintained both multilineage differentiation and self-renewal activity. This indicates symmetric self-renewal division of HSCs at some stage during culture (Figure 6C). Therefore, there are at least 2 classes of HSCs, each of which behaves differently in SFSD culture: maintaining HSCs and multiplying HSCs. Identification of the differences between these 2 populations with respect to surface markers, state of gene expression, or other behaviors such as status of interaction with stromal cells in vitro, would greatly facilitate the development of an optimal culture system for ex vivo expansion. Assessment of the biologic function of individual human HSCs would also provide a powerful tool for the characterization of cellular and molecular determinants that govern their reconstitution ability.
Detection of self-renewal and multi-lineage differentiation by multiplied SRCs. (A) Experimental protocol for the detection of self-renewal and multilineage differentiation by multiplied SRCs. (B) PCR tracings of individual clones in bone marrow cells from mice that underwent transplantation with SFSD-cultured cells are shown in column 1. The bone marrow cells were then transplanted into secondary mice. PCR tracings of individual clones in CD19+EGFP+ and CD33+EGFP+ cells from the secondary mice are shown in column 2. Each clone was detected by amplifying a unique genomic-proviral junction sequence with primer pairs located in the genome (clone number shown in Table S1) and vector (LTR3). The numbers at the top indicate each mouse. The numbers on the left indicate each SRC clone.
Detection of self-renewal and multi-lineage differentiation by multiplied SRCs. (A) Experimental protocol for the detection of self-renewal and multilineage differentiation by multiplied SRCs. (B) PCR tracings of individual clones in bone marrow cells from mice that underwent transplantation with SFSD-cultured cells are shown in column 1. The bone marrow cells were then transplanted into secondary mice. PCR tracings of individual clones in CD19+EGFP+ and CD33+EGFP+ cells from the secondary mice are shown in column 2. Each clone was detected by amplifying a unique genomic-proviral junction sequence with primer pairs located in the genome (clone number shown in Table S1) and vector (LTR3). The numbers at the top indicate each mouse. The numbers on the left indicate each SRC clone.
Schematic demonstration of the fate of HSCs in vitro. The fates of HSCs in SFSD culture are shown as depletion (A), maintenance (B), and expansion (C). Our data indicate that 73% of HSCs were maintained and 27% expanded in SFSD culture.
Schematic demonstration of the fate of HSCs in vitro. The fates of HSCs in SFSD culture are shown as depletion (A), maintenance (B), and expansion (C). Our data indicate that 73% of HSCs were maintained and 27% expanded in SFSD culture.
New classes of SRCs have been recently identified.13,28-30 While we focused on long-term repopulating cells (LTRCs) in this study, it is of interest to determine what other types of SRCs also expanded in the SDSF culture system. Rapid-SRCs (R-SRCs) identified in Lin-CD34+CD38lowCD36- cells repopulate the myeloid and erythroid lineage within 2 weeks of iBMT.28 Because we did not analyze at 2 weeks after transplantation and the long-term fate of R-SRCs has not been shown, we could not determine their contribution in this study. Short-term repopulating cells (STRCs) with lymphomyeloid differentiation potential were identified by transplanting CD34+CD38+ cells into NOD/SCID-β2 microglobulin-null mice.29 Since we did not examine the presence of STRCs in NOD/SCID mice at 10 weeks after iBMT, we could not exclude the possibility that the 27 clones that did not transmit to secondary hosts in experiment 3 (Table 2) were STRCs. While we believe that most of the clones identified in this study were LTRCs, clonal analysis at an early time point after transplantation is required to determine the magnitude of expansion of STRCs and R-SRCs. Further study would also reveal the extent of clonal exhaustion of LTRCs during expansion culture.
Collectively, our data demonstrate that current ex vivo expansion conditions result in reliable stem cell expansion, and the clonal tracking we have employed is the only reliable method that can be used in the development of clinically appropriate expansion methods.
Prepublished online as Blood First Edition Paper, January 3, 2006; DOI 10.1182/blood-2005-08-3108.
Supported by grants from the Ministry of Education, Science, and Culture, Japan; and a Research Grant on Human Genome, Tissue Engineering (H17-014) from the Japanese Ministry of Health, Labor, & Welfare, Tokyo, Japan.
K.A. designed and performed research and wrote the paper; T.Y., T.S., H. Miyatake, H. Matsuzawa, and M.O. performed research; H. Miyoshi and T.T. contributed vital new reagents; S.K. and T.H. analyzed data.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank members of the animal facility and the center for regenerative medicine of Tokai University, especially Mayumi Nakagawa and Tomoko Uno, and members of the Tokai Cord Blood Bank for their assistance. We also thank John Dick for critical reading of the manuscript.