Abstract
Somatic hypermutation and class-switch recombination in germinal centers critically depend on activation-induced cytidine deaminase (AID). Deregulation of AID may lead to the aberrant activation or persistence of both genetic processes, thus contributing to the pathogenesis of B-cell lymphomas by mistargeted mutagenesis or recombination. The Epstein-Barr virus (EBV) establishes an asymptomatic latent infection in more than 90% of the human population, but it has also been linked to lymphomagenesis. A cooperative relationship of EBV and the germinal center reaction during the establishment of viral persistence has been postulated, but the contribution of EBV latent genes to the respective genetic events remains to be investigated in detail. In the present study, we show that activation of the EBV growth program has a clear inhibitory effect on AID expression, due to a negative effect of the master transcription factor of this program, EBNA2. This mechanism may counterbalance AID induction by the LMP1 protein, in order to prevent deleterious genetic changes during EBV-induced B-cell growth. EBNA2-mediated AID inhibition also provides a molecular explanation for the previously observed differences in somatic hypermutation activity in EBV-associated lymphoproliferative diseases, thus pointing to a crucial mechanism of EBV-mediated regulation of genomic integrity.
Introduction
The maturation of humoral immunity during acute infections depends on the germinal center reaction. Here, B cells proliferate and perform somatic hypermutation to introduce random point mutations into the variable region of the immunoglobulin (Ig) genes, as well as class-switch recombination to exchange the Ig constant region. Activation-induced cytidine deaminase (AID) is a key factor of these genetic events1,2 and is even capable of inducing these processes in non-B cells if ectopically expressed.3,4 AID acts during the initial phase of both processes by generating DNA lesions in the respective gene loci. As AID overexpression has been implicated in untargeted mutagenesis and tumorigenesis,5,6 the expression and activity of AID need to be tightly regulated in B cells in order to prevent lymphomagenesis.
The Epstein-Barr virus (EBV) is a human herpesvirus that establishes asymptomatic latent infection in more than 90% of the human population.7 It is, however, also associated with several human B-cell malignancies8 and is by itself capable of growth transforming human B cells in vitro. In the resultant lymphoblastoid cell lines (LCLs), 9 latent viral genes are expressed—3 latent membrane proteins (LMPs) and 6 EBV nuclear antigens (EBNAs). Among them, LMP1 and LMP2A are constitutively active signaling molecules that mimic the physiological signals of CD40 and the B-cell receptor, respectively. EBNA1 is required for maintenance and replication of the viral episome in human cells. The transcription factor EBNA2 is a master switch for many viral promoters used in LCLs and can also be considered a constitutively active analog of a cellular signal, as it acts similarly to the active form of Notch in stimulating transcription through binding to the transcriptional repressor Rbp-Jκ.9
Besides this so-called latency III (or growth program) of LCLs, other forms of EBV latent gene expression have been described in tumors.8 In the latency I program in Burkitt lymphoma (BL) cells, only EBNA1 is expressed, while EBNA2-dependent gene expression is apparently counterselected.10 The malignant Hodgkin/Reed-Sternberg cells in Hodgkin lymphoma express the latency II program in which LMP1 and LMP2A can be detected in addition to EBNA1. Restricted expression of viral genes has also been described in nonmalignant B cells during viral persistence and appears to be correlated to their differentiation stage.11 On this basis, it has been suggested that EBV uses the normal B-cell differentiation process of the germinal center reaction and uses differential expression of viral genes mimicking important physiological signals in order to establish viral latency in the memory B-cell compartment. In these long-lived memory B cells, barely any expression of viral genes is detectable, and this latency program likely forms the basis for immune escape and long-term EBV persistence.12
Several observations in normal as well as pathological situations have raised questions about the relationship of EBV and germinal centers. While in tonsillar B cells during latent infection, EBNA2 and promoter use typical of the growth program have been found only in naive B cells,11 EBV-driven proliferation in infectious mononucleosis (IM) affects mainly memory B cells.13 Also, while during viral persistence a tonsillar population of EBV-infected cells with a germinal center phenotype expresses the latency II program including LMP1,11 the expression of LMP1 has been shown to negatively interfere with the formation of germinal centers in transgenic mice.14 Finally, EBV+ cells can scarcely be detected in germinal centers in normal tonsils, and the rare EBV+ cells in germinal centers in IM have been shown not to participate in somatic hypermutation characterizing the germinal center reaction.15
A similar multifaceted association of EBV with the genetic events of the germinal center has been described. LMP1 is capable of inducing CD40-independent switch recombination in mouse and human cells,14,16 and it has been shown to induce AID expression in cell lines. On the other hand, LCLs express LMP1 but are not typically performing ongoing switch recombination and are mostly IgM+.17 Also, while ongoing somatic hypermutation has been described in some EBV-driven B-cell lymphoproliferations such as posttransplantation lymphoproliferative disease (PTLD) and in the EBV+ B cells proliferating on the background of angioimmunoblastic lymphadenopathy with disproteinaemia (AILD),18,19 EBV-infected cells proliferating in vivo in IM as well as in vitro are characterized by a clear absence of hypermutation capacity.13,20
To understand how EBV affects the normal as well as malignant development of B cells, it is therefore important to determine how EBV, the germinal center reaction, and the respective genetic events are interrelated. As a contribution to these issues, we have here studied the effect of EBV latent genes on the expression of AID in vitro and in vivo.
Materials and methods
Cell culture and conditional gene expression
The human EBV– B-lymphoma cell line BJAB/K3,21 the human EBV– BL cell line BL41/K3,21 and the estrogen-dependent LCL EREB2-522 were cultured in RPMI-1640 medium containing 10% fetal bovine serum (FBS), penicillin, streptomycin, sodium pyruvate, and glutamine (all from Invitrogen, Frederick, MD) and maintained at 37°C in a humidified 5% CO2 atmosphere. BJAB/K3 and BL41/K3 were cultured in the presence of 800 μg/mL geniticin (Gibco, Grand Island, NY). β-Estradiol (Sigma, Poole, United Kingdom) was added to the cell culture medium at a final concentration of 1 μM.
Quantitative reverse-transcription–polymerase chain reaction (RT-PCR)
Total RNA was isolated from whole-cell lysates with the RNeasy-Kit (Qiagen, Hilden, Germany), and cDNA was generated with the first-strand cDNA synthesis-Kit (Roche, Mannheim, Germany) using oligo-p(dT)15 primers. For quantitative PCR in the Light cycler Sybr Green system (Roche), the following primers were used for AID (hAIDRT2: GTGACATTCCTGGAAGTTGC and hAIDRT3: CCAACCTCAGTCTGAGGATCTTC), for CD19 (CD19fw: CTCCTTCTCCAACGCTGAGT and CD19rv: TGGAAGTGTCACTGGCATGT), for CD21 (hCD21RT1: AGATCCTAAGAGGCCGAATGG and hCD21RT2: CACATAGCCAGGGTTACAGC), and for Ig heavy chain (hmuHCRT1: CTGACCTTCCAGCAGAATGCG and hmuHCRT2: AAGTAGACATCGGGCCTGTGC). mRNA copy numbers were determined in duplicate, and calculated and normalized to CD19 according to the instructions of the manufacturer. CD19 was used for normalization as array kinetics analyses showed no variation in its levels in EREB2-5 cells upon EBNA2 regulation.
Western blot and antibodies
Cells (3 × 106) were lysed in Laemmli SDS sample buffer, and protein concentrations were measured by the BioRad DC protein assay (Biorad, Reinach, Switzerland). Equal amounts of proteins were loaded on a denaturing SDS-PAA-Gel, separated, transferred to a PVDF membrane (Amersham Biosciences, Buckinghamshire, United Kingdom), and incubated with the 5G9 antibody for AID,23 an anti-LMP1 antibody (Dako, Glostrup, Denmark), or an anti-GAPDH antibody (Abcam, Cambridge, United Kingdom).
In situ hybridization and immunohistochemistry
Paraffin wax–embedded tonsil samples from 5 patients with acute infectious mononucleosis and from 2 virus carriers were retrieved from the files from the Institute for Pathology, Erlangen. All tissue samples had been submitted for diagnostic purposes and were studied in accordance with national ethical principles. In situ hybridization for the detection of the small EBV-encoded RNAs (EBERs) was done using 35S-labeled single-stranded RNA probes as described previously.24 For the immunohistochemical detection of AID expression, paraffin sections were dewaxed, rehydrated, and subjected to antigen retrieval as described.23 Bound primary antibody was detected using tyramide signal amplification (TSA) and streptavidin biotinylated alkaline phosphatase complex (ABC-AP; Dako).23 For double-staining immunofluorescence and confocal laser microscopy, dewaxed paraffin sections were simultaneously incubated with the AID-specific antibody in combination with reagents directed against LMP1 or EBNA2 (Dako). Subsequently, FITC-labeled goat anti–rat Fab fragment (dianova, Hamburg, Germany) and Cy5-labeled rat anti–mouse F(ab)2 fragment (dianova) were added for 30 minutes at room temperature in the dark. Images shown in Figure 3A-B were visualized using a Leica DMLB microscope (Leica, Solms, Germany), equipped with a 40×/0.75 numerical aperture (NA) objective (Figure 3A) or a 20×/0.40 NA objective (Figure 3B), and a Leica DC200 camera. Confocal images in Figure 3C-D were visualized using a Leica TCS SP2/Leica DM RXE confocal microscope equipped with an HCXPL APO 63×/1.32-0.6 NA oil objective. Immersion oil with a diffraction index of 1.515 (Merck, Darmstadt, Germany) was used. Images were acquired and processed using Leica confocal software version 2.61 build 1537.
Results
To investigate the effect of latency III genes of EBV on AID expression, we used a lymphoblastoid cell line in which the function of EBNA2 (and hence the latency III program including LMP1 and LMP2A) is controlled by estrogen-dependent nuclear translocation of an ER-EBNA2 fusion protein (EREB2-5, Figure 1A).22 The cells were incubated with or without estrogen for 3 days, and AID expression was quantified by real-time RT-PCR and Western blot. Remarkably, activation of the latency III genes by addition of estrogen caused a clear decrease in AID expression at both the RNA and protein levels (Figure 1B, samples 1 and 2, and Figure 1C). Evidently, at least one of the latency III genes must have a pronounced inhibitory effect on AID expression.
In order to elucidate the nature of this inhibitor, we performed a kinetics experiment with the EREB2-5 cell line. Three days after withdrawal of estrogen, EBNA2-dependent gene expression was activated by estrogen re-addition, and AID mRNA expression levels were monitored at different time points. As shown in Figure 1B, AID levels dropped sharply during the first 4 hours of EBNA2 activation, and then increased transiently with a slight delay to the typical transient peak22 of appearance of the EBNA2 target gene LMP1 (Figure 1D), which induces AID upon overexpression in EREB2-5 (not shown) and other B cells.16 The final steady-state AID expression levels were clearly below the expression seen in the absence of estrogen, confirming the net inhibitory effect of the latency III program on AID. The sharp drop in AID expression immediately after EBNA2 activation, long before the appearance of EBNA2 target genes, suggested that this may represent a direct negative effect of EBNA2 on AID expression.
Effect of EBV latent gene expression on AID expression. (A) Schematic representation of generation of EREB2-5 cells from primary umbilical cord blood B cells. (B) Quantitative RT-PCR of AID mRNA levels normalized to CD19 in EREB2-5 cells cultured with or without estrogen for 3 days, followed by EBNA2 reinduction and analysis at the indicated time points. mRNA copy numbers were determined in duplicate; mean and variance of the 2 values are shown. (C) Western blot analysis of AID protein expression in cells cultured with and without estrogen. (D) LMP1 protein expression in the cells analyzed in panel B. ns indicates nonspecific band.
Effect of EBV latent gene expression on AID expression. (A) Schematic representation of generation of EREB2-5 cells from primary umbilical cord blood B cells. (B) Quantitative RT-PCR of AID mRNA levels normalized to CD19 in EREB2-5 cells cultured with or without estrogen for 3 days, followed by EBNA2 reinduction and analysis at the indicated time points. mRNA copy numbers were determined in duplicate; mean and variance of the 2 values are shown. (C) Western blot analysis of AID protein expression in cells cultured with and without estrogen. (D) LMP1 protein expression in the cells analyzed in panel B. ns indicates nonspecific band.
To directly test this assumption in a system lacking other viral proteins, we used derivatives of an EBV– Burkitt lymphoma cell line (BL41) as well as a Burkitt-like cell line lacking the c-myc translocation (BJAB), both of which carry a transfected estrogen-regulatable EBNA2 fusion protein.21 Upon activation of EBNA2 by estrogen addition, a sharp drop in AID mRNA levels was observed in both cell lines (Figure 2A,D), concomitant with the previously reported EBNA2-mediated repression of IgM heavy chain (Figure 2B,E 21 ). The addition of estrogen to BL cells or LCLs lacking the ER-EBNA2 fusion protein did not adversely affect AID expression (data not shown). For BL41, a decrease in AID protein levels (Figure 2C) could also be shown, while for BJAB, AID protein levels were below detection limit. This confirms that EBNA2 alone, in the absence of any other viral genes, is capable of inhibiting AID expression. Evidently, EBNA2 function may inhibit AID expression in several different transformed cell lines—LCLs, BLs, and BJAB—suggesting that the mechanism of inhibition is independent of the transformation or differentiation status of the cells.
EBNA2 alone is sufficient for suppression of AID expression. Quantitative RT-PCR of AID mRNA levels (A,D) and Ig μ heavy chain (B,E), each normalized to CD19 in BL41/K3 (A-B) and BJAB/K3 (D-E) cells, at the indicated time points after addition of estrogen. (C) Western blot analysis of AID protein expression in BL41/K3 cells with or without estrogen. (A-B, D-E) mRNA copy numbers were determined in duplicate; mean and variance of the 2 values are shown.
EBNA2 alone is sufficient for suppression of AID expression. Quantitative RT-PCR of AID mRNA levels (A,D) and Ig μ heavy chain (B,E), each normalized to CD19 in BL41/K3 (A-B) and BJAB/K3 (D-E) cells, at the indicated time points after addition of estrogen. (C) Western blot analysis of AID protein expression in BL41/K3 cells with or without estrogen. (A-B, D-E) mRNA copy numbers were determined in duplicate; mean and variance of the 2 values are shown.
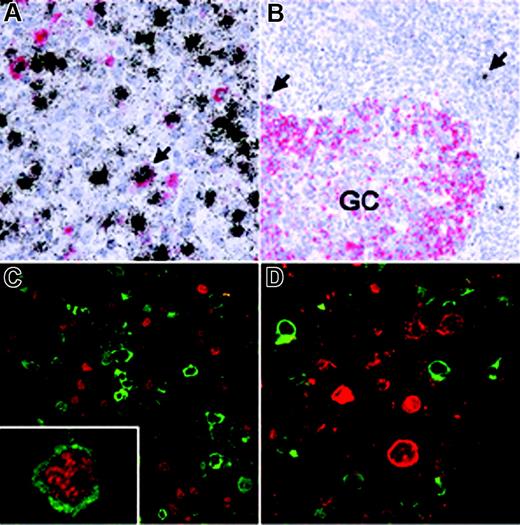
To assess how EBV affects AID expression in vivo, we analyzed EBV-infected cells in IM which is characterized by an EBV-driven proliferation of normal B cells. First, we combined in situ hybridization for the EBER transcripts expressed by all EBV-infected cells with immunohistochemistry using the monoclonal anti-AID antibody EK2-5G9. The vast majority of EBV-infected proliferating cells in IM did not express AID (Figure 3A; Table 1), while EBV– germinal center cells present in the same sections were AID positive. This is in line with a net inhibitory effect of the EBV growth program on AID expression. However, variable, usually small numbers of extrafollicular cells showed AID expression and most of these scored EBER positive, suggesting either that AID-positive extrafollicular cells are preferentially infected by the virus or that EBV may allow or induce AID expression in a minor subpopulation of the B cells in vivo (eg, via a certain viral gene expression pattern). An analogous staining of reactive tonsils from individuals with persistent EBV infection also revealed that EBV-infected cells during virus persistence barely ever express AID (Figure 3B; Table 1).
AID expression in EBV-infected cells in vivo. (A) Double-labeling EBER-specific in situ hybridization (black grains) and AID immunohistochemistry (red staining) of a tonsil from a patient with infectious mononucleosis (IM) shows that most EBV-infected cells are AID negative. Conversely, a significant number of AID-positive cells were also EBV+ (eg, arrow). (B) In a hyperplastic tonsil from a patient with persistent EBV infection, AID expression is detected in germinal center (GC) cells, while scattered extrafollicular EBV+ cells (arrows) are AID negative. (C) Double-labeling immunofluorescence of an IM tonsil shows that most EBNA2-positive cells (red) are AID negative (green) and vice versa, while only few EBNA2-positive cells coexpressing AID were identified (inset). (D) Double-labeling immunofluorescence also reveals that AID (green) and LMP1 (red) are expressed in nonoverlapping cell populations.
AID expression in EBV-infected cells in vivo. (A) Double-labeling EBER-specific in situ hybridization (black grains) and AID immunohistochemistry (red staining) of a tonsil from a patient with infectious mononucleosis (IM) shows that most EBV-infected cells are AID negative. Conversely, a significant number of AID-positive cells were also EBV+ (eg, arrow). (B) In a hyperplastic tonsil from a patient with persistent EBV infection, AID expression is detected in germinal center (GC) cells, while scattered extrafollicular EBV+ cells (arrows) are AID negative. (C) Double-labeling immunofluorescence of an IM tonsil shows that most EBNA2-positive cells (red) are AID negative (green) and vice versa, while only few EBNA2-positive cells coexpressing AID were identified (inset). (D) Double-labeling immunofluorescence also reveals that AID (green) and LMP1 (red) are expressed in nonoverlapping cell populations.
The influence of activation of the EBV growth program on AID expression in vivo was investigated by double staining for AID and EBNA2 or LMP1. As shown in Figure 3C and Table 1, most cells expressing EBNA2 clearly scored negative for AID expression, in line with the in vitro data. Notably, the majority of LMP1-expressing cells in IM is also AID negative (Figure 3D; Table 1), implying that in EBV-infected B cells in vivo the inhibitory effect of the EBNA2-driven growth program on AID expression is also dominant over the previously described positive effect of LMP1. As EBV-infected cells in IM change the expression pattern of EBV latent genes during proliferation, the low number of AID-positive cells expressing latency III genes (Figure 3C [inset] and 3D; Table 1) may reflect the delay between activation of viral genes and complete degradation of the AID protein. Furthermore, isolated cells expressing a latency II pattern (EBNA2–/LMP1+) have been described in IM.25 Alternatively, AID expression in some EBV-infected cells in vivo may be initiated or sustained by other mechanisms, such as CD40 signaling or cytokines. We conclude that AID is not expressed in most B cells that are driven into growth by the expression of EBV latency III genes, even though many of these cells phenotypically resemble extrafollicular blasts (eg, by CD30 expression). Accordingly, activation-induced AID induction (which is likely caused by LMP1-mediated NFκB activation) is prevented in cells expressing EBNA2, implying that the negative effect of this key transcription factor of the growth program on AID expression is also dominant in vivo.
Discussion
The present study highlights a complex mechanism of AID regulation by EBV in infected B cells, and thereby also provides a molecular explanation for the established discrepancies in the activity of somatic hypermutation in various EBV-associated B-cell proliferations. We show that the effect of EBV latency III genes on AID is dominated by EBNA2 activity, which exerts a substantial AID inhibition that counteracts AID induction by the LMP1 protein.
Considering the strong dosage dependence of AID activity,26 its modulation by EBNA2 is expected to have profound effects on the activity of Ig diversification processes. Indeed, a compilation of in vivo data on EBNA2 expression and somatic hypermutation activity in the known EBV-associated lymphoproliferations reveals a compelling inverse correlation. While during proliferation of the mostly EBNA2-positive EBV+ cells in IM no ongoing hypermutation can be detected even if they enter germinal centers,13,15 the EBNA2-negative EBV+ cells in AILD and in Burkitt lymphoma are characterized by substantial hypermutation activity.18 An even more striking link is seen in EBV+ cells in PTLD: no ongoing hypermutation could be observed in the EBNA2-expressing proliferating cells, while it is clearly detectable in EBNA2-negative cases.19,27 Even though in some cases the induction of AID by other stimuli may exceed the inhibitory potential of EBNA2,28,29 the negative interference of EBNA2 with immunoglobulin diversification also provides an additional explanation for its counterselection during the pathogenesis of Burkitt lymphoma,10 as it would prevent both translocation and hypermutation of the c-myc gene. Notably, the down-regulation of the translocated c-myc allele by substantial EBNA2 activity may have adversely affected previous experimental approaches to test the effect of EBV latent genes on somatic hypermutation29 and precludes the respective in vitro experiments in the presently available human hypermutation models.
In certain EBV-infected cells that express little or no EBNA2, the expression of LMP1 or other viral genes may potentially induce hypermutation or switch recombination in vivo by causing AID up-regulation,16 in particular in PTLD and AILD. It remains a major open question, though, whether the establishment of these different latency programs in B cells is triggered by physiological activation signals,30 their mimicry by viral genes, or other mechanisms. EBV+ cells in IM alter their gene expression programs during proliferation, suggesting that in vivo intrinsic oscillations of promoter use and latent gene expression contribute to immune escape.13 Considering the significant impact of alternative latency programs on lymphomagenesis, it will be most important to understand how they eventually become fixed in EBV+ cells during their aberrant proliferation, and can thus contribute to deregulated AID activity.
The question why EBV has adopted the strategy of EBNA2-mediated AID inhibition in the physiological setting is presently open to speculation. The EBNA2-driven growth program certainly plays an essential role in the establishment of EBV persistence in humans, but in the physiological context its duration will be highly restricted by the immunogenicity of the proteins involved, implying that any associated deleterious effects might interfere with viral penetrance. Deleterious mutations occurring during hypermutation in the Ig genes might initially be compensated for by LMP2A but would be fatal for the cells if LMP2A is down-regulated in memory B cells.31 Similar concerns may be raised for other genetic aberrations caused by uncontrolled hypermutation or class switching, as the potential malignant transformation of B cells would clearly counteract the establishment of asymptomatic latency. Accordingly, the inhibition of AID by EBV during the growth program might in fact be highly beneficial for the virus as well as for the host.
In conclusion, our study implies that EBNA2, by inhibiting AID, negatively interferes with somatic hypermutation and class-switch recombination, the hallmark genetic events of the germinal center. As LMP1 expression has also been shown to negatively interfere with the germinal center formation, the idea of a cooperative relationship of the Epstein-Barr virus and the germinal center reaction remains to be proven.
Authorship
S.T., L.M., M.B., and S.K. performed research and analyzed the data; B.K. and E.K. contributed vital reagents; G.N. and B.J. designed research and wrote the paper.
The authors declare no competing financial interests.
Prepublished online as Blood First Edition Paper, August 1, 2006; DOI 10.1182/blood-2006-05-021303.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank André Kutzera for technical assistance; all members of the B.J., B.K., and Ursula Zimber-Strobl labs for help and discussion; Julia Rastelli, Ursula Zimber-Strobl, and Ralf Küppers for critical reading of the paper; and Dirk Eick and Georg Bornkamm for generous support.
This work was supported by the Wilhelm Sander foundation (grant 2003.046.1) (B.J.) and the Deutsche Forschungsgemeinschaft (grant Nie312/3-1) (G.N.).