Abstract
Because of a lack of specific clonality markers, information on lineage involvement and cell of origin of acute myeloid leukemia with normal karyotype (AML-NK), is missing. Because Nucleophosmin (NPM) gene is frequently mutated in AML-NK and causes aberrant NPM cytoplasmic localization (NPMc+), it was used as an AML lineage clonality marker. Clonal NPM exon 12 mutations were detected in myeloid, monocytic, erythroid, and megakaryocytic cells but not in fibroblasts or endothelia that were laser-microdissected from 3 patients with NPMc+ AML. Aberrant cytoplasmic expression of mutated NPM proteins was identified with anti-NPM antibodies in 2 or more myeloid hemopoietic cell lineages in 99 (61.5%) of 161 of NPMc+ AML paraffin-embedded bone marrow biopsies; lymphoid involvement was excluded in 3 investigated cases. These findings suggest that NPMc+ AML derives from either a common myeloid or earlier progenitor. Immunohistochemical studies show that varying combinations and ratios of NPMc+ leukemic cells from distinct lineages are responsible for heterogeneity within each French-American-British (FAB) classification type and for NPMc+ AML falling into different FAB categories. These findings question the value of FAB criteria in subdividing the WHO category of “AML not otherwise characterized” and suggest that, for clinical use, NPMc+ AML be provisionally regarded as a separate AML with prognostic significance.
Introduction
Clonal cell lineage involvement in myelodysplastic syndrome and acute myeloid leukemia (AML) carrying recurrent genetic abnormalities can be successfully investigated by a number of techniques, including cytogenetics, fluorescence in situ hybridization (FISH), FISH combined with immunophenotyping, and polymerase chain reaction (PCR) on purified cell populations, which are all able to detect leukemia-specific molecular alterations.1-7 Unfortunately, no specific genetic markers are as yet available for a significant proportion of AML. Under these circumstances, cell lineage clonality can be investigated by analysis of X-chromosome inactivation patterns,8 but this technique has not been applied widely to AML. Thus, no or scarce information is available on cell lineage involvement or cell of origin in most AMLs, especially those with normal karyotype (AML-NK),9 which lack specific clonality markers and account for 40% to 50% of de novo adult AML.10 All genetically poorly defined AMLs, including AML-NK, are now included into the category of “acute myeloid leukemia not otherwise characterized” of the World Health Organization (WHO) classification.11 This large, poorly characterized subgroup of AML is currently defined according to criteria of the French-American-British (FAB) classification12 (with some modification), which is based on morphologic and cytochemical features of the leukemic cells and degree of maturation. Given its frequency and extreme heterogeneity, AML not otherwise characterized clearly needs to be defined better.
We recently identified mutations of the NPM gene exon 12 as the most common and specific genetic lesion associated with AML-NK,13 being observed in 50% to 60% of cases, as confirmed to date in more than 3500 patients with AML.14-19 At the NPM protein C-terminus, these mutations modify critical tryptophan(s) and create a new nuclear export signal (NES) motif, which act together to aberrantly localize NPM leukemic mutants in the cytoplasm20,21 —hence the term NPMc+ (cytoplasmic-positive) AML.13 As the NPM mutant protein dislocates the NPM wild-type allele into the cytoplasm20 and prevents the p19(Arf) tumor suppressor from initiating a p53 response that induces cell-cycle arrest,22,23 NPM mutations appear critical in NPMc+ AML pathogenesis.
In this study, we tested mutated and cytoplasmic NPM as a clonality marker in a large cohort of patients with NPMc+ AML and used it to assess lineage involvement and cell of origin and to determine the validity of FAB criteria in defining the WHO category of acute myeloid leukemia not otherwise characterized.11 To address these issues, we screened laser-microdissected cells of different lineages from 3 patients with NPMc+ AML for the presence of NPM exon 12 mutations and sought cytoplasmic-mutated NPM proteins in 161 NPMc+ AML paraffin-embedded bone marrow biopsies using anti-NPM antibodies, including reagents that specifically recognize mutant NPM proteins on Western blotting and immunohistochemistry.20,24 Immunohistochemical results were then correlated with FAB criteria adopted in the WHO classification.
We found that clonal NPM mutations affect different cell lineages in about 60% of NPMc+ AML, suggesting that either a common myeloid or an earlier progenitor without the ability to differentiate into lymphoid lineages is involved in NPMc+ AML. These findings also led us to question the FAB criteria that currently define the large WHO category of acute myeloid leukemia not otherwise characterized,11 as they emerged as inapplicable to NPMc+ AML.
Materials and methods
Tumor samples
NPM mutations were investigated in 3 NPMc+ AML-NK where frozen bone marrow biopsies were available for laser-capture microdissection of leukemic cells. Data on NPM exon 12 mutations in the bulk bone marrow population were available from patient 1 (heterozygous mutation B) and patient 3 (heterozygous mutation A)13 (Table 1).
Because laser microdissection techniques are technically demanding and require frozen, undecalcified bone marrow trephines, which, in this series, were available only in 3 patients, mutated NPM proteins were then sought by immunohistochemistry in paraffin-embedded bone marrow biopsies from 161 patients with NPMc+ AML (88 from the Gruppo Italiano Malattie Ematoligiche dell'Adulto/European Organisation for Research and Treatment of Cancer [GIMEMA/EORTC] AML12 trial; 73 not enrolled in the trial). Eighty cases showed normal karyotype; no mitoses were available in 8 cases. The 73 patients not included in the trial were not investigated by cytogenetics. Approval for the study was provided by the institutional review board of each center participating in the GIMEMA LAM99P/AML12 EORTC trial. Informed consent was provided by patients at each participating center according to the Declaration of Helsinki.
Mutation analysis of NPM was available in 75 of 161 patients, 30 of whom had also been investigated for FLT3 mutations (ITD and D835). As negative control, 25 samples of AML-NK lacking NPM mutations were used.13 All bone marrow biopsies were fixed in B5 and decalcified in EDTA, as previously described.13 Immunohistochemical results were correlated with currently used FAB criteria in the WHO classification.11
NPM mutation analysis on laser-microdissected cells
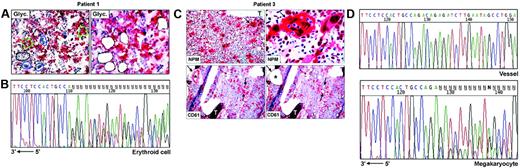
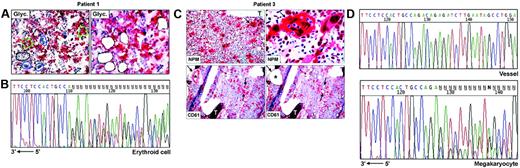
Frozen sections (9 μm thick) from undecalcified bone marrow biopsies25 were attached on membrane slides (PALM Microlaser Technologies, Hamburg, Germany) and air-dried overnight at room temperature. Antibodies against NPM are not suitable for detecting cytoplasmic NPM in frozen sections.13 Therefore, for selecting cells to microdissect, glycophorin A (Figure 1A) and CD61 (Figure 1C) were used, respectively, as markers of erythroid and megakaryocytic cells; myeloid cells were recognized by morphology and glycophorin A and CD61 negativity. Megakaryocytes may contain other cells (most frequently polymorphonuclear leucocytes) in the cytoplasm as a result of emperipolesis,26,27 a phenomenon in which cells enter the megakaryocyte cytoplasm and leave it after several hours.28 To avoid contamination of microdissected megakaryocytes with other cells, we therefore selected only megakaryocytes that did not show emperipolesis, taking care to destroy their cytoplasm with the laser beam and to isolate only the nuclei. Paraffin samples from the 3 patients with NPMc+ AML used in this study showed cytoplasmic NPM in all leukemic cells of different lineages (Figure 1C, top left); thus, the risk of isolating normal residual cells was negligible.
Single megakaryocytes and groups of 5 to 15 erythroid or myeloid cells were microdissected using a pulsed ultraviolet laser (Olympus Micro-Beam with PALM “inside” technology; Olympus, Tokyo, Japan) (Figure 1A,C), laser pressure catapulted into a cap coated with 20 to 40 μL Tris-HCl 10 mM, pH 7.4, and centrifuged into DNAse-free tubes coated with 1% BSA-10 mM Tris. Pools of 10 to 50 myeloid, erythroid, or megakaryocytic cells were collected (total, 6-10 pools, corresponding to ∼ 300 cells/lineage). Pools of 30 to 50 fibroblasts from areas adjacent to bone trabeculae (patient 2, not shown) or CD61+ endothelial cells (patient 3, Figure 1C) served as negative controls for the somatic origin of the mutations and for monitoring contamination by adjacent cells during the isolation procedures. Cells were cryopreserved at –80°C until analysis.
NPM mutation analysis
Pooled cells in 20 μL volume were incubated with Proteinase K (0.25 μg/μL) for 4 hours at 50°C, followed by 10 minutes at 95°C for enzyme inactivation. A nested PCR strategy was used to amplify a genomic fragment spanning the NPM exon 12 sequence.13,20 For the first amplification round, oligonucleotides NPM1-ex12F (5′-GCCAAATCTGGCCAACTCTA-3′) and NPM1-ex12R3′ (5′-TTTTACAAGACTATTTGCCATTCC-3′) (0.2 μM each) were used in a 25-μL reaction volume containing Expand High Fidelity (Roche Applied Science, Indianapolis, IN) buffer, 1.5 mM MgCl2, 200 μM each dNTP and 1.5 U Taq DNA Polymerase. After an initial denaturation step at 95°C for 5 minutes, the reaction was carried on for 35 cycles at 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 1 minute, ending with a final extension step at 72°C for 7 minutes. For the second amplification round, 1.5 μL of the first reaction mix was used as template. The reaction was done in 50 μL volume containing Expand High Fidelity buffer, 1.5 mM MgCl2, 200 μM each dNTP, 0.2 μM each primer (NPM1-ex12F3′,5′-TTAACTCTCTGGGGTAGAATGAA-3′, and NPM1-ex12R5′,5′-CAAGACTATTTGCCATTCCTAAC-3′) and 2.5 U Taq DNA polymerase. Cycling conditions were 95°C for 5 minutes, followed by 35 cycles at 95°C for 30 seconds, 62°C for 30 seconds, and 72°C for 1 minute, and a final extension step at 72°C for 7 minutes. To prevent DNA contamination, working areas were designated for pre-PCR and post-PCR manipulation, and multiple water samples were used as negative control.
PCR products were purified using the QIAQUICK purification kit (QIAGEN, Hilden, Germany) and sequenced directly using primer NPM1-1112R (5′-CCTGGACAACATTTATCAAACACGGTA-3′) and NPM-ex12R5′. All sequences were compared with the germ line NPM cDNA sequence (GenBank accession no. NM_002520, version 4).
Antibodies
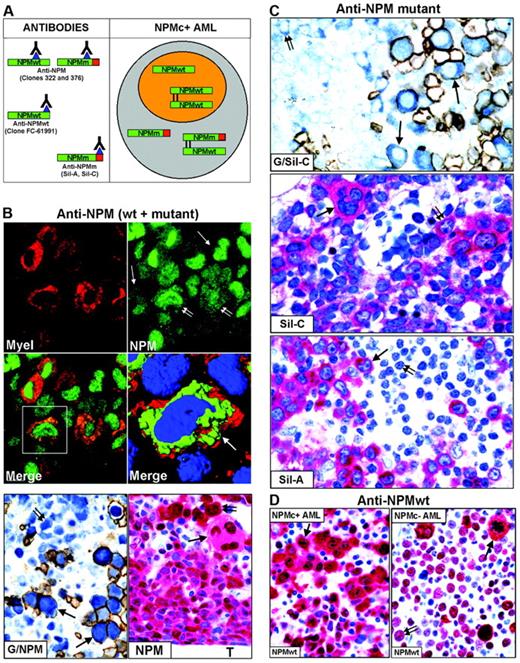
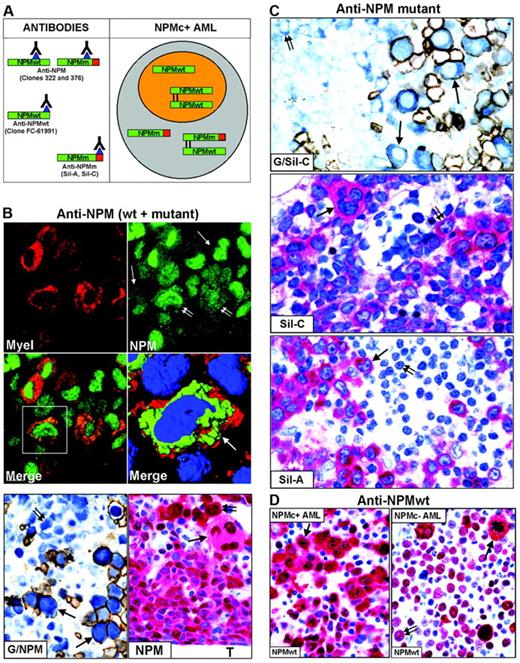
Nucleophosmin was detected in paraffin sections using the following antibodies: monoclonal anti-NPM antibodies (clones 322 and 376), recognizing both wild-type and mutant NPM13,20,29 (Figure 2A, left); polyclonal antibodies (Sil-A and Sil-C),20,24 which detect mutated but not wild-type NPM (Figure 2A, left); and a monoclonal antibody (clone FC-61991) that recognizes wild-type but not mutated NPM (Figure 2A, left). Antibodies directed against nucleolin/C23 (Santa Cruz Biotechnology, Heidelberg, Germany), myeloperoxidase, glycophorin A, CD61, CD68 (macrophage-restricted), and CD34 (all from Dako, Glostrup, Denmark) were also used. For immunofluorescence studies, secondary goat anti–mouse Alexa 488 and goat anti–mouse Alexa 568 conjugated antibodies were used (Molecular Probes, Eugene, OR).
Immunohistochemical studies
Paraffin sections from all bone marrow samples were subjected to antigen retrieval (microwaving in 1 mM EDTA buffer, pH 8.0)13 and incubated with primary antibodies. The antigen-antibody reaction was revealed using the immunoalkaline phosphatase (APAAP) technique13,30 ; double staining for glycophorin A/NPM was performed using a sequential APAAP/immunoperoxidase procedure.31 In selected NPMc+ AML cases, double immunofluorescence for myeloperoxidase/NPM was carried out, as previously described.32
Lineage involvement criteria
Immunohistologic findings were related to morphologic features, according to the WHO classification.11 Involvement of myeloid and monocytic cell lineages was easy to detect because NPMc+ myeloid and monocytic cells usually represented the majority of the leukemic population. The following criteria were used to define involvement of erythroid and megakaryocytic series: erythroid cells were considered as involved when even a few surface glycophorin A-positive cells expressing cytoplasmic NPM and nuclear C23 were detected. Cytoplasmic expression of NPM (not related to the presence of NPM mutations) is sometimes seen in megakaryocytes in normal and pathologic bone marrows. Therefore, criteria for megaryocyte involvement was represented by greater than 50% megakaryocytes expressing cytoplasmic NPM in the presence of nuclear C23. Erythroid and/or megakaryocytic involvement was confirmed by Sil-A and Sil-C antibody detection of mutated NPM when antigens survived fixation or decalcification
Results
NPM mutations target different cell lineages in NPMc+ AML
To investigate whether NPM exon 12 mutations are clonally represented in multiple hematopoietic lineages, we performed PCR amplification and direct sequencing of genomic DNA from laser-microdissected cells in 3 NPMc+ AMLs showing immunohistochemical evidence of cytoplasmic NPM in different cell lineages. In patients 1 and 2, leukemic cells from myeloid, erythroid, and megakaryocyte lineages were analyzed (Table 1). Patient 3 displayed myeloid and megakaryocytic involvement at immunohistochemistry. However, the small clusters of NPMc+ myeloid blasts seen in paraffin sections were hard to recognize in frozen sections; thus, only megakaryocytes were microdissected (Table 1; Figure 1C). Fibroblasts or vessels were also isolated as negative control from patients 2 and 3 (Table 1), whereas no control could be microdissected from patient 1 (Table 1).
For each lineage, 3 to 4 independent pools of cells were investigated in the 3 patients, corresponding to a total of 31 samples; of these, 26 (84%) were successfully amplified. Lack of amplification in the remaining 5 samples was probably due to loss of DNA during laser microdissection or to incomplete proteinase K digestion. Sequencing analysis revealed the presence of heterozygous, clonal NPM exon 12 mutations in all patients (Table 1). Patient 1 carried a 4-bp (base pair) insertion (CATG) at position 960_963, corresponding to mutation B,13 which was detected in each sample amplified (3 myeloid, 3 erythroid, 4 megakaryocyte) (Table 1; Figure 1B), confirming the results obtained by mutational analysis of the bone marrow bulk population. In patient 2, 6 of 8 samples amplified (2 myeloid, 2 erythroid, and 2 megakaryocyte) displayed a TCTG duplication at position 960_963, corresponding to mutation “A,” the most frequent in NPMc+AML.13 Conversely, only wild-type sequences were detected in the remaining 2 samples (1 myeloid, 1 erythroid) (Table 1). However, this patient carried a heterozygous single base-pair deletion (ΔT1146) in the untranslated sequences of the same exon, which was also observed in paired DNA from normal fibroblasts and corresponds to a frequent, previously described germ line polymorphism.17 The deletion T1146 cosegregated with the allele lacking the TCTG duplication (data not shown) and could therefore be used as a marker for allelic representation. Indeed, only one allele (ie, the wild-type) was detected in these 2 samples, indicating that the absence of NPM exon 12 mutations was due to lack of amplification of the second allele, as occasionally observed in single-cell–based approaches. Patient 3, from whom only cells of megakaryocytic lineage could be analyzed, carried mutation A, which was detected in all 4 samples sequenced (Figure 1C-D, bottom). No somatic NPM mutations were found in fibroblasts or endothelial cells microdissected from the same section in 2 patients (2 and 3, respectively) with available material, excluding contamination from adjacent cells (Table 1; Figure 1D, top).
Thus, in each patient, the same mutation was found in different cell types of the myeloid lineage, documenting their clonal origin and validating immunohistochemical data of aberrant cytoplasmic NPM expression.
NPMc+ AML cells of different lineages harbor cytoplasmic-mutated NPM proteins
To determine whether cells of different lineages in NPMc+ AML carry NPM-mutated proteins, we immunostained for NPM paraffin-embedded biopsies from 161 patients with NPM+ AML.
Reactivity with antibodies recognizing both wild-type and mutated NPM proteins. These antibodies reacted strongly with 161 of 161 biopsies (Table 2). Aberrant cytoplasmic (in addition to nuclear) NPM expression was detected in 2 or more cell lineages in 99 (61.5%) of 161 cases, whereas the remaining cases showed involvement of only one cell lineage (myeloid, myelo-monocytic, or erythroid). C23/nucleolin positivity was nucleus restricted in 161 of 161 cases (not shown). No difference in frequency and type of lineage involvement emerged between NPMc+ AML with normal karyotype (n = 80 of 161) and NPMc+ AML for which cytogenetics was not available (n = 81 of 161). In 75 of 161 cases in which NPM mutation analysis had been performed on whole bone marrow samples,21 mutation type and presence or absence or frequency of multilineage involvement were not correlated (not shown). No difference in the frequency and type of lineage involvement was observed in the group of NPM mutated/FLT3 unmutated (n = 15) versus NPM mutated/FLT3-ITD (n = 10) versus NPM mutated/FLT3-D835 (n = 5) samples.
NPM exon 12 mutations are found in NPMc+ leukemic cells of different lineages. (A) Patient 1. Bone marrow frozen sections are stained for glycophorin A. The cells selected for analysis are shown before (left) and after laser microdissection (right, empty areas), and are labeled as follows: 1 (small group of glycophorin A-negative myeloid blasts); 2, 3, and 4 (single megakaryocytes); 5 and 6 (small groups of glycophorin A-positive erythroid precursors). APAAP; hematoxylin counterstain, 400×. (B) Sequencing analysis of NPM exon 12 in microdissected erythroid cells from patient 1. The heterozygous insertion of 4 nucleotides generates a shift in the reading frame. The arrow indicates the sequence orientation. (C) Patient 3. (Top, left) Bone marrow paraffin section showing numerous NPMc+ megakaryoytes (arrow) and small clusters of NPMc+ myeloid blasts; T indicates a bone trabecula. APAAP; hematoxylin counterstain, 200×. (Top, right) Higher magnification from the same case showing NPMc+ megakaryocyte (arrow) with marked emperipolesis (double arrows). APAAP; hematoxylin counterstain, 1000×. (Bottom, left) CD61+ megakaryocytes selected for laser microdissection are indicated (arrows); CD61+ endothelial cells of a blood vessel (asterisk) were used as negative control. APAAP; hematoxylin counterstain, 200×. (Bottom right) Empty areas correspond to microdissected megakaryocytes after laser catapulting. APAAP; hematoxylin counterstain, 200×. (D) Chromatograms of NPM exon 12 sequences obtained from genomic DNA of vessel endothelial cells (control) (top) and megakaryocytes (bottom), microdissected from patient 3. Although a wild-type sequence is detected in the control DNA (vessel), the leukemic megakaryocytes show a heterozygous insertion of 4 nucleotides, creating a shift in the reading frame. The arrow indicates the sequence orientation.
NPM exon 12 mutations are found in NPMc+ leukemic cells of different lineages. (A) Patient 1. Bone marrow frozen sections are stained for glycophorin A. The cells selected for analysis are shown before (left) and after laser microdissection (right, empty areas), and are labeled as follows: 1 (small group of glycophorin A-negative myeloid blasts); 2, 3, and 4 (single megakaryocytes); 5 and 6 (small groups of glycophorin A-positive erythroid precursors). APAAP; hematoxylin counterstain, 400×. (B) Sequencing analysis of NPM exon 12 in microdissected erythroid cells from patient 1. The heterozygous insertion of 4 nucleotides generates a shift in the reading frame. The arrow indicates the sequence orientation. (C) Patient 3. (Top, left) Bone marrow paraffin section showing numerous NPMc+ megakaryoytes (arrow) and small clusters of NPMc+ myeloid blasts; T indicates a bone trabecula. APAAP; hematoxylin counterstain, 200×. (Top, right) Higher magnification from the same case showing NPMc+ megakaryocyte (arrow) with marked emperipolesis (double arrows). APAAP; hematoxylin counterstain, 1000×. (Bottom, left) CD61+ megakaryocytes selected for laser microdissection are indicated (arrows); CD61+ endothelial cells of a blood vessel (asterisk) were used as negative control. APAAP; hematoxylin counterstain, 200×. (Bottom right) Empty areas correspond to microdissected megakaryocytes after laser catapulting. APAAP; hematoxylin counterstain, 200×. (D) Chromatograms of NPM exon 12 sequences obtained from genomic DNA of vessel endothelial cells (control) (top) and megakaryocytes (bottom), microdissected from patient 3. Although a wild-type sequence is detected in the control DNA (vessel), the leukemic megakaryocytes show a heterozygous insertion of 4 nucleotides, creating a shift in the reading frame. The arrow indicates the sequence orientation.
Except for one case of M6b33 without an increase in myeloblasts (Table 2), the majority (50%-75%) of NPMc+ leukemic cells were of myeloid and/or monocytic lineage in M1, M2, M4, M5a, and M6 (Figure 2B); in M5b, cytoplasmic NPM was observed in 30% to 75% of leukemic cells, usually representing the most immature cells (not shown); more differentiated monocytic leukemic cells were weakly positive or negative. In cases showing erythroid involvement, NPM cytoplasmic positivity was strongest in proerythoblasts than in any other leukemic cell (Figure 2B, G/NPM and NPM); more mature erythroid precursors were usually weakly positive or negative (Figure 2B, G/NPM), in line with NPM down-regulation during differentiation of erythroid and other hemopoietic progenitors.34-37 NPMc+ erythroid cells ranged from isolated (best identified by double glycophorin A/NPM staining) (Figure 2B, G/NPM) through variably sized clusters to most marrow cells (M6 cases). Strongly NPMc+/glycophorin A-negative proerythroblasts were occasionally observed (not shown), unsurprisingly, because glycophorin A may be weakly expressed or absent in most immature erythroid precursors. In cases with megakaryocytic involvement, NPMc+ megakaryocytes accounted for at least 50% of all megakaryocytes (Figure 2B, NPM), a proportion of which displayed emperipolesis (Figure 3B, NPM).
In 3 NPMc+ AMLs, mixed (B and T) lymphoid nodules showed nucleus-restricted NPM expression (Figure S1, available at the Blood website; see the Supplemental Figure link at the top of the online article), suggesting NPM is not mutated in lymphoid cells. The great majority of NPMc+ AML cases (158 of 161) were CD34– (not shown).
As expected, 25 AML-NK samples without NPM mutations (negative control) showed nucleus-restricted NPM and C23 expression.13
Reactivity with specific anti-NPM mutant antibodies. To confirm that NPMc+ cells of different lineages, labeled by antibodies recognizing both wild-type and mutated NPM, carry mutated NPM proteins, we immunostained the 161 AML biopsies with specific anti-NPM mutants antibodies, Sil-A and Sil-C.
Mutated NPM proteins are found in NPMc+ leukemic cells of different lineages. (A, left) Epitopes recognized by anti-NPM antibodies. Clones 322 and 376 do not distinguish between wild-type (NPMwt) and mutant NPM (NPMm); clone FC-61991 recognizes NPMwt but not NPMm; the Sil-A and Sil-C antibodies recognize NPMm but not NPMwt. The red square area in NPMm indicates the mutated C-terminal portion of nucleophosmin. (Right) Schematic representation of subcellular distribution of NPMwt, NPMm, and NPMwt/NPMm heterodimer molecules in nuclear (orange) and cytoplasmic (gray) compartments of NPMc+ AML. (B, top) Double immunofluorescence staining of paraffin section for myeloperoxidase (Myel: red, cytoplasmic) and nucleophosmin (NPM: green, nuclear plus cytoplasmic) from a AML-M6 (×1000); single arrows indicate NPMc+ erythroid blasts and double arrows NPMc+ myeloid blasts. (Merge) Myeloid blasts (square box) show cytoplasmic colocalization of myeloperoxidase and NPM (×1000). (B, middle) Three-dimensional reconstruction at confocal microscope of the cell in the square box (arrow) (myeloperoxidase, red; NPM, green; nucleus, blue). (B, bottom) Double immunoenzymatic staining (G/NPM) of AML-M6. Leukemic proerythroblasts (arrows) are double-stained for NPM (clone 376) in blue (nucleus plus cytoplasm) and glycophorin in brown (surface). Myeloid blasts (double arrow) are glycophorin A-negative and show nuclear plus cytoplasmic NPM positivity. APAAP/Immunoperoxidase; no counterstain, ×1000. Multilineage involvement in NPMc+ AML (figure indicated as NPM). Nuclear plus cytoplasmic expression of NPM (clone 376) is seen in leukemic myelo-monocytic cells adjacent to bone trabecula (T), occasional strongly positive proerythroblasts (double arrow) and megakaryocytes (arrow); APAAP; hematoxylin counterstain, ×1000. (C, top) Proerythroblasts (arrows) are double-stained for NPM mutant (Sil-C) in blue (cytoplasmic-restricted) and glycophorin in brown (surface). Myeloid blasts (double arrow) are glycophorin-negative and show cytoplasmic-restricted expression of NPM mutant (APAAP/Immunoperoxidase; no counterstain, ×1000). (Middle) Cytoplasmic-restricted expression of mutant NPM protein in myeloblasts, cluster of proerythoblasts (double arrow), and isolated megakaryocyte (arrow). APAAP; hematoxylin counterstain, ×1000. (Bottom) Myeloblasts (arrow) show cytoplasmic-restricted expression of mutant NPM, whereas lymphoid cells (double arrow) are Sil-A negative (APAAP; hematoxylin counterstain, ×1000). (D) Wild-type NPM expression (clone FC-61991) is nuclear plus cytoplasmic (arrow) in leukemic cells of NPMc+ AML (left) but nucleus-restricted in NPMc– AML (right, double arrow) with exception of a mitotic figure (arrow). APAAP; hematoxylin counterstain, ×1000.
Mutated NPM proteins are found in NPMc+ leukemic cells of different lineages. (A, left) Epitopes recognized by anti-NPM antibodies. Clones 322 and 376 do not distinguish between wild-type (NPMwt) and mutant NPM (NPMm); clone FC-61991 recognizes NPMwt but not NPMm; the Sil-A and Sil-C antibodies recognize NPMm but not NPMwt. The red square area in NPMm indicates the mutated C-terminal portion of nucleophosmin. (Right) Schematic representation of subcellular distribution of NPMwt, NPMm, and NPMwt/NPMm heterodimer molecules in nuclear (orange) and cytoplasmic (gray) compartments of NPMc+ AML. (B, top) Double immunofluorescence staining of paraffin section for myeloperoxidase (Myel: red, cytoplasmic) and nucleophosmin (NPM: green, nuclear plus cytoplasmic) from a AML-M6 (×1000); single arrows indicate NPMc+ erythroid blasts and double arrows NPMc+ myeloid blasts. (Merge) Myeloid blasts (square box) show cytoplasmic colocalization of myeloperoxidase and NPM (×1000). (B, middle) Three-dimensional reconstruction at confocal microscope of the cell in the square box (arrow) (myeloperoxidase, red; NPM, green; nucleus, blue). (B, bottom) Double immunoenzymatic staining (G/NPM) of AML-M6. Leukemic proerythroblasts (arrows) are double-stained for NPM (clone 376) in blue (nucleus plus cytoplasm) and glycophorin in brown (surface). Myeloid blasts (double arrow) are glycophorin A-negative and show nuclear plus cytoplasmic NPM positivity. APAAP/Immunoperoxidase; no counterstain, ×1000. Multilineage involvement in NPMc+ AML (figure indicated as NPM). Nuclear plus cytoplasmic expression of NPM (clone 376) is seen in leukemic myelo-monocytic cells adjacent to bone trabecula (T), occasional strongly positive proerythroblasts (double arrow) and megakaryocytes (arrow); APAAP; hematoxylin counterstain, ×1000. (C, top) Proerythroblasts (arrows) are double-stained for NPM mutant (Sil-C) in blue (cytoplasmic-restricted) and glycophorin in brown (surface). Myeloid blasts (double arrow) are glycophorin-negative and show cytoplasmic-restricted expression of NPM mutant (APAAP/Immunoperoxidase; no counterstain, ×1000). (Middle) Cytoplasmic-restricted expression of mutant NPM protein in myeloblasts, cluster of proerythoblasts (double arrow), and isolated megakaryocyte (arrow). APAAP; hematoxylin counterstain, ×1000. (Bottom) Myeloblasts (arrow) show cytoplasmic-restricted expression of mutant NPM, whereas lymphoid cells (double arrow) are Sil-A negative (APAAP; hematoxylin counterstain, ×1000). (D) Wild-type NPM expression (clone FC-61991) is nuclear plus cytoplasmic (arrow) in leukemic cells of NPMc+ AML (left) but nucleus-restricted in NPMc– AML (right, double arrow) with exception of a mitotic figure (arrow). APAAP; hematoxylin counterstain, ×1000.
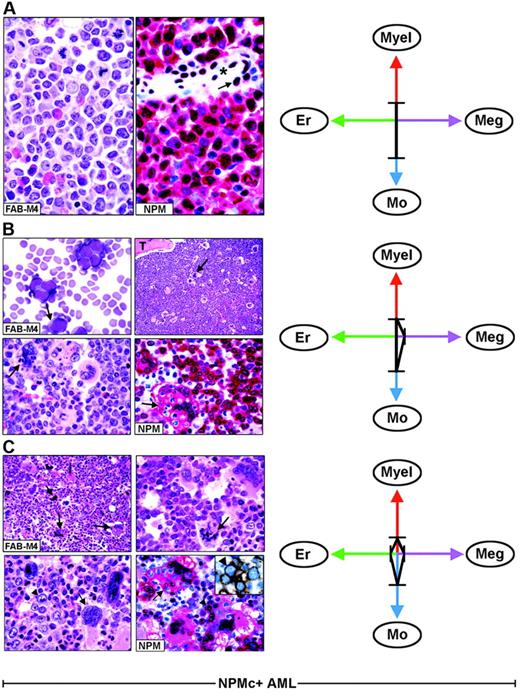
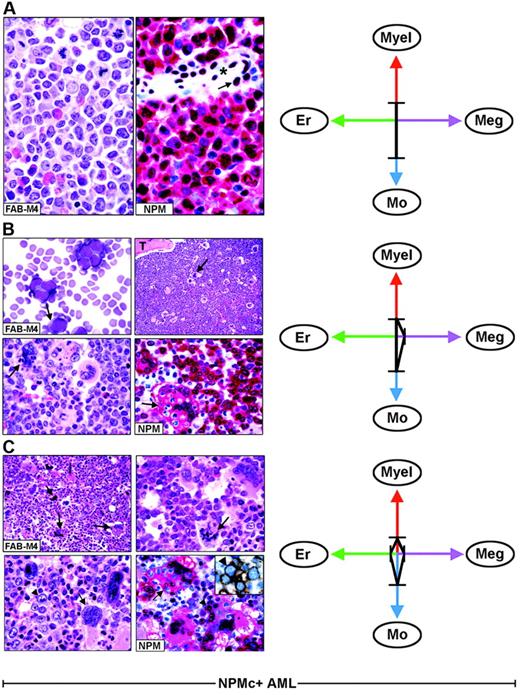
Each FAB category contains a mixture of leukemic cells of different lineages. (A) Hematoxylin and eosin (left) and NPM staining (right, APAAP; hematoxylin counterstain, ×1000) of an AML-M4, showing nuclear plus cytoplasmic NPM expression only in myeloid and monocytic cells. The arrow indicates the nucleus-restricted positivity in endothelial cells of a blood vessel (asterisk). (B) AML-M4 with myelo-monocytic and megakaryocytic involvement. (Top left) Myelo-monocytic cells in a marrow smear. (May-Grünwald-Giemsa, ×1000) The bone marrow biopsy from the same case shows an increased number of megakaryocytes (top right, arrow, hematoxylin and eosin, ×1000) displaying marked emperipolesis (bottom left, arrow hematoxylin and eosin, ×1000); T indicates a bone trabecula. (Bottom right) NPM (clone 376) is nuclear and cytoplasmic (red) in virtually all myelo-monocytic cells and megakaryocytes (arrow). (C) Bone marrow biopsy from AML-M4 with multilineage involvement, showing heterogeneous morphologic features in different areas. (Top left, hematoxylin and eosin ×400) Increased number of dysplastic megakaryocytes (arrows). (Top, right, hematoxylin and eosin, ×1000) Area with mixture of myelo-monocytic cells and displastic megakaryocytes (arrow). (Bottom, left, hematoxylin and eosin, ×1000) Area with mixture of proerythroblasts (arrowhead) and dysplastic megakaryocytes (arrow). (Bottom right, APAAP; hematoxylin counterstain, × 1000) NPM expression (clone 376) is detected in the nucleus and cytoplasm (red) of proerythroblasts (arrowhead) and megakaryocytes (arrow). The same NPM staining pattern (not shown) is also seen in myelomonocytic leukemic cells in other areas of the biopsy. The inset shows double immunostaining for surface glycophorin A (brown) and NPM (blue) in proerythroblasts from the same biopsy (immunoperoxidase/fast blue APAAP, no counterstain, ×1000). (Right panels). The ratios and type of lineages involved in each case (A-C) are schematically illustrated by the 4-axis diagram. Myel indicates myeloid; Mo, monocytic; Er, erythroid; Meg, megakaryocytic.
Each FAB category contains a mixture of leukemic cells of different lineages. (A) Hematoxylin and eosin (left) and NPM staining (right, APAAP; hematoxylin counterstain, ×1000) of an AML-M4, showing nuclear plus cytoplasmic NPM expression only in myeloid and monocytic cells. The arrow indicates the nucleus-restricted positivity in endothelial cells of a blood vessel (asterisk). (B) AML-M4 with myelo-monocytic and megakaryocytic involvement. (Top left) Myelo-monocytic cells in a marrow smear. (May-Grünwald-Giemsa, ×1000) The bone marrow biopsy from the same case shows an increased number of megakaryocytes (top right, arrow, hematoxylin and eosin, ×1000) displaying marked emperipolesis (bottom left, arrow hematoxylin and eosin, ×1000); T indicates a bone trabecula. (Bottom right) NPM (clone 376) is nuclear and cytoplasmic (red) in virtually all myelo-monocytic cells and megakaryocytes (arrow). (C) Bone marrow biopsy from AML-M4 with multilineage involvement, showing heterogeneous morphologic features in different areas. (Top left, hematoxylin and eosin ×400) Increased number of dysplastic megakaryocytes (arrows). (Top, right, hematoxylin and eosin, ×1000) Area with mixture of myelo-monocytic cells and displastic megakaryocytes (arrow). (Bottom, left, hematoxylin and eosin, ×1000) Area with mixture of proerythroblasts (arrowhead) and dysplastic megakaryocytes (arrow). (Bottom right, APAAP; hematoxylin counterstain, × 1000) NPM expression (clone 376) is detected in the nucleus and cytoplasm (red) of proerythroblasts (arrowhead) and megakaryocytes (arrow). The same NPM staining pattern (not shown) is also seen in myelomonocytic leukemic cells in other areas of the biopsy. The inset shows double immunostaining for surface glycophorin A (brown) and NPM (blue) in proerythroblasts from the same biopsy (immunoperoxidase/fast blue APAAP, no counterstain, ×1000). (Right panels). The ratios and type of lineages involved in each case (A-C) are schematically illustrated by the 4-axis diagram. Myel indicates myeloid; Mo, monocytic; Er, erythroid; Meg, megakaryocytic.
Both antibodies gave the same reactivity pattern; however, because of antigen denaturation, they reacted with only 98 of 161 cases. Notably, in the 98 evaluable samples, they labeled similar percentages and cell lineage types as antibodies recognizing both wild-type and mutated NPM. The most important difference was that Sil-A and Sil-C antibody reactivity was consistently cytoplasmic-restricted (Figure 2C; G/Sil-C, Sil-C, and Sil-A), in keeping with previous findings that NPM leukemic mutants are localized only in the cytoplasm.20 These antibodies were particularly useful to show the presence of NPM mutated proteins in isolated or small clusters of erythroid precursors (Figure 4, panel M1, Sil-A) and to confirm that megakaryocytes belong to the leukemic clone (Figure 2C, Sil-C).
In 2 cases, mixed (B and T) lymphoid nodules (Figure 2C, Sil-A), showed no NPM-mutated proteins in the lymphoid compartment, indicating NPM is not mutated in lymphoid cells. None of 25 AML-NKs lacking NPM mutations reacted with Sil-A and Sil-C (not shown).
Thus, specific Sil-A and Sil-C antibodies prove leukemic cells of different lineages carry mutated NPM proteins, as predicted by detection of cytoplasmic NPM with ordinary monoclonal antibodies (recognizing both wild-type and mutated NPM).
NPMc+ AML cells of different lineages display aberrant cytoplasmic expression of wild-type NPM
To assess whether NPM mutants recruit wild-type NPM into the cytoplasm of cells from different lineages, we immunostained 25 NPMc+ AML-NK biopsies showing multilineage involvement with an antibody detecting only wild-type NPM (Figure 2A, anti-NPMwt, left).20 Antigenicity was preserved in 10 of 25 cases; the NPM expression pattern (nuclear plus cytoplasmic), the percentage and the type of immunostained cell lineages overlapped with the staining patterns of ordinary monoclonal anti-NPM antibodies (clones 322 and 376), indicating that the wild-type NPM protein was partially dislocated into the cytoplasm (Figure 2D, left).20,22 Antigenicity was preserved in 12 of 25 AML-NKs without NPM mutations, all showing identical nucleus-restricted positivity for wild-type NPM (Figure 2D, right) and C23/nucleolin (not shown).
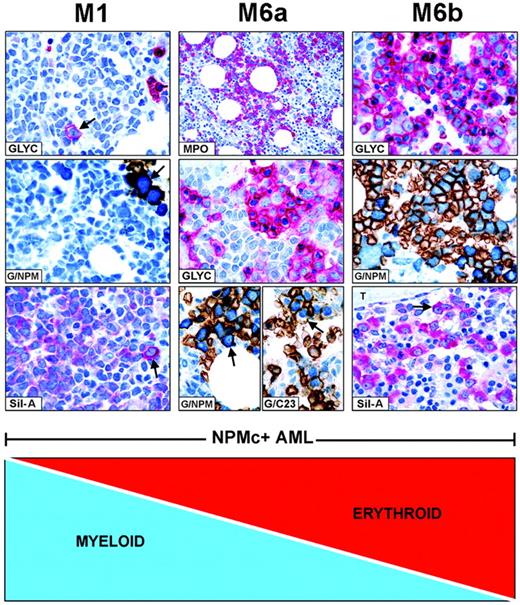
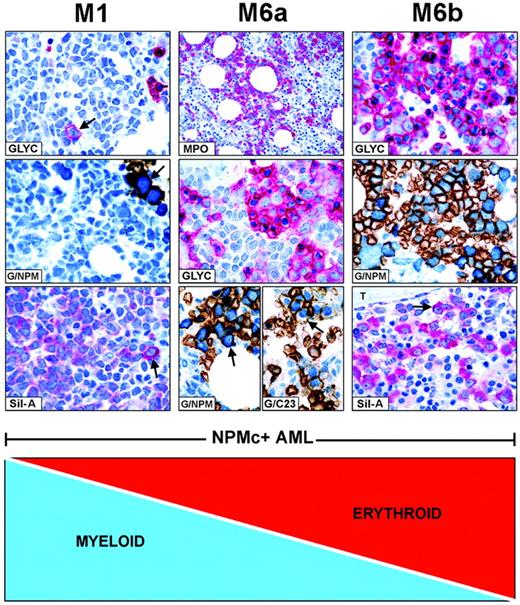
FAB categories are created by mixtures of NPMc+leukemic cells of different lineages. Combination of myeloid and erythroid cells is taken as an example of how FAB categories are created in NPMc+ AML. (Left panel, M1) NPMc+ AML of M1-type showing numerous glycophorin A-negative myeloid blasts (blue) and rare glycophorin A-positive (red) proerythroblasts (GLYC, arrow; APAAP; hematoxylin counterstain, ×1000) that express cytoplasmic NPM, as shown by double staining for glycophorin A(brown)/NPM (blue) (G/NPM, arrow; immunoperoxidase/fast blue APAAP, no counterstain, ×1000), and labeling in red (cytoplasmic-restricted) of the mutant NPM (Sil-A, arrow; APAAP; hematoxylin counterstain, ×1000). (Middle panel, M6a) NPMc+ AML of M6a type characterized by equal representation of myeloperoxidase-positive myeloid blasts (MPO, red; APAAP; hematoxylin counterstain, ×400) and glycophorin A-positive erythroid cells (GLYC, red; APAAP; hematoxylin counterstain, ×1000). Erythroid cells double stain for surface glycophorin A (brown)/cytoplasmic NPM (blue) (G/NPM, arrow) and for surface glycophorin A (brown)/nuclear C23 (blue) (G/C23, arrow; immunoperoxidase/fast blue APAAP, no counterstain, ×1000). (Right panel, M6b) NPMc+ AML of M6b type showing a predominant population of erythroid precursors (GLYC, red; APAAP; hematoxylin counterstain, ×1000) expressing cytoplasmic NPM as documented by double staining for surface glycophorin A (brown)/cytoplasmic NPM (blue) (G/NPM; immunoperoxidase/fast blue APAAP, no counterstain, ×1000), and by labeling in red (cytoplasmic-restricted) for mutant NPM (Sil-A, arrow; APAAP; hematoxylin counterstain, ×1000); T indicates a bone trabecula.
FAB categories are created by mixtures of NPMc+leukemic cells of different lineages. Combination of myeloid and erythroid cells is taken as an example of how FAB categories are created in NPMc+ AML. (Left panel, M1) NPMc+ AML of M1-type showing numerous glycophorin A-negative myeloid blasts (blue) and rare glycophorin A-positive (red) proerythroblasts (GLYC, arrow; APAAP; hematoxylin counterstain, ×1000) that express cytoplasmic NPM, as shown by double staining for glycophorin A(brown)/NPM (blue) (G/NPM, arrow; immunoperoxidase/fast blue APAAP, no counterstain, ×1000), and labeling in red (cytoplasmic-restricted) of the mutant NPM (Sil-A, arrow; APAAP; hematoxylin counterstain, ×1000). (Middle panel, M6a) NPMc+ AML of M6a type characterized by equal representation of myeloperoxidase-positive myeloid blasts (MPO, red; APAAP; hematoxylin counterstain, ×400) and glycophorin A-positive erythroid cells (GLYC, red; APAAP; hematoxylin counterstain, ×1000). Erythroid cells double stain for surface glycophorin A (brown)/cytoplasmic NPM (blue) (G/NPM, arrow) and for surface glycophorin A (brown)/nuclear C23 (blue) (G/C23, arrow; immunoperoxidase/fast blue APAAP, no counterstain, ×1000). (Right panel, M6b) NPMc+ AML of M6b type showing a predominant population of erythroid precursors (GLYC, red; APAAP; hematoxylin counterstain, ×1000) expressing cytoplasmic NPM as documented by double staining for surface glycophorin A (brown)/cytoplasmic NPM (blue) (G/NPM; immunoperoxidase/fast blue APAAP, no counterstain, ×1000), and by labeling in red (cytoplasmic-restricted) for mutant NPM (Sil-A, arrow; APAAP; hematoxylin counterstain, ×1000); T indicates a bone trabecula.
These findings prove that, in NPMc+ AML, cells of different lineages carry in the cytoplasm both mutated and wild-type NPM proteins.
Cytoplasmic NPM, multilineage involvement, and AML classification
Immunohistologic findings were used to assess FAB criteria in the current WHO classification of AML.11 Table 2 shows the correlation between lineage involvement in NPMc+ AML and FAB categories; 99 (61.5%) of 161 cases with involvement of 2 or more lineages were distributed as follows: M0-M1 (8 of 21, 38.1%), M2 (11 of 24, 45.8%), M4 (46 of 56, 82.1%), M5a (7 of 20, 35.0%), M5b (18of 30, 60.0%), and M6 (9 of 10, 90.0%).
As illustrated by the 4-axis diagram in Figure 3 (right panels), different combinations and diverse ratios of myeloid, monocyte, erythroid, and megakaryocyte cells, belonging to the leukemic clone, underlie heterogeneity within each FAB type and caused NPMc+ AML to fall within several FAB categories.
In NPMc+ AML, each FAB category comprises heterogeneous combinations of NPMc+ leukemic cells of different lineages. As an example, Figure 3 illustrates 3 AML cases classified as myelomonocytic leukemia (FAB-M4) (panels A-C). In one patient, immunohistochemistry detected only NPMc+ myelo-monocytic cells (Figure 3A); in the second patient, we observed NPMc+ myelo-monocytes and megakaryocytes (Figure 3B); the third patient had a mixture of NPMc+ myelo-monocytes, megakaryocytes, and erythroid cells that varied in different areas of the biopsy (Figure 3C). All variations in the ratio of NPMc+ cells of different lineages may occur. This also applies to other FAB categories (Table 2).
In NPMc+AML, diverse combinations of NPMc+ leukemic cells of various lineages give rise to different FAB categories. For example, as illustrated in Figure 4, patients with many NPMc+ myeloid cells and rare NPMc+ erythroid precursors are morphologically classified as M0, M1, or M2 (M1 given as example in the left panels). A progressive increase in NPMc+ erythroid over myeloid cells results in M6a (Figure 4, middle panel) and M6b at the other end of the spectrum (Figure 4, right panel). All variations in the ratio of NPMc+ myeloid/NPMc+ erythroid cells are possible. In NPMc+ AML, the continuum also applies to M4 and M5 categories (not shown).
These findings highlight the inadequacy of FAB categories in defining NPMc+ AML.
Discussion
In this study, we found that clonal NPM mutations occur in 2 or more cell lineages in about 60% of NPMc+ AML, suggesting derivation from either a common myeloid progenitor or an earlier progenitor without the ability to differentiate into lymphoid lineages. Our findings also show that FAB criteria are inapplicable to NPMc+ AML, thus questioning current WHO definition criteria for the category of acute myeloid leukemia not otherwise characterized.11
Mutated NPM is a unique cell lineage clonality marker in AML, which also permits the study of the poorly characterized subgroup of AML-NK. FISH cannot detect NPM mutations, and searching for them in microdissected cells is technically demanding. A very promising approach is to search for NPM mutations in AML subpopulations at various differentiation stages after separation by fluorescence-activated cell sorting (FACS). Because of the lack of fresh leukemic cells for FACS analysis, we used immunohistochemistry on paraffin-embedded samples to study lineage involvement in AML. This technique is simple and cheap, and it also has the advantage, compared with FACS studies, of allowing cytoplasmic NPM to be correlated with optimal cytologic details. Combining immunohistochemistry with bone marrow biopsy was invaluable, because it served to identify cytoplasmic NPM even in isolated or clusters of erythroid or megakaryocyte cells, to assess dysplastic features, and to define distribution of leukemic infiltrates (eg, relation with bone trabeculae or other structures).
The most suitable reagents for labeling routine bone marrow biopsies are antibodies recognizing both wild-type and mutant NPM,13,29 because the epitopes recognized by these reagents are strongly resistant to fixatives. These antibodies predict the presence of mutated NPM in cells of different lineages, by detecting aberrant cytoplasmic NPM expression. This finding was corroborated by immunostaining with specific anti-NPM mutant antibodies and by molecular data from laser-microdissected cells from 3 NPMc+ AMLs, with 2 patients displaying mutation A and one mutation B. These mutations are predicted to encode for NPM proteins carrying, at their C-terminus, mutated tryptophan(s) and a new NES motif, which are both crucial for nuclear export and aberrant NPM cytoplasmic accumulation in leukemic cells.20
To date, information on lineage involvement in AML is scarce and usually limited to small series of patients.1,2,4,6,38-40 In AML with recurrent chromosomal abnormalities, such as t(15; 17), t(8;21), or inv(16), only a single-cell lineage appears to be involved.41-44 Multilineage involvement is frequently observed in patients with AML carrying the inv(3)(q21q26).40 Because of the lack of specific clonality markers, no information on cell lineage is available in AML-NK. Cuneo et al9 could not detect any cryptic chromosome aberrations using FISH techniques. In this study, we report a high frequency of involvement of 2 or more cell lineages in about 60% of NPMc+ AML. Because cytoplasmic or mutated NPM is closely associated with normal karyotype,13 our findings represent the first demonstration of high frequency of multilineage involvement in a large group of AML-NK.
Notably, all our cases with multilineage involvement were de novo AML, which concurs with previous observations that cytoplasmic or mutated NPM is very rare in secondary AML.13 This is a novel finding, because Bernell et al,45 using FISH combined with standard morphology (May-Grünwald-Giemsa/FISH), have shown that multilineage involvement is more frequent in secondary than in de novo AML. However, our findings are not in conflict with that study, because the 6 de novo AMLs they investigated carried cytogenetic abnormalities, which are usually mutually exclusive with cytoplasmic or mutated NPM. Interestingly, multilineage involvement has also been detected in de novo AML in which the NPM gene fuses with the MLF1 gene as result of the t(3:5) translocation.46 In our NPMc+ AML cases, there was no correlation between types of NPM mutations and type or frequency of lineage involvement, consistent with the observation that, despite molecular heterogeneity, all NPM mutants share the same alterations at their C-terminus. These molecular alterations are responsible for the cytoplasmic accumulation of NPM mutants, which may represent an important event in promoting the leukemic process. Multilineage involvement also appears to be an intrinsic feature of NPMc+ AML, independent of the presence of additional genetic alterations, such as FLT3-ITD.
Although 40% of the NPMc+ AML cases in this study showed no immunohistochemical evidence of multilineage involvement, lack of detection may be due to technical limitations. The number of NPMc+ erythroid and megakaryocytic cells might be too low to be detected and, as multilineage involvement is sometimes found in isolated areas of a tissue section but not in others, timing and sample size could underlie lack of detection. Thus, the fraction of NPMc+ AML cases with multilineage involvement may be even higher than the 60% shown in this study. Because patients in our series were heterogeneous in terms of age and treatment, the pattern of lineage involvement could not be correlated with clinical outcome. A large cohort of uniformly treated patients enrolled in multicenter clinical trials is required to investigate this issue.
The cell of origin of NPMc+ AML remains unknown. Our findings that mutated NPM is expressed in myeloid, monocytic, erythroid, and megakaryocytic cells but not in lymphoid elements suggest derivation of NPMc+ AML from either a common myeloid precursor or an earlier progenitor without the ability to differentiate into lymphoid lineages. This view is in keeping with the distinctive gene expression signature of NPMc+ AML, which is characterized by up-regulation of genes involved in stem cell maintenance (eg, most HOX genes).18,47 Negativity for CD34, a consistent feature of NPMc+ AML,13 might reflect CD34 down-regulation because of leukemic transformation or derivation from the few lineage marker-negative CD34–/CD38– hematopoietic stem cells in the bone marrow.48 The nature of the NPMc+ AML cell of origin will need to be addressed in depth by analyzing flow-sorted progenitor populations and their engraftment potential in mice, and then interpreting the results in light of the recently revised road map for adult blood lineage development.49 Regardless of the cell of origin, NPM mutations and mutant-driven cytoplasmic dislocation of wild-type NPM operate in cells of different lineages, possibly contributing to leukemogenesis.
Our immunohistologic findings clearly show that combinations and diverse ratios of NPMc+ leukemic clone-derived myeloid, monocytic, erythroid, and megakaryocyte cells are responsible for the wide morphologic spectrum observed in NPMc+ AML. Other factors that may contribute to this feature include bone marrow microenvironment conditions favoring proliferation of one cell lineage over another at the time the biopsy is taken, sampling (ie, leukemic cells of different lineages may combine at different ratios in different areas of the same biopsy), or additional as yet unrecognized secondary genetic alterations.
These results affect profoundly the current WHO classification of AML which incorporates morphologic, immunophenotypic, genetic, and clinical features “in an attempt to define entities that are biologically homogeneous and that have clinical relevance.”11 The WHO classification of AML encompasses 4 major categories: (1) AML with recurrent genetic abnormalities, usually occurring in younger persons and associated with relatively good response to therapy and outcome; (2) AML with multilineage dysplasia, more frequent in older patients, often carrying unfavorable cytogenetics; (3) therapy-related AML, a poor prognostic subgroup; and (4) AML not otherwise characterized, encompassing cases that do not fulfil criteria for inclusion in the other groups. Unfortunately, 60% to 70% of all de novo AMLs fall within the last category, which, as it includes AML with normal karyotype (ie, 40%-50% of all de novo adult AML), embraces a greater percentage of AML cases than the sum total of the other 3 categories. Thus, AML not otherwise characterized clearly needs to be better defined.
Because NPMc+ AML is mutually exclusive with recurrent chromosomal abnormalities and is closely associated with normal karyotype,13 it presently falls into the WHO category of acute myeloid leukemia not otherwise characterized, which is subclassified according to FAB criteria (ie, predominant cell type and degree of maturation).11 As we show in this study, FAB categories cannot be applied to NPMc+ AML, where varying combinations and ratios of NPMc+ cells of different lineages (all belonging to the leukemic clone) are frequently observed and give rise to a wide morphologic spectrum. Furthermore, NPM mutations predominate over FAB criteria, because NPMc+ AML displays distinctive features which are independent of FAB type13 (ie, age distribution, high frequency of FLT3-ITD mutations), distinctive gene-expression profiling,47 better response to induction therapy,13 and survival.16-19 Consequently, including NPMc+ AML in the WHO category of AML not otherwise characterized, where FAB serve as major diagnostic criteria, seems inappropriate. Indeed, the inappropriateness of FAB criteria may extend to other AML types included in the category of AML not otherwise characterized (eg, AML-NK devoid of NPM mutations).
On the basis of these findings, we propose the term AML not otherwise characterized be restricted to AML for which no genetic lesions have been so far identified and question FAB morphology as suitable criteria for subclassification. When updating the WHO classification, we suggest that, because of its frequency (about one third of all AML) and its distinctive biologic and clinical features,13 as well as its association with NPM mutations (likely a primary genetic event19 that remains stable during the course of the disease15,50 ), NPMc+ AML be considered as a provisional separate AML type for clinical use (pending elucidation of the role of mutated NPM in leukemogenesis). When combined with mutational analysis of FLT3-ITD, this would help identify patients with better prognosis within the heterogenous category of AML-NK.16-19 Undoubtedly, future classification schemes of AML, especially the category with normal karyotype, will have to take into consideration a number of genetic and molecular criteria, including newly discovered AML-associated genetic lesions and distinctive molecular signatures highlighted by gene expression profiling.51,52
Authorship
Contribution: B.F. had the original idea for the study and wrote the manuscript; L.P. was responsible for the mutational analysis of laser-microdissected cells and contributed to writing the manuscript; A.L., M.P.M., and N.B. produced and characterized the anti-NPM mutant antibodies; R.P., A.T., and M.C. carried out all immunohistochemical studies in paraffin-embedded samples; B.B., A.P., and E.T. produced and characterized the ordinary anti-NPM monoclonal antibodies; I.N. and R.M. performed the confocal microscopy analysis; C.M. was involved in the original identification of NPM mutations; and G.M., G.S., N.C., F.D.R., S.P., F.M., and M.F.M. were responsible for the patients enrolled in the GIMEMA/EORTC AML12 trial and provided the AML bone marrow samples that were investigated in this study.
Conflict-of-interest disclosure: B.F. and C.M. have applied for a patent related to the work that is described in the present work. The remaining authors declare no competing financial interests.
Prepublished online as Blood First Edition Paper, August 22, 2006; DOI 10.1182/blood-2006-06-026716.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank Dr Geraldine Boyd for editing the paper and Mrs Claudia Tibido for helpful secretarial assistance. This work was supported by the Italian Association for Cancer Research (AIRC). L.P. is a Julie Gould Scholar. N.B. is recipient of a fellowship from the Federazione Italiana per la Ricerca sul Cancro (FIRC).