Severe congenital neutropenia (SCN) and cyclic neutropenia (CyN) are sporadic or inherited hematologic disorders of myelopoiesis. Heterozygous mutations in the gene encoding neutrophil elastase (ELA2) have been reported in both diseases. We used an inducible system to express a panel of ELA2 mutations and found for almost all mutants disruption of intracellular neutrophil elastase (HNE) protein processing at different levels. This disruption resulted in cytoplasmic accumulation of a nonfunctional protein, thereby preventing its physiologic transport to azurophil granules. Furthermore, the secretory capacity of the mutant proteins was greatly diminished, indicating alteration of the regulated and the constitutive pathways. Through analysis of primary granulocytes from SCN patients carrying ELA2 mutations, we found an identical pattern of intracellular accumulation of mutant HNE protein in the cytoplasm. Moreover, cells expressing mutant HNE protein exhibited a significant increase in apoptosis associated with up-regulation of the master ER chaperone BiP, indicating that disturbance of intracellular trafficking results in activation of the mammalian unfolded protein response.
Introduction
Cyclic neutropenia (CyN) and severe congenital neutropenia (SCN) are hematologic disorders characterized by early-stage maturation arrest of myelopoiesis.1,2 These diseases are found to follow an autosomal-dominant inheritance or to arise sporadically, with the latter presumably based on new dominant mutations. Recently, the molecular cause of CyN or SCN in some patients has been identified as a heterozygous mutation in the gene encoding neutrophil elastase.3 Meanwhile, more than 40 different mutations have been identified in patients with neutropenia, with mutations detectable in 35% of patients with severe congenital neutropenia and in 44% of patients with cyclic neutropenia.4
Human neutrophil elastase is a serine protease predominantly stored in azurophil granules of neutrophil granulocytes, which are formed during the promyelocyte phase. Functionally, the protein mainly attacks Gram-negative bacteria, but it is also able to degrade a great variety of different proteins such as fibronectin, collagen, elastin, and cytokines, including granulocyte–colony-stimulating factor (G-CSF).5,6 The activity of neutrophil elastase can be antagonized by multiple endogenous inhibitors, including α1-antitrypsin, SLPI, and elafin.7
As a typical lysosomal protein, HNE is produced as a precursor and an inactive peptide that is converted to the mature protein after proteolytic processing of the N- and C-termini.8,9 The signal peptide (N-terminus) is removed cotranslationally, and the zymogen containing a 2-amino-acid propeptide is processed amino terminally. The protein acquires complex N-linked high-mannose oligosaccharide side chains that are rapidly converted to complex structures in the medial cisternae of the Golgi complex.10 After glycosylation, most of the protein is constitutively secreted in an enzymatically inactive form,11 whereas only a portion of the HNE proteins is directed to azurophil granules for regulated secretion. Recent data suggest that adaptor protein 3 (AP3) may be involved in shuttling HNE from the trans-Golgi to granules.12 Furthermore, Benson et al12 suggest that disruption of either neutrophil elastase or its supposed cargo protein, AP3, perturbs the intracellular trafficking of neutrophil elastase to azurophil granules, implying that this may be involved in the pathogenesis of congenital neutropenias associated with ELA2 mutations.
In the present study, we generated an inducible cell system based on the monoblastlike cell line U937, allowing controlled expression of different ELA2 mutations. By applying this system, we observed the disruption of intracellular HNE protein trafficking that resulted in cytoplasmic accumulation of a nonfunctional protein. Our studies were complemented by the analysis of intracellular HNE protein localization in primary granulocytes from SCN patients. Again, we observed cytoplasmic HNE protein accumulation. This protein accumulation functionally resulted in apoptosis, suggesting that mutant proteins induce cellular stress response mechanisms.
Patients, materials, and methods
Samples from healthy volunteers and patients with SCN or CyN
Peripheral blood of patients with severe congenital neutropenia or cyclic neutropenia and from healthy control donors was collected according to the guidelines approved by the Hannover Medical School institutional review board. Informed consent was provided according to the Declaration of Helsinki. Patients were treated with daily subcutaneous injections of 1 to 10 μg/kg body weight of recombinant human G-CSF (rh-G-CSF; Neupogen; Amgen, Munich, Germany). Healthy donors were untreated or treated with 5 μg/kg body weight of rh-G-CSF for 2 days before blood was drawn. Polymorphonuclear and mononuclear cells were separated by a Ficoll-Hypaque density gradient (Amersham Biosciences, Freiburg, Germany), including hypotonic lysis of erythrocytes. Subsequently, polymorphonuclear cells were harvested and processed for subcellular fractionation or immunofluorescence.
Cell culture
U937T cells originated from the human monoblast-like cell line U937 and were stably transfected with the tet-VP16 gene under control of the tet-operator promoter13 (generously provided by G. Grosveld, Department of Genetics, St Jude Children's Research Hospital, Memphis, TN). Cells were grown in RPMI 1640 medium (Life Technologies, Karlsruhe, Germany) supplemented with 10% heat-inactivated fetal calf serum (FCS; Life Technologies), 100 U/mL penicillin, 100 μg/mL streptomycin (PAA Laboratories, Cölbe, Germany), 2 mM l-glutamine (PAA Laboratories), 0.5 μg/mL puromycin (Sigma, Taufkirchen, Germany), and 1 μg/mL tetracycline (Sigma). 293T cells (ATCC; LGC Promochem, Wesel, Germany) were cultured in MEM (Life Technologies) with 10% FCS, 100 U/mL penicillin, 100 μg/mL streptomycin, 1 × nonessential amino acids (Life Technologies), and 1 mM sodium pyruvate (Life Technologies). HT 1080 cells (ATCC) were maintained in 10% FCS-containing DMEM (Life Technologies) supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, 1 × nonessential amino acids, 1 mM sodium pyruvate, and 2 mM l-glutamine. HeLa cells (ATCC) were cultured in Eagle MEM (CytoGen, Sinn, Germany) complemented with 10% FCS without added antibiotics.
Construction of HNE-expressing vectors
cDNA encoding neutrophil elastase (ELA2) was reverse transcribed from total RNA of HL60 cells and cloned into the BamHI restriction enzyme site of expression vector pLXIN (BD Clontech, Heidelberg, Germany). The integrity of the wild-type ELA2 sequence was verified by sequencing. Site-directed mutagenesis with the use of oligonucleotide cassettes (Quikchange site-directed mutagenesis kit; Stratagene, Amsterdam, The Netherlands) was used to generate each of the neutrophil elastase mutations. The integrity of the reading frame and the desired mutations were verified by sequencing. To create the tet-off system, all constructs were individually recloned into the BamHI site of pRevTRE expression vector (BD Clontech). Details on plasmid constructions and primer sequences are available upon request.
Viral vector production and retroviral cell transduction
Retroviral particles were produced by triple transfection of 293T cells using the calcium-phosphate method (CalPhos; BD Clontech) with the following plasmids: pRevTRE (containing wild-type or mutated ELA2 or without any insert as a control), gag-pol, and VSV-G (vesicular stomatitis virus G-protein). Gag-pol and VSV-G plasmids were generously provided by Ted Friedman (Center for Molecular Genetics and Department of Pediatrics, University of California, San Diego, CA). Viral titers of hygromycin-resistant polyclonal cell populations were estimated with the use of a standard titration assay performed on HT 1080 cells and were routinely in the range of 1 × 105 cfu/mL.
For transduction, 4 × 105 U937T cells were plated into Retronectin-coated 6-well plates (Takara, Gennevilliers, France). Cells were transduced with the addition of retroviral supernatant 24 hours after cell plating, which was repeated once after 24 hours. Cells were selected in medium containing hygromycin (PAA Laboratories). Single-cell clones were isolated from stably transduced cell pools using limited dilution. Ten independent single clones per mutation were examined for tetracycline-dependent expression of neutrophil elastase by Western blot and RNA analysis.
Inducible gene expression in U937T cells
To induce tetracycline-dependent expression of neutrophil elastase, cells were washed 3 times with phosphate-buffered saline (PBS) and seeded at 3 × 105 cells/mL in RPMI medium containing 10% FBS (tet-system–approved FBS; BD Clontech), 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM L-glutamine. To remove remaining tetracycline, cells were washed after 3 hours and 24 hours of incubation. Cells were transferred to fresh medium on day 3 and harvested on day 4.
Transient transfection of HeLa cells
For transient transfection, 4 × 104 adherent HeLa cells were cultured on glass coverslips in 24-well plates. Cells were transfected using the calcium-phosphate method (CalPhos; BD Clontech) with pLXIN vector containing wild-type or mutant ELA2 sequences. Forty-eight hours after transfection, cells were fixed with 2% paraformaldehyde in PBS, pH 7.4, and were processed for immunofluorescence labeling. For tunicamycin assays, cells were incubated with medium containing 2 μg/mL tunicamycin (Sigma) for 16 hours before fixation.
Deglycosylation experiments
The presence of N-linked glycans in the various constructs of neutrophil elastase was analyzed by treating total cell lysates with endoglycosidase H (New England Biolabs, Frankfurt/Main, Germany) or N-glycosidase F (New England Biolabs). For this purpose, cell lysates were resuspended in denaturation buffer (New England Biolabs) and heated to 95°C for 7 minute, followed by addition of the enzyme. After incubation at 37°C for 90 minutes, samples were analyzed by Western blotting.
Subcellular fractionation
Differential centrifugation was modified based on a protocol allowing isolation of subcellular fractions.14 Briefly, 0.5 to 2 × 108 cells were resuspended in hypotonic lysis buffer (HLB: 1 mM EDTA, 10 mM phenyl phosphate, 10 mM β-glycerol phosphate, 10 mM sodium fluoride) containing protease inhibitors (100 mM phenylmethylsulfonyl fluoride, 2.8 mg/mL aprotinin, 2 mg/mL pepstatin A, and 2 mg/mL leupeptin (all from Sigma). After incubation on ice for 25 minutes, cells were disrupted with 15 strokes in a Dounce glass homogenizer. The homogenate was centrifuged at 500g for 15 minutes to enrich nuclei (P1). The fraction enriched for mitochondria, lysosomes, and endoplasmic reticulum (P2) was pelleted at 10 800g for 15 minutes. The resultant supernatant was centrifuged at 100 000g for 60 minutes to obtain a pellet enriched for plasma membranes, Golgi membranes, and secretory vesicles (P3). The remaining supernatant (S) contained soluble cytosolic proteins. All pellets were washed once in HLB containing protease and subsequently were lysed in RIPA buffer (10 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.5% Na-deoxycholate, 0.1% SDS) containing protease inhibitors. Protein concentrations were determined using the Bradford assay (BioRad, Munich, Germany). Finally, all isolated fractions were characterized by Western blot analysis.
Western blot analysis
To separate proteins, cell pellets were lysed in RIPA buffer containing protease inhibitors. Protein concentrations were determined using Bradford reagent. Proteins (25 μg or 50 μg) were resolved electrophoretically on denaturing polyacrylamide gels and transferred onto nitrocellulose membranes (Schleicher & Schüll, Dassel, Germany) using a semidry-transfer system (Biometra, Göttingen, Germany). Nonspecific interactions were blocked by preincubation of the membranes with blocking solution (Western Breeze; Invitrogen, Karlsruhe, Germany). Primary antibodies directed against the following proteins were used: neutrophil elastase (C-17; Santa Cruz, Heidelberg, Germany) or anti–neutrophil elastase (Merck Biosciences Calbiochem, Schwalbach/TS, Germany), β-actin (clone AC-40; Sigma), nup 153 (Progen, Heidelberg, Germany), nucleoporin15,16 (BD PharMingen, Heidelberg, Germany), lamp-2 (BD PharMingen), bcl-2 (Dako Cytomation; Hamburg, Germany), PARP-1 (Santa Cruz), calnexin (BD Biosciences; Heidelberg, Germany), GM 130 (BD Biosciences), CD4517 (kindly provided by B. Schraven, University of Magdeburg, Magdeburg, Germany), and clathrin (Progen). All antibodies were diluted in PBS containing 0.5% Tween 20 and 2% nonfat dried milk. After incubation with secondary antibody (Santa Cruz) conjugated to horseradish peroxidase, membranes were developed with the use of a chemiluminescent substrate (ECL advanced chemiluminescence detection system; Amersham Biosciences). For quantification, band intensities were measured by using National Institutes of Health (NIH) Image software, and protein signals were normalized to β-actin levels.
Preparation of cell extracts and enzymatic assays
A modified protocol was used to prepare cell extracts.9 Briefly, 1 × 108 cells were washed in PBS followed by 2 freeze-thaw cycles. Subsequently, the pellet was resuspended in 450 μL of 100 mM Tris-Cl, pH 8.5, containing 1 M MgCl2 and 0.1% Triton X-100. After sonification for 30 seconds (Branson Sonifier 250; Branson Ultrasonics, Eemnes, The Netherlands) to obtain complete homogenization of the cells, 1 mL of 5 mM Tris-Cl, pH 8.5, containing 1 M NaCl and 0.1% Triton X-100 was added. The lysate was centrifuged at 15 000g and 4°C for 2 hours to pellet DNA. Of the remaining lysate, 40 μL was mixed with 53 μL of 200 mM Tris-Cl, pH 8.5, containing 1 M NaCl and one of the following substrates (all substrates were dissolved in 1-methyl-2 pyrrolidine from Sigma): 50 mM suc-Ala-Ala-Ala-pNA (Bachem, Bubendorf, Germany) or 10 mM suc-Tyr-Leu-Val-pNA (Bachem). The mixture was incubated for 30 minutes (suc-Ala-Ala-Ala-pNA) or 5 minutes (suc-Tyr-Leu-Val-pNA) at 40°C. The reaction was stopped by the addition of 107 μL soybean trypsin inhibitor (200 μg/mL; Sigma). Absorbance at 405 nm (suc-Tyr-Leu-Val-pNA) or 410 nm (suc-Ala-Ala-Ala-pNA) was determined using Spectra max 340 PC (Molecular Devices, Munich, Germany). A standard curve was obtained by measuring the activities of a serial dilution of porcine pancreatic elastase in parallel (Sigma). Relative activity was calculated according to the following equation: ratio of relative HNE activity (activity (+ind) – activity (–ind)) to relative HNE expression (HNE protein (+ind) – HNE protein (–ind) determined by Western blot). Each sample was analyzed in at least 2 independent experiments with duplicate samples, and data are given as mean ± SD.
Detection of secreted neutrophil elastase
Secretion of neutrophil elastase was determined by analysis of the amount of HNE protein present in cell culture supernatants of U937T cells after induction of tetracycline-dependent HNE expression. On day 3 after promoter induction, 6 × 107 cells were seeded in 100 mL complete medium. Supernatants were harvested after 24 hours. Cellular proteins were harvested in parallel to analyze the total HNE protein amount by Western blot. HNE amounts in supernatants were determined with the human Elastase ELISA Test Kit (HyCult Biotechnology b.v.; Uden, The Netherlands) according to the manufacturer's instructions. Absorbance at 450 nm was determined using Spectra max 340 PC (Molecular Devices). Relative secretion was calculated with the following equation: ratio of relative HNE secretion (HNE sup [+ind] – HNE sup [–ind]) to relative HNE expression (HNE protein [+ind] – HNE protein [–ind] determined by Western blot). Each sample was analyzed in at least 2 independent experiments with duplicate samples, and data are given as mean ± SD.
Immunofluorescence labeling and confocal laser microscopy
Forty-eight hours after transfection, HeLa cells were fixed with 2% paraformaldehyde. Polymorphonuclear cells were adhered on poly-l-lysine–coated glass slides for 30 minutes at 37°C in 0.2% BSA-PBS followed by fixation with 2% paraformaldehyde (Sigma). Cells were permeabilized with 0.5% Triton-X 100 (Sigma) in PBS for 5 minutes. Unspecific binding sites were blocked with 10% human serum. Incubations with primary and secondary antibodies were carried out in PBS with 10% human serum.18 The following primary antibodies were used: anti–neutrophil elastase (Calbiochem), anti–neutrophil elastase C-17 (Santa Cruz), anti–lamp-3 (Santa Cruz), anticalnexin (Acris; Hiddenhausen, Germany), anti-MPO (Abcam, Hiddenhausen, Germany), and antigiantin (Hiss Diagnostics, Freiburg, Germany). Secondary antibodies conjugated with rhodamine Red-X, Cy2, or TRITC were from Jackson ImmunoResearch Laboratories (Hamburg, Germany). After each incubation, unbound antibodies were removed by 3 rinses with PBS. Nuclei were counterstained with TO-PRO-3 (Molecular Probes, Invitrogen). Glass slides were mounted with Moviol 4-88 Reagent (Calbiochem) containing DABCO (Sigma). Confocal microscopy was performed with a Leica TCS SP2 AOBS (Leica Microsystems, Bensheim, Germany) equipped with a 40 ×/1.25 NA oil-immersion objective lens; oil was used as the imaging medium. Digital images were saved as .tiffs, inserted, and processed in Adobe Photoshop (Adobe Systems, San Jose, CA).
Apoptosis assay
U937T cells expressing wild-type (WT) and mutated (MT) HNE were cultured in the absence of tetracycline to induce transgene expression. On day 2, cells were transferred to enriched medium (OptiMEM 1; Invitrogen) without added serum to avoid protease inhibitor activity. On day 3, 10 μM camptothecin (Sigma) was added to induce apoptosis. The rate of apoptosis was measured by flow cytometry (FACSCalibur; BD Biosciences) using an annexin V assay (Roche Applied Science, Mannheim, Germany) and was evaluated using CellQuest Pro software (BD Biosciences). Each sample was analyzed in at least 4 independent assays. Apoptosis values of each sample were normalized to those of WT + CPT, and data are given as mean ± SD.
RNA isolation and real-time PCR
Total RNA was extracted from cells with the RNeasy kit (Qiagen, Hilden, Germany). RNAs were quantified photometrically. Before cDNA synthesis, RNA samples were DNase digested to avoid potential contamination with genomic DNA. cDNA was prepared by reverse transcription of total RNA using random hexamer oligonucleotides (MWG Biotech, Ebersberg, Germany) and Superscript II (Life Technologies).
For quantitative real-time reverse transcription–polymerase chain reaction (RT-PCR) analysis, the iCycler technology (BioRad) was applied according to the manufacturer's instructions using TaqMan PCR core reagents (Eurogentec, Seriang, Belgium). For BiP, predeveloped TaqMan assay reagents (PDARs; Applied Biosystems, Foster City, CA) were used according to the manufacturer's instructions. Means of BiP expression were normalized to GAPDH levels using GAPDH predeveloped TaqMan assay reagents (Applied Biosystems). Input amounts were optimized, resulting in threshold values between 20 and 35 cycles. Each sample was analyzed in at least 2 independent assays with duplicate samples, and data are given as mean ± SD. Relative expression levels were calculated using the comparative Ct method. Therefore, the target PCR Ct values were normalized to the Ct value of the reference gene (GAPDH) by subtracting the GAPDH Ct value from the target Ct value. The relative expression level for each target PCR was calculated using the equation: relative expression = 2–(Ct(target)–Ct(GAPDH)) × 100.
Results
Mutations in HNE proteins lead to disruption of intracellular processing and alteration of proteolytic activity and secretion in U937T cells
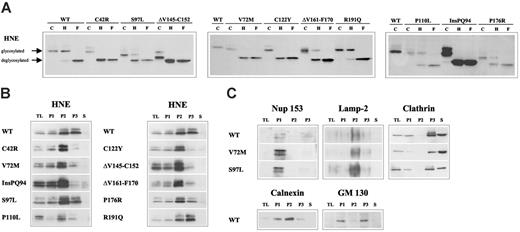
To analyze the functional consequences of HNE mutations, we overexpressed wild-type (WT) or mutated (MT) ELA2 genes (Figure 1) from a tetracycline-responsive promoter in the human monoblast-like cell line U937. First, we monitored glycosylation as a marker for intracellular processing and transport of the protein. The WT-HNE protein was partially resistant to endoglycosidase H digestion, demonstrating correct processing up to the medial Golgi complex. In contrast, 5 of the 10 mutants were sensitive to endoglycosidase H, demonstrating that the mutant proteins were not properly processed (Figure 2A). Next, we determined the intracellular HNE localization in subcellular fractions after differential centrifugation of cell lysates (Figure 2B). As expected, WT-HNE was mainly detected in the fractions containing lysosomes and ER (P2) and secretory and membrane vesicles, including Golgi membranes (P3) (Figure 2B-C). Six of 10 mutants demonstrated differences in subcellular HNE distribution compared with WT, suggesting perturbation of HNE protein trafficking. The loss of complex glycosylation was associated with significant reduction of HNE protein in the fraction containing Golgi membranes (P3). Only one mutant (ΔV161-F170) showed intracellular mislocalization despite an apparently normal glycosylation.
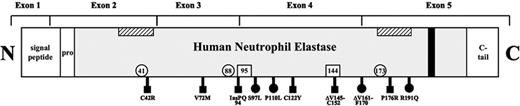
Scheme of HNE. Exon boundaries of the neutrophil elastase sequence, including the amino-terminal signal, the propeptide, and the carboxy-terminal tail. ▧, predicted, hypothetical transmembrane domains12 ; ▪ hypothetical tyrosine-based sorting signal; □, locations of 3 residues forming the catalytic site (His-41, Asp-88, Ser-173); □ locations of the Asn that are N-glycosylated (Asn-95, Asn-144); ▪ mutations analyzed in the present study and causing SCN; and • mutations causing SCN or CyN.
Scheme of HNE. Exon boundaries of the neutrophil elastase sequence, including the amino-terminal signal, the propeptide, and the carboxy-terminal tail. ▧, predicted, hypothetical transmembrane domains12 ; ▪ hypothetical tyrosine-based sorting signal; □, locations of 3 residues forming the catalytic site (His-41, Asp-88, Ser-173); □ locations of the Asn that are N-glycosylated (Asn-95, Asn-144); ▪ mutations analyzed in the present study and causing SCN; and • mutations causing SCN or CyN.
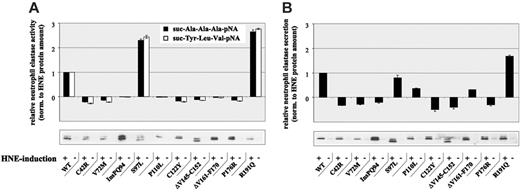
To further analyze the secretory pathway, we determined proteolytic activity and secretion capacity of U937T cells expressing WT- or MT-HNE proteins (Figure 3). In cells expressing WT-HNE or the mutant S97L or R191Q, a clear induction of proteolytic activity was found on promoter induction for both synthetic substrates tested (Figure 3A). In contrast, all other mutants had no proteolytic activity on transgene induction, confirming that these mutations were associated with alterations in protein function. These findings are supported by Li and Horowitz,19 who also measured reduced proteolytic activity for most of the mutants.
Analysis of protein secretion revealed the up-regulation of HNE secretion on transgene induction in WT, S97L-, and R191Q-expressing cells (Figure 3B). Again, all other mutants demonstrated a suppression (P110L and ΔV161-F170) or even a lack of HNE secretion, supporting our hypothesis of disruption of the secretory pathway.
Disruption of subcellular localization of MT-HNE proteins in U937T cells. (A) Glycosylation of WT-HNE or MT-HNE in U937T cells. Whole cell lysates of U937T-WT-HNE or -MT-HNE cells after induction of transgene expression were incubated with endoglycosidase H (H) or N-glycosidase F (F) or without enzyme (control; C) and were analyzed by immunoblot using anti-HNE (29-34 kDa). Resistance to digestion by endoglycosidase H indicates that HNE has reached the medial Golgi compartment. (B) Presence of HNE protein (29-34 kDa) in subcellular fractions of U937T cell lysates obtained by differential centrifugation: P1 (nuclear and nuclear-associated proteins), P2 (mitochondria, lysosomes, ER), P3 (plasma membrane, secretory and other vesicles, Golgi complex), and S (soluble cytosolic proteins). TL indicates total cell lysate. (C) Localization of cellular marker proteins in subcellular fractions obtained by differential centrifugation for WT-HNE and the mutants V72M and S97L. The nuclear pore complex protein nup 153 (190 kDa) defines P1. Lamp-2 (120 kDa) is a lysosomal protein specifying the lysosomal fraction (P2). To characterize fractions P3 and S, clathrin (180 kDa), which may be membrane associated or soluble, was used. Fractionation of the ER and the Golgi complex were defined with the use of calnexin (90 kDa) and GM 130 (130 kDa).
Disruption of subcellular localization of MT-HNE proteins in U937T cells. (A) Glycosylation of WT-HNE or MT-HNE in U937T cells. Whole cell lysates of U937T-WT-HNE or -MT-HNE cells after induction of transgene expression were incubated with endoglycosidase H (H) or N-glycosidase F (F) or without enzyme (control; C) and were analyzed by immunoblot using anti-HNE (29-34 kDa). Resistance to digestion by endoglycosidase H indicates that HNE has reached the medial Golgi compartment. (B) Presence of HNE protein (29-34 kDa) in subcellular fractions of U937T cell lysates obtained by differential centrifugation: P1 (nuclear and nuclear-associated proteins), P2 (mitochondria, lysosomes, ER), P3 (plasma membrane, secretory and other vesicles, Golgi complex), and S (soluble cytosolic proteins). TL indicates total cell lysate. (C) Localization of cellular marker proteins in subcellular fractions obtained by differential centrifugation for WT-HNE and the mutants V72M and S97L. The nuclear pore complex protein nup 153 (190 kDa) defines P1. Lamp-2 (120 kDa) is a lysosomal protein specifying the lysosomal fraction (P2). To characterize fractions P3 and S, clathrin (180 kDa), which may be membrane associated or soluble, was used. Fractionation of the ER and the Golgi complex were defined with the use of calnexin (90 kDa) and GM 130 (130 kDa).
Mutations in neutrophil elastase lead to cytoplasmic protein accumulation
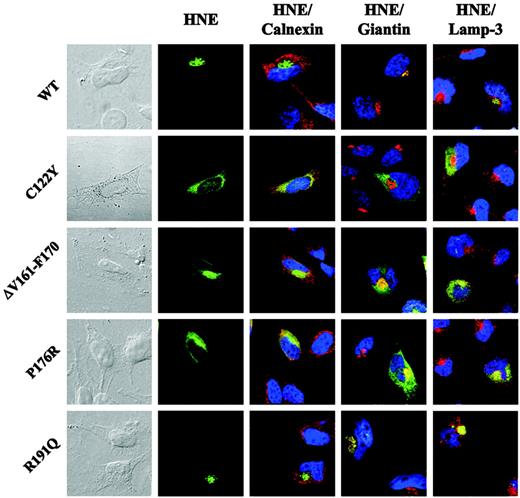
To define the subcellular distribution of the mutant proteins more precisely, we used immunofluorescence microscopy. Given that U937 cells contain only a small ring of cytoplasm around their nuclei, we used a different human cell line (HeLa) and transiently transfected ELA2 WT or MT sequences. Most of the WT-HNE colocalized with Golgi structures (Figure 4 and Figure S1, which is available on the Blood website; see the Supplemental Figures link at the top of the online article). Mutant HNE proteins displayed a distinct intracellular localization compared with WT. Mutant C122Y colocalized with neither the Golgi complex nor with the lysosomal marker lamp-3 (Figures 4, S2). Instead, the protein was localized in the entire cytoplasm, where it partially colocalized with calnexin, a marker of ER. The remaining portion of the mutant protein might have been retrotranslocated to the cytosol or targeted to other domains of the ER. Mutant ΔV161-F170, which displayed normal glycosylation but altered intracellular localization and loss of proteolytic activity and secretion in U937T cells, partially colocalized with calnexin and the Golgi marker giantin (Figures 4, S3). In addition, as already found for mutant C122Y, significant accumulation of the protein was found in other parts of the cytoplasm, indicating a post-Golgi trafficking defect. Analysis of mutant P176R revealed an intracellular HNE localization similar to that of ΔV161-F170 (Figures 4, S4). These findings suggest that MT-HNE proteins accumulate in cytoplasmic structures and that this mistargeting is associated with loss of function (proteolytic activity and secretion). Interestingly, R191Q, a mutant that displayed physiologic protein processing and function, demonstrated HNE protein localization comparable with that of WT-HNE (Figures 4, S5). Further details for each mutation are listed in Table 1.
Altered proteolytic activity and secretion of MT-HNE in U937T cells. (A) Proteolytic activity of WT-HNE or MT-HNE in U937T cells using the chromogenic substrates suc-Ala-Ala-Ala-pNA (▴) and suc-Tyr-Leu-Val-pNA (□). Relative activity of HNE was defined as the difference between activity with and without promoter induction and was normalized to the amount of HNE protein synthesized (determined by immunoblotting). Data are given as mean ± SD. (B) Secretion of WT-HNE or MT-HNE from U937T cells. Relative secretion was defined as the difference between secretion with and without promoter induction and was normalized to the amount of total HNE protein synthesized (determined by immunoblotting). Data are given as mean ± SD.
Altered proteolytic activity and secretion of MT-HNE in U937T cells. (A) Proteolytic activity of WT-HNE or MT-HNE in U937T cells using the chromogenic substrates suc-Ala-Ala-Ala-pNA (▴) and suc-Tyr-Leu-Val-pNA (□). Relative activity of HNE was defined as the difference between activity with and without promoter induction and was normalized to the amount of HNE protein synthesized (determined by immunoblotting). Data are given as mean ± SD. (B) Secretion of WT-HNE or MT-HNE from U937T cells. Relative secretion was defined as the difference between secretion with and without promoter induction and was normalized to the amount of total HNE protein synthesized (determined by immunoblotting). Data are given as mean ± SD.
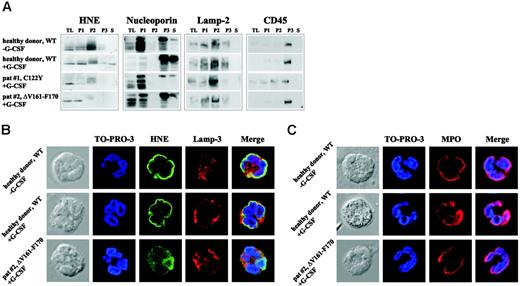
Intracellular mistrafficking and cytoplasmic accumulation of mutant HNE proteins in granulocytes from SCN patients
To address the significance of these phenotypic changes for primary cells, we analyzed intracellular HNE protein localization in neutrophils from patients with SCN carrying defined ELA2 mutations. Because SCN patients are treated by daily subcutaneous injections of G-CSF, untreated and G-CSF–treated healthy donors were used as controls. In healthy control cells, WT-HNE protein was found mainly in fraction P2, which contains the azurophil granules (Figure 5A). Lesser amounts were found in fractions P1 and P3. In both SCN patients, HNE protein was not detected in P3, reflecting secretory and membrane vesicles. Furthermore, a faint fuzzy band of higher molecular weight was present in fraction P2, which might have corresponded to unprocessed variants or protein bound to interacting factors, possibly located in the ER. A similar pattern was found in additional samples collected from patients with SCN or CyN carrying distinct ELA2 mutations (data not shown). Next, immunofluorescence microscopy was used to determine subcellular protein localization. In granulocytes from healthy controls with and without G-CSF treatment, HNE protein was predominantly perinuclear. Only small amounts of the protein were detected in other parts of the cytoplasm (Figure 5B). The perinuclear localization of WT-HNE may represent protein within the intracellular processing cascade localizing to the ER or the Golgi complex, or both. In granulocytes from 2 SCN patients carrying the mutations C122Y (patient 1)20 or ΔV161-F170 (patient 2), perinuclear staining of HNE protein was not present (C122Y; Figure S6) or was only partially present (ΔV161-F170; Figure 5B). Instead, most HNE was identified in a diffuse cytoplasmic, possibly cytosolic, distribution. Only minor colocalization was detected with a marker protein of azurophil granules, lamp-3, indicating a trafficking defect to granules. To determine whether mislocalization was restricted to MT-HNE, we examined the distribution of another azurophil granule protein, myeloperoxidase (MPO).10 In contrast to HNE, MPO was found in a perinuclear pattern not only in the granulocytes of healthy controls but also in those of SCN patients (Figures 5C, S6), indicating selective mistargeting of mutant elastase.21
Mutations in ELA2 lead to cytoplasmic accumulation of MT-HNE proteins in transiently transfected HeLa cells. Subcellular localization of WT- or MT-HNE protein in HeLa cells. Cells were labeled with anti-HNE (green) and anticalnexin (ER marker; red), antigiantin (Golgi marker; red), and anti–lamp-3 (lysosomal marker; red). TO-PRO-3 was used to counterstain the nuclei (blue). Yellow displays colocalization of HNE protein with organelle-specific markers (Figures S1-S5).
Mutations in ELA2 lead to cytoplasmic accumulation of MT-HNE proteins in transiently transfected HeLa cells. Subcellular localization of WT- or MT-HNE protein in HeLa cells. Cells were labeled with anti-HNE (green) and anticalnexin (ER marker; red), antigiantin (Golgi marker; red), and anti–lamp-3 (lysosomal marker; red). TO-PRO-3 was used to counterstain the nuclei (blue). Yellow displays colocalization of HNE protein with organelle-specific markers (Figures S1-S5).
Induction of cellular stress responses in U937T cells expressing mutant HNE proteins
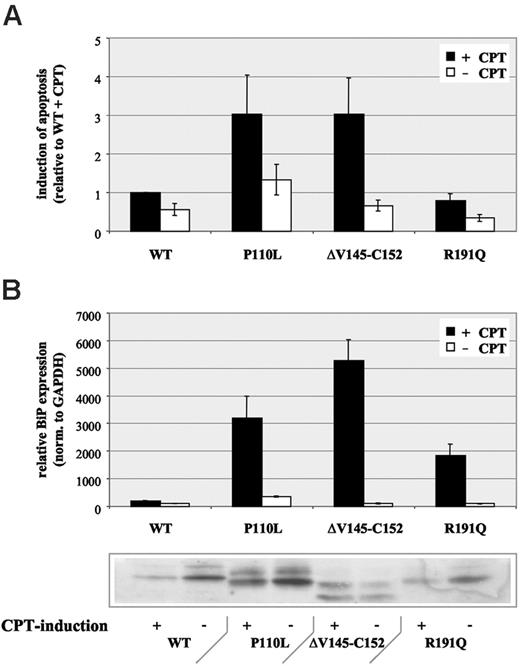
Next, we were interested in studying the phenotypic consequence of such cytoplasmic protein accumulation instead of transport to granules or vesicles for regulated or constitutive secretion. Through analysis of apoptosis, we found an increase of camptothecin-dependent apoptosis for those HNE mutations with protein accumulation and loss of function (P110L, ΔV145-C152), whereas a mutant with physiologic HNE transport and function (R191Q) demonstrated no increased apoptosis compared with WT protein (Figure 6A).
Based on these findings, we hypothesized that HNE mutations may not properly fold, thereby inducing a specific cellular stress response termed unfolded protein response (UPR).22 This pathway can lead characteristically to retrotranslocation of mutant proteins to the cytosol or their accumulation in the ER. Typically, UPR induction is associated with up-regulated transcription of ER chaperones.22 Therefore, we analyzed expression of the ER chaperone BiP (GRP 78). Consistent with our hypothesis, we found a moderate up-regulation in MT-HNE–expressing cells compared with WT-HNE–expressing controls (Figure 6B). After induction of apoptosis with camptothecin, BiP expression was significantly up-regulated (18- to 53-fold) in MT-HNE–expressing cells only (Figure 6B). Surprisingly, R191Q, 1 of the 2 mutants with no functional changes (no detectable trafficking defect, no loss of function) demonstrated a clear induction of BiP expression.
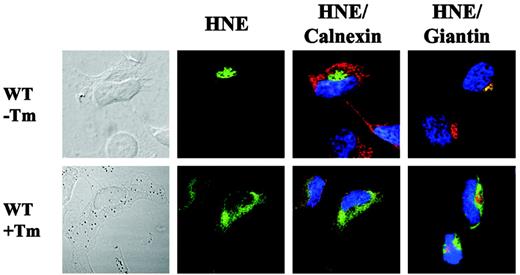
Finally, we were interested in analyzing subcellular WT-HNE localization after synthetic induction of the UPR pathway by tunicamycin, a specific inhibitor of N-glycosylation in ER.23 The addition of tunicamycin resulted in partial colocalization with the ER and cytoplasmic accumulation of WT-HNE in transfected HeLa cells (Figure 7). This localization was similar to that displayed by mutant C122Y. Thus, we propose that intracellular misprocessing of mutant HNE may induce the unfolded protein stress response.
Intracellular mistrafficking and cytoplasmic accumulation of MT-HNE proteins in neutrophil granulocytes of patients with SCN. (A) Localization of HNE protein (29-34 kDa) in subcellular fractions of primary polymorphonuclear cell lysates obtained by differential centrifugation. Analysis of the following fractions: P1 (nuclear and nuclear-associated proteins), P2 (mitochondria, lysosomes, ER), P3 (plasma membrane, secretory and other vesicles, Golgi complex), and S (soluble cytosolic proteins). TL indicates total cell lysate. A healthy donor without G-CSF treatment and a healthy donor who received G-CSF subcutaneously for 2 days served as controls. Both SCN patients received daily treatment with G-CSF. Each patient carries a heterozygous mutation in ELA2 (patient 1, C122Y; patient 2, ΔV161-F170). Localization of cellular marker proteins in subcellular fractions was obtained by differential centrifugation. Nucleoporin (62 kDa) is a nuclear and cytoplasmic pore complex protein, thus defining P1, P3, and S. Lamp-2 (120 kDa) is a lysosomal membrane protein that fractionates mainly in P2, and CD45 (180-220 kDa) is a plasma membrane–associated protein mainly localized in fraction P3. (B) Subcellular localization of WT-HNE in granulocytes of healthy donors with or without rh-G-CSF treatment or of MT-HNE in granulocytes of a patient with SCN and carrying a heterozygous mutation in ELA2 (patient 2, ΔV161-F170). Cells were stained with anti-HNE (green) and lamp-3 (lysosomal marker; red). TO-PRO-3 was used to counterstain the nuclei (blue). Yellow represents colocalization of HNE protein with lamp-3. (C) Subcellular localization of MPO in granulocytes of healthy donors with or without rh-G-CSF treatment or in granulocytes of a patient with SCN and carrying a heterozygous mutation in ELA2 (patient 2, ΔV161-F170). Cells were stained with anti-MPO (red). TO-PRO-3 was used to counterstain the nuclei (blue).
Intracellular mistrafficking and cytoplasmic accumulation of MT-HNE proteins in neutrophil granulocytes of patients with SCN. (A) Localization of HNE protein (29-34 kDa) in subcellular fractions of primary polymorphonuclear cell lysates obtained by differential centrifugation. Analysis of the following fractions: P1 (nuclear and nuclear-associated proteins), P2 (mitochondria, lysosomes, ER), P3 (plasma membrane, secretory and other vesicles, Golgi complex), and S (soluble cytosolic proteins). TL indicates total cell lysate. A healthy donor without G-CSF treatment and a healthy donor who received G-CSF subcutaneously for 2 days served as controls. Both SCN patients received daily treatment with G-CSF. Each patient carries a heterozygous mutation in ELA2 (patient 1, C122Y; patient 2, ΔV161-F170). Localization of cellular marker proteins in subcellular fractions was obtained by differential centrifugation. Nucleoporin (62 kDa) is a nuclear and cytoplasmic pore complex protein, thus defining P1, P3, and S. Lamp-2 (120 kDa) is a lysosomal membrane protein that fractionates mainly in P2, and CD45 (180-220 kDa) is a plasma membrane–associated protein mainly localized in fraction P3. (B) Subcellular localization of WT-HNE in granulocytes of healthy donors with or without rh-G-CSF treatment or of MT-HNE in granulocytes of a patient with SCN and carrying a heterozygous mutation in ELA2 (patient 2, ΔV161-F170). Cells were stained with anti-HNE (green) and lamp-3 (lysosomal marker; red). TO-PRO-3 was used to counterstain the nuclei (blue). Yellow represents colocalization of HNE protein with lamp-3. (C) Subcellular localization of MPO in granulocytes of healthy donors with or without rh-G-CSF treatment or in granulocytes of a patient with SCN and carrying a heterozygous mutation in ELA2 (patient 2, ΔV161-F170). Cells were stained with anti-MPO (red). TO-PRO-3 was used to counterstain the nuclei (blue).
Discussion
Neutrophil elastase belongs to the family of serine proteases stored in azurophil granules of neutrophil granulocytes. Mutations in the gene encoding neutrophil elastase have been associated with cyclic and severe congenital neutropenia, a hematologic disorder characterized by early-stage maturation arrest of myelopoiesis.3,4 At present, more than 40 ELA2 mutations have been identified in patients with these disorders. Recent studies suggest incorrect intracellular trafficking of the mutant proteins12 that may be associated with accelerated apoptosis.24 However, the underlying molecular mechanisms and the significance of these changes in primary granulocytes of patients with SCN or CyN remain unclear.
Increased apoptosis in U937T cells expressing MT-HNE proteins. (A) Induction of apoptosis with camptothecin (CPT) in U937T cells expressing wild-type HNE (WT) or MT-HNE proteins (P110L, ΔV145-C152, R191Q). Evaluation of apoptosis by quantification of annexin V-positive cells by flow cytometry. Data are given as mean ± SD. (B) Real-time PCR analysis of relative BiP (GRP78) RNA expression in cells expressing wild-type HNE (WT) or MT-HNE proteins (P110L, ΔV145-C152, R191Q) with or without CPT treatment. Immunoblot using anti-HNE (29-34 kDa) confirms induction of transgene expression in corresponding cell lysates. Data are given as mean ± SD.
Increased apoptosis in U937T cells expressing MT-HNE proteins. (A) Induction of apoptosis with camptothecin (CPT) in U937T cells expressing wild-type HNE (WT) or MT-HNE proteins (P110L, ΔV145-C152, R191Q). Evaluation of apoptosis by quantification of annexin V-positive cells by flow cytometry. Data are given as mean ± SD. (B) Real-time PCR analysis of relative BiP (GRP78) RNA expression in cells expressing wild-type HNE (WT) or MT-HNE proteins (P110L, ΔV145-C152, R191Q) with or without CPT treatment. Immunoblot using anti-HNE (29-34 kDa) confirms induction of transgene expression in corresponding cell lysates. Data are given as mean ± SD.
In the present study, we have investigated the intracellular processing, localization, and functional consequences of mutant HNE expression using an inducible cell expression system based on the monoblast-like cell line U937. In summary, our data suggest that, depending on the particular mutation, intracellular trafficking is interrupted at different compartments, such as before (C42R, V72M, InsPQ94, C122Y, ΔV145-C152) or after (ΔV161-F170) entering the Golgi complex or during trafficking to lysosomes or secretory vesicles (P110L, P176R). This mistargeting was associated with loss of function of the mutant HNE protein (proteolytic activity and secretion). To precisely define subcellular localization of mutant HNE proteins, we next performed immunofluorescence staining and confocal laser microscopy in HeLa cells expressing individual ELA2 mutations. In these cells, MT-HNE proteins accumulated in cytoplasmic structures outside the lysosomal compartment. Taken together, our data suggest that intracellular processing and trafficking of mutant HNE proteins is interrupted after ER, resulting in cytoplasmic protein accumulation. Only 2 mutants, S97L and R191Q, exhibited physiologic intracellular trafficking and HNE protein localization, with the mutant protein retaining its secretory ability and its proteolytic activity.
The significance of this mislocalization was confirmed in primary granulocytes of patients with SCN or CyN carrying ELA2 mutations. Consistent with our cell model, the mutant proteins accumulated in the cytoplasm in compartments other than ER or Golgi. To our knowledge, this is the first report demonstrating MT-HNE mislocalization in primary granulocytes of patients with chronic neutropenia, confirming the importance of intracellular mistrafficking of the mutant proteins for the neutropenic phenotype.
Interestingly, in a recent model, it was proposed that HNE is a transmembrane protein that would be targeted to azurophil granules through the cytosolic adaptor protein 3 (AP3), which might bind to its hypothetical cytosolic tail.12 Consequently, mutations disrupting the hypothetical transmembrane domains were interpreted to result in different intracellular trafficking that would lead to a predominant granular localization. All the mutants presented in our study were located within or around the predicted hypothetical transmembrane domains. According to this model, they should have been directed to the azurophil granules only. In accordance with Benson et al,12 we could not detect any mutant HNE at the plasma membrane with immunofluorescence microscopy (data not shown). However, our data indicated an inability to target the mutant proteins to lysosomal or azurophil granules and a loss of proteolytic activity and constitutive secretion. Instead, we found these nonfunctional mutant HNE proteins accumulated in the cytoplasm.
By analyzing the functional consequences, we found MT-HNE protein accumulation to be associated with an increased sensitivity to apoptosis. These data are consistent with recent findings indicating increased apoptosis in bone marrow cells from patients with congenital neutropenia25,26 or in mutant HNE expressing HL-60 cells.24 Interestingly, blockage of protein transport resulting in cytoplasmic accumulation associated with the induction of apoptosis is a process that has been observed after induction of the unfolded protein response (UPR).27 This pathway is induced by misfolded or unfolded proteins and is involved in several human diseases, among them Alzheimer disease and osteogenesis imperfecta.28,29 In view of this stress response, we analyzed expression of the master chaperone of the ER, the Hsp70 family member BiP, whose expression is specifically induced upon UPR activation.22 Consistent with this hypothesis, induction of apoptosis by MT-HNE proteins was associated with the upregulation of BiP, suggesting that accumulation of folding-incompetent HNE proteins can activate UPR signaling. Furthermore, induction of the ER stress response by tunicamycin resulted in a subcellular WT-HNE distribution resembling that of mutant HNE proteins.
Cytoplasmic accumulation of WT-HNE in cells after ER stress induction. Subcellular localization of WT-HNE protein in HeLa cells with or without ER stress induction by tunicamycin (Tm). Cells were labeled with anti-HNE (green) and anticalnexin (ER marker; red) or antigiantin (Golgi marker; red). TO-PRO-3 was used to counterstain the nuclei (blue). Yellow represents colocalization of HNE protein with the organelle-specific markers.
Cytoplasmic accumulation of WT-HNE in cells after ER stress induction. Subcellular localization of WT-HNE protein in HeLa cells with or without ER stress induction by tunicamycin (Tm). Cells were labeled with anti-HNE (green) and anticalnexin (ER marker; red) or antigiantin (Golgi marker; red). TO-PRO-3 was used to counterstain the nuclei (blue). Yellow represents colocalization of HNE protein with the organelle-specific markers.
UPR activation aims to reduce levels of misfolded proteins by enhancing their folding to the native state and promoting transit to the distal secretory pathway.30 The cell death pathway is induced upon prolonged UPR activation only if protein accumulation cannot be prevented. Based on this mechanism, we propose that all mutations initially activate UPR-responding proteins, even those (S97L, R191Q) that show a pattern of intracellular processing and protein function comparable to WT-HNE. These distinct mutant proteins will successfully be folded and secreted upon UPR-mediated elevation of the folding capacity by ER chaperones. In contrast, the other mutations, despite enhanced ER chaperone synthesis, will not be properly folded. These unfolded proteins accumulate and induce prolonged UPR activation with initiation of the apoptotic pathway. The identification of the UPR may allow the development of specific therapeutic interventions. This avenue has already been opened up for different protein-misfolding diseases.31 Small molecules act as chemical or pharmacologic chaperones by binding to the native state of these mutant proteins, thereby rescuing them from misfolding and intracellular mistrafficking.
In summary, our data suggest a new mechanism by which ELA2 mutations may impair myeloid precursor cell differentiation, resulting in chronic neutropenia. ELA2 mutations may cause incorrect folding of the mutant proteins that will consequently remain in the cytoplasm and thus induce UPR-associated programmed cell death. Given the peak of HNE synthesis at the promyelocyte stage,11,32 our experimental data may explain the maturation arrest of promyelocytes associated with congenital neutropenia.
Prepublished online as Blood First Edition Paper, March 21, 2006; DOI 10.1182/blood-2005-11-4689.
Supported by the Deutsche Forschungsgemeinschaft (grant DFG-KFO 110/1-1); the Elterverein zur Unterstützung der Behandlung krebskranker Kinder Hannover e.V. (C.B.); the Dieter-Schlag-Stiftung (C.B.); and Bundesministerium für Bildung und Forschung (BMBF) Network Congenital Bone Marrow Failure Syndromes (grant 01GM0307).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank all the patients who participated in this study for their support. We thank R. Bauerfeind, head of the MHH core facility for confocal laser microscopy, for his support and helpful discussions. We thank I. Fernandez-Munoz and A. Vietmeyer for their excellent technical assistance and G. Teicke and W. Hofmann for editorial assistance.