Abstract
ADAMTS13 biosynthesis appeared to occur mainly in hepatic stellate cells, but detection of ADAMTS13 mRNA in many other tissues suggests that vascular endothelium may also produce ADAMTS13. We showed that ADAMTS13 mRNA and protein were detectable in human umbilical vein endothelial cells, aortic endothelial cells, and endothelium-derived cell line (ECV304). ADAMTS13 in cell lysate or serum-free conditioned medium cleaved von Willebrand factor (VWF) specifically. ADAMTS13 and VWF were localized to the distinct compartments of endothelial cells. Moreover, ADAMTS13 was preferentially sorted into apical domain of ECV304 and Madin-Darby canine kidney (MDCK) cells. Apical sorting of ADAMTS13 depended on the CUB domains and their association with lipid rafts. A mutation in the second CUB domain of ADAMTS13 (4143-4144insA), naturally occurring in patients with inherited thrombotic thrombocytopenic purpura, resulted in a significant reduction of ADAMTS13 secretion and a reversal of its polarity in MDCK cells. These data demonstrated that ADAMTS13 is synthesized and secreted from endothelial cells; the apically secreted ADAMTS13 from endothelial cells may contribute significantly to plasma ADAMTS13 proteases. The data also suggest a critical role of the CUB domains and a novel cargo-selective mechanism for apical sorting of a soluble ADAMTS protease in polarized cells.
Introduction
ADAMTS13, a member of adisintegrin and metalloprotease with thrombospondin type 1 repeats (ADAMTS) family, limits platelet-rich thrombi by cleaving von Willebrand factor (VWF) between the Tyr1605 and Met1606 residues. VWF is synthesized in vascular endothelial cells and megakaryocytes or platelets, and normally stored in Weibel-Palade bodies1 of endothelial cells or the α-granules of platelets.2,3 Upon stimulation, VWF is secreted as unusually large (UL) multimers that may remain associated with the endothelial cell surface.4-6 These UL-VWF multimers may be the preferred substrate for ADAMTS13.4,5 Inability to cleave these UL-VWF multimers to smaller sizes on the endothelial cell surface may result in an accumulation of UL-VWF, leading to enhanced platelet adhesion and aggregation, and thrombotic thrombocytopenic purpura (TTP),7,8 characterized by profound thrombocytopenia, microangiopathic hemolytic anemia, and organ failure.7,8
ADAMTS13 appears to be synthesized in liver,9-11 particularly in hepatic stellate cells,12-14 which reside in the space of Disse adjacent to the hepatocytes. However, the contribution of hepatic stellate cells in regulation of plasma ADAMTS13 levels remains to be determined. We have recently shown that despite a dramatic increase in ADAMTS13 expression in hepatic stellate cells upon activation by administration of carbon tetrachloride, plasma ADAMTS13 activity is not proportionally increased in rats.14 In addition, patients with liver cirrhosis or with significant proliferation and activation of hepatic stellate cells exhibit variable or reduced levels of plasma ADAMTS13 activity.15 Therefore, plasma ADAMTS13 protease may be derived from other sources as well.
Recent reports have shown that ADAMTS13 is synthesized and released from human megakaryocytes and platelets.16,17 ADAMTS13 mRNA is detected in almost every organ tissue by reverse-transcriptase–polymerase chain reaction (RT-PCR),9-11,18,19 suggesting that the vascular endothelium may also be the source of plasma ADAMTS13. Considering the enormous surface area of vascular endothelial beds, a small fraction of endothelial cells producing ADAMTS13 at any given time may contribute significantly to plasma levels of ADAMTS13 activity. Consequently, any perturbation of endothelial cell function or ADAMTS13-VWF interactions at the site of their syntheses by inflammatory cytokines or drugs may play an important role in the pathogenesis of TTP20,21 and perhaps other thrombotic complications.22
ADAMTS13 consists of signal peptide, propeptide, and metalloprotease domains, followed by disintegrin domain, first thrombospondin type 1 repeat (TSP1), Cys-rich domain, and spacer domain. The carboxyl terminus of ADAMTS13 has 7 additional TSP1 repeats and 2 CUB (C1r/C1s, urinary EGF, and bone morphogenetic protein) domains.9-11 The proximal carboxyl terminus of ADAMTS13 appears to determine substrate recognition and specificity, whereas the middle and distal carboxyl termini may be dispensable at least under static conditions.23-26 However, the CUB domains of ADAMTS13 may be critical for docking of ADAMTS13 to VWF under flow.27 Of more interest, mutations in the CUB domains result in severe deficiency of plasma ADAMTS13 activity.9,28,29 In vitro studies have shown that some of the mutants exhibit secretion defect.30,31 For example, one of the mutations occurs in the second CUB domain, inserts an “A” in between the nucleotides 4143 and 4144 of ADAMTS13 (4143-4144insA), deletes the second CUB domain, and appends REQPG after S1381.31 Such a mutant exhibits a significant impairment in secretion while retaining a comparable proteolytic activity of the wild-type ADAMTS13.31 These data suggest that in addition to ligand binding, the CUB domains may play a critical role in biosynthesis and secretion of ADAMTS13 protease.
Here we report the biosynthesis and intracellular sorting of ADAMTS13 protease in human vascular endothelial cells and canine kidney tubular epithelial cells. The data demonstrate that ADAMTS13 is synthesized and secreted from human vascular endothelial cells in culture and in situ; the apically secreted ADAMTS13 from endothelial cells may contribute significantly to plasma ADAMTS13 protease. The data also suggest a critical role of the CUB domains and a novel cargo-selective mechanism for apical sorting of a soluble ADAMTS protease in polarized cells.
Materials and methods
Plasmid constructs
Plasmids containing full-length (FL) and CUB domain–deleted ADAMTS13 (delCUB) were generated as described previously.10,23,26 Construct 4143-4144insA that deleted the second CUB domain and appended REQPG after S138131 was created by PCR with a forwarding primer (5′-GGCACGGACTGCACTGAGTTTTCCTGGGG-3′) and a reverse primer (5′-GCCTGGCTGCTCTCTTGACTCCCAGTAGAG-3′) and pcDNA3.1-FL-V5-His as a template. A chimeric protein consisting of a prothrombin signal peptide (SP) and an enhanced green fluorescent protein (GFP) with or without 2 CUB domains of ADAMTS13 was created by PCR using pS-GFP32 and pcDNA3.1-FL-V5-His23 as templates. The fusion products were purified from an agarose gel and ligated into pcDNA 3.1 V5-His TOPO (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendation. This cloning strategy appends the following vector-encoded amino acid sequence onto the ADAMTS13 sequence: a linker region (KGNSADIQHSGGRSSLEGPRFE), the V5 epitope (GKPIPNPLLGLDST), a tripeptide (RTG), and a 6xHis-tag as described.10,23,26 All PCR reactions were performed with Turbo Hotstart Pfu DNA polymerase (Stratagene, La Jolla, CA) in the absence or presence of 7.5% dimethyl sulfate (DMSO). The accuracy of all constructions was confirmed by DNA sequencing.
Cell culture
Primary human umbilical vein endothelial cells (HUVECs) were isolated from fresh umbilical cord veins by digestion with collagenase (Sigma, St Louis, MO).33 Primary human aortic endothelial cells (HAECs), human endothelium-derived cell line (ECV304),34 and human hepatic stellate cell line (LX-2)35 were obtained from Drs Peter Davies (University of Pennsylvania, Philadelphia, PA), Harry Ischiropoulos (The Children's Hospital of Philadelphia, Philadelphia, PA), and Scott Friedman (Cornell University, New York, NY), respectively. Madin-Darby canine kidney II (MDCK II) and African green monkey kidney (COS7) cells were from American Type Culture Collection (ATCC, Manassas, VA). HAECs and HUVECs were cultured in EBM-2 medium containing 2% fetal bovine serum (Cambrex Bio Science, Walkersville, MD). ECV304, LX-2, and MDCK II cells were cultured in Dulbecco modified Eagle medium (DMEM; Invitrogen) in the presence of 10% FetalPlex (Genimi BioProducts, Brier, WA).
RT-PCR
Total RNAs were extracted and purified with TRIzol reagent (Invitrogen) from cells according to the manufacturer's recommendations. RT-PCR was performed with 0.3 to 0.5 μg total RNA and specific pairs of primers that amplify the signal peptide, metalloprotease domain, spacer domain, and second CUB domain of ADAMTS13 (Table 1) with SuperScript RT-PCR reagent (Invitrogen). Each pair of primers spanned at least one intron of the ADAMTS13 gene, therefore eliminating the possibility of amplifying contaminated genomic DNA. The SuperScript RT-PCR reagent was supplemented by platinum Tag polymerase and 6% DMSO in the reaction. The amplified PCR products were analyzed by electrophoresis on 1% agarose gel and visualized by staining with ethidium bromide. The PCR products were cloned into pCR2.1 TOPO (Invitrogen) and sequenced to confirm that they were the fragements of the ADAMTS13 gene.
Stable transfection
Western blotting
The cell lysates or conditioned media (approximately 30-fold concentrated) were separated on 8% Tris-glycine SDS-gel under denaturing and reducing conditions. The proteins were transferred onto nitrocellulose membranes (Bio-Rad, Hercules, CA), blocked with 2.5% nonfat milk in 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.05% Tween 20 (TBST) at room temperature for 30 minutes, then incubated overnight with rabbit anti–ADAMTS13 IgG (2 μg/mL; kindly provided by Dr Fritz Scheiflinger at Baxter BioSciences, Orth, Austria) or mouse anti-V5 antibody (1:5000; Invitrogen) or mouse anti–GFP IgG (1:10 000; Berkeley Antibody, Richmond, CA) in TBST containing 2.5% nonfat milk. The membrane was washed 3 times with TBST and incubated for 2 hours with peroxidase-conjugated anti–rabbit IgG (DAKO, Carpinteria, CA) or anti–mouse IgG (1:5000; DAKO). The bound secondary antibody was detected by SuperSignal chemiluminescent ECL system (Pierce, Rockford, IL).23 For quantification, the luminograms were scanned, and the relative amount of proteins secreted into the apical and basolateral domains of the cells was determined by densitometry using NIH ImageJ (Bethesda, MD), and results were expressed as the percentage of apically secreted protein of the total.
Immunofluorescent microscopy
Cells grown on a chamber slide or cryosections of umbilical cord veins were washed 3 times with PBS and fixed with ethanol–acetic acid (9:1, vol/vol) at –20°C for 10 minutes. The cells or tissue sections were incubated with rabbit anti–ADAMTS13 IgG (20 g/mL) or mouse anti–VWF IgG (1:200) (DAKO) in PBS with 0.5% bovine serum albumin (BSA) at 4°C overnight. After being washed with PBS 3 times, the cells or tissue sections were incubated with Cy3-conjugated goat anti–rabbit IgG (1:100) or Cy2-conjugated anti–mouse IgG (1:100) (Jackson ImmunoResearch Lab, West Grove, PA). The nuclei were stained with 4′, 6-diamidino-2-phenylindole (DAPI) in mounting medium (Vector Laboratories, Burlingame, CA).23 The cells were examined under either the Nikon (Melville, NY) inversed fluorescent microscope (or confocal microscope). The digital images were processed by MetaVue software version 6.2 (UI, Downingtown, PA).
ADAMTS13 activity
Metabolic labeling
Cells grown on 10-cm dishes were washed with PBS and incubated with methionine- and cysteine-free DMEM for 30 minutes at 37°C to deplete intracellular pool of methionine and cysteine. The cells were then labeled for 2 hours with 100 μCi (3.7 MBq) 35S-methionine/cysteine in serum-free Opti-MEM medium. The cells and conditioned medium were collected at 0 and 24 hours after addition of the cell-type–specific medium (serum-free). The cells were lysed with 25 mM MES, pH 7.5, 50 mM NaCl, 0.5% SDS, 1% NP40, and 0.1% protease inhibitor cocktail (Sigma), and conditioned media were concentrated with centricon-10 (Millipore, Billerica, MA). The newly synthesized and 35S-methionine/cysteine-labeled ADAMTS13 protein was precipitated with rabbit anti–ADAMTS13 IgG (1:1000) and protein A-Sepharose 4B (BD Biosciences, San Jose, CA). The eluate was electrophoretically separated on 10% SDS–polyacrylamide gel that was then fixed. After being incubated with Amplify solution (BD Biosciences) for 30 minutes,40 the gel was dried on filter paper and exposed to X-ray film at –80°C for 3 hours.
Analysis of the polarity of ADAMTS13 secretion
The cells were grown on 6-well Transwell filter units (pore size, 0.4 μm) as illustrated in Figure 3D (Fisher Scientific, Pittsburgh, PA) in DMEM containing 10% FetalPlex. After cells reached confluency and formed the tight monolayer as determined by transmembrane resistance with an Ohmmeter (World Precision Instruments, Sarasota, FL) as described previously,32,40 the culture media in both apical and basolateral domains of the cells were removed and replaced with 2 mL fresh Opti-MEM serum-free medium or DMEM containing 10% FetalPlex or 2 mL fresh serum-free Opti-MEM. After 24 hours to 48 hours of culture, the conditioned media were collected.32,40 The V5-His–tagged recombinant full-length ADAMTS13 or the mutants in the conditioned media were concentrated either by 50 μL TALON Co2+-agarose (BD Biosciences) in the presence of 50 mM Tris-HCl, pH 8.0, or by filtration with Centricon-10 as previously described.23,26
Raft association
The specific membrane domains (ie, lipid rafts) were isolated as described previously.32,40 Briefly, MDCK cells expressing V5-His–tagged recombinant ADAMTS13 or various mutants grown on 10-cm dishes were lysed with 0.8 mL MBS (20 mM MES, pH 6.5, 150 mM NaCl) containing 1% Triton X-100 and 0.1% protease inhibitor cocktail (Sigma) on ice with a gentle agitation for 30 minutes. The cell lysate (1 mL) was well mixed with 1 mL of 80% of sucrose in MBS buffer, pH 6.5, and transferred to 12-mL ultracentrifugation tube. On top of the 40% sucrose–cell lysate mixture, 4 mL of 30% sucrose in MBS buffer and subsequent 6 mL of 5% sucrose in MBS buffer were gently loaded. After being centrifuged at 260 576g with SW41 rotor (Beckman, Fullerton, CA) for 16 hours, the protein was fractionated by removing 1.2 mL each from the top to the bottom, and precipitated with 10% trichloroacetic acid on ice for 30 minutes. The pellets were redissolved with 0.2 N sodium hydroxide. Samples (50 μL) of trichloroacetic acid–concentrated fractions were analyzed by SDS–polyacrylamide gel electrophoresis (PAGE) and immunoblotting with either monoclonal anti–V5 IgG (1:5000) or monoclonal anti–caveolin-1 IgG (1:2000) (BD Biosciences).
Cholesterol depletion
Monolayer MDCK cells expressing full-length ADAMTS13-V5-His on a 6-well Transwell filter unit were treated with lovastatin (40 μg/mL) and methyl-β-cyclodextrin (250 μg/mL) in DMEM medium containing 10% FetalPlex for 48 hours. The transmembrane resistance was measured prior to and after the drug treatment to confirm the cell viability and integrity of the cell monolayer. The conditioned media were collected from the apical and basolateral domains of the cells, and concentrated by Talon Co2+-resin (BD Biosciences). ADAMTS13 was determined by anti–V5 IgG as described under “Western blotting.”
Results
ADAMTS13 is produced in human vascular endothelial cells
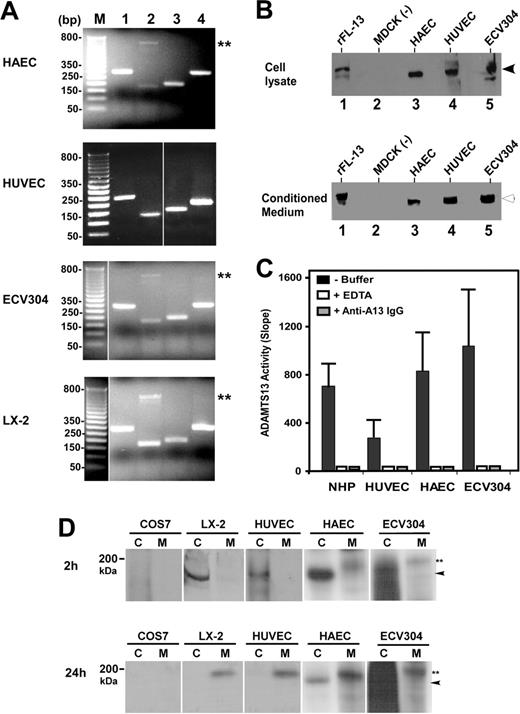
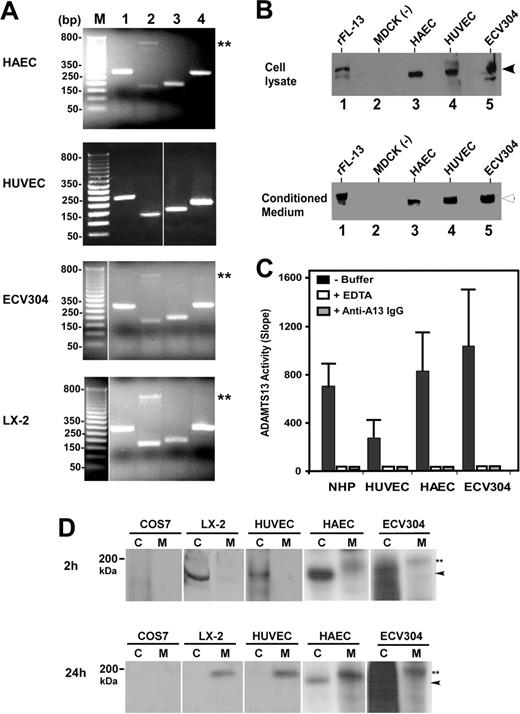
To determine whether ADAMTS13 is synthesized in vascular endothelial cells, we performed RT-PCR, Western blotting, proteolytic activity, and metabolic labeling assays on primary arterial, venous, and endothelium-derived cell lines. RT-PCR amplified the signal peptide, the metalloprotease domain, the spacer domain, and the second CUB domain of ADAMTS13 from total RNAs isolated from HUVECs, HAECs, and ECV304 cells (Table 1). The sizes of the amplified ADAMTS13 cDNA fragments from human endothelial cells were as expected and identical to those amplified from LX-2 cells (Figure 1A). LX-2 is derived from primary human hepatic stellate cells.35 Like primary human,12 mouse,13 and rat hepatic stellate cells,14 LX-2 produces a full-length ADAMTS13 protease that cleaves VWF specifically (data not shown). The control reactions without RNA template were all negative. All PCR products were sequenced and thus confirmed to be the fragments of human ADAMTS13 cDNA (data not shown).
When cell lysates or serum-free conditioned media of HUVECs, HAECs, and ECV304 were immunoblotted with rabbit anti–human ADAMTS13 IgG, a full-length ADAMTS13 protease was detected (Figure 1B). The molecular weights of the full-length ADAMTS13 in the cell lysates and conditioned media were approximately 175 kDa and approximately 190 kDa (Figure 1B), respectively, similar to that of recombinant ADAMTS13-V5-His expressed in stably transfected MDCK cells (Figure 1B). The ADAMTS13 protease secreted from all 3 human endothelial cells was capable of cleaving FRETS-VWF73 substrate, specifically (Figure 1C). The proteolytic cleavage of FRETS-VWF73 by ADAMTS13 in the conditioned media was completely inhibited by the addition of 10 mM EDTA or 40 μg/mL rabbit anti–human ADAMT13 IgG (Figure 1C). The specificity of the cleavage at the Tyr-Met bond was further verified by cleavage of GST-VWF73-H and plasma VWF substrate (data not shown).
To rule out the possibility that RT-PCR may be too sensitive to amplify a minute ADAMTS13 mRNA that is not translated into protein, and that the ADAMTS13 protein detected by Western blotting in the cell lysate and serum-free conditioned medium may be in fact derived from the previous culture medium containing 10% FetalPlex via endocytosis, we performed 35S-methanine/cysteine metabolic labeling on primary HUVECs, HAECs, and ECV304 cells. The newly synthesized ADAMTS13 was clearly detectable in the cell lysate (approximately 175 kDa) after 2 hours of labeling and in the conditioned medium (approximately 190 kDa) after 2 hours of labeling plus 24 hours of incubation in complete serum-free medium containing 25 μCi/mL (9.25 MBq) 35S-methionine/cysteine (Figure 1D). The similar bands were also detected in LX-2 cells stably expressing full-length ADAMTS13-V5-His (positive control), but not in COS7 cells (negative control) (Figure 1D). These data clearly support the hypothesis that ADAMTS13 protease is synthesized and secreted from human vascular endothelial cells.
Expression of ADAMTS13 in human vascular endothelial cells. (A) RT-PCR detects ADAMTS13 mRNA. The amplified cDNA fragments correspond to the signal peptide (lane 1), metalloprotease domain (lane 2), spacer domain (lane 3), and the second CUB domain (lane 4) of ADAMTS13. M indicates a 1-kb DNA maker. Double asterisks indicate nonspecific band. (B) Western blot detects full-length ADAMTS13 protein in both cell lysates and conditioned media of HAECs, HUVECs, and ECV304. MDCK cells transfected with (rFL-13) or without (–) ADAMTS13-V5-His were used as positive or negative control. The arrows indicate the intact full-length ADAMTS13 proteins. (C) ADAMTS13 protease in normal human plasma (NHP) or secreted from HUVECs, HAECs, and ECV304 cleaves FRETS-VWF73 specifically, which is completely inhibited by 10 mM EDTA (+ EDTA) and 40 μg/mL rabbit anti–ADAMTS13 IgG (+ anti–A13 IgG). The relative proteolytic activity (slope, fluorescent units/min) was shown as means ± SD from 3 independent experiments. (D) Metabolic labeling and detection of newly synthesized ADAMTS13 in the cell lysate (C) and conditioned medium (M) of HUVECs, HAECs, and ECV304 at 2 hours and 24 hours. The COS7 cells and LX-2 cells transfected with ADAMTS13-V5-His were used as negative and positive controls, respectively. The arrowheads indicate intracellular ADAMTS13 in cell lysate, whereas double asterisks indicate the secreted ADAMTS13 in the conditioned medium. The vertical white lines indicate areas of noncontiguous lanes assembled.
Expression of ADAMTS13 in human vascular endothelial cells. (A) RT-PCR detects ADAMTS13 mRNA. The amplified cDNA fragments correspond to the signal peptide (lane 1), metalloprotease domain (lane 2), spacer domain (lane 3), and the second CUB domain (lane 4) of ADAMTS13. M indicates a 1-kb DNA maker. Double asterisks indicate nonspecific band. (B) Western blot detects full-length ADAMTS13 protein in both cell lysates and conditioned media of HAECs, HUVECs, and ECV304. MDCK cells transfected with (rFL-13) or without (–) ADAMTS13-V5-His were used as positive or negative control. The arrows indicate the intact full-length ADAMTS13 proteins. (C) ADAMTS13 protease in normal human plasma (NHP) or secreted from HUVECs, HAECs, and ECV304 cleaves FRETS-VWF73 specifically, which is completely inhibited by 10 mM EDTA (+ EDTA) and 40 μg/mL rabbit anti–ADAMTS13 IgG (+ anti–A13 IgG). The relative proteolytic activity (slope, fluorescent units/min) was shown as means ± SD from 3 independent experiments. (D) Metabolic labeling and detection of newly synthesized ADAMTS13 in the cell lysate (C) and conditioned medium (M) of HUVECs, HAECs, and ECV304 at 2 hours and 24 hours. The COS7 cells and LX-2 cells transfected with ADAMTS13-V5-His were used as negative and positive controls, respectively. The arrowheads indicate intracellular ADAMTS13 in cell lysate, whereas double asterisks indicate the secreted ADAMTS13 in the conditioned medium. The vertical white lines indicate areas of noncontiguous lanes assembled.
Localization of ADAMTS13 and VWF in endothelial cells
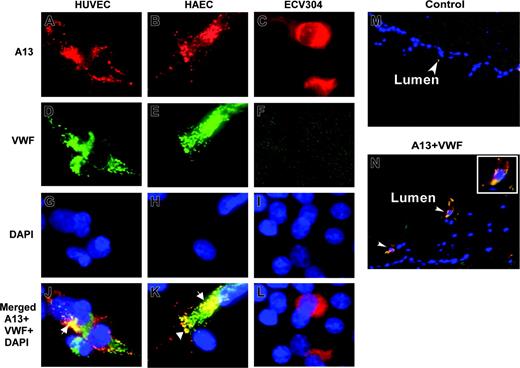
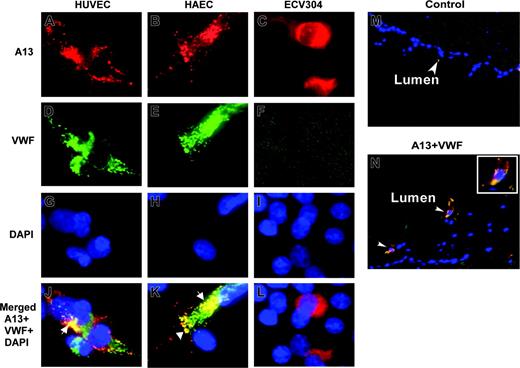
To determine whether ADAMTS13 and VWF are colocalized in endothelial cells, we performed immunofluorescent microscopy on HUVECs, HAECs, and ECV304 cells. ADAMTS13 and VWF appeared to be only partially overlapped, primarily in the perinuclear region (Figure 2J,K, white arrows). Little, if any, ADAMTS13 was detected in the Weibel-Palade bodies of HUVECs and HAECs, in which VWF is normally stored and abundantly stained (Figure 2J,K). ECV304 cells adopted epithelial-like phenotype and lose the ability to express VWF (Figure 2F). The data suggest that ADAMTS13 and VWF are localized to the distinct compartments in the endothelial cells.
ADAMTS13 antigen is detected in the endothelial cells of umbilical veins
To demonstrate whether ADAMTS13 is also produced in vivo, we performed immunohistochemistry on the cryosections of umbilical cord veins. ADAMTS13 antigen was positive in the endothelial cells of fresh umbilical veins that coexpressed VWF (Figure 2N). The control sections incubated with only secondary antibodies were negative (Figure 2M). This result suggests that ADAMTS13 may be synthesized in the vascular endothelial cells in vivo as well.
ADAMTS13 is sorted apically in polarized cells
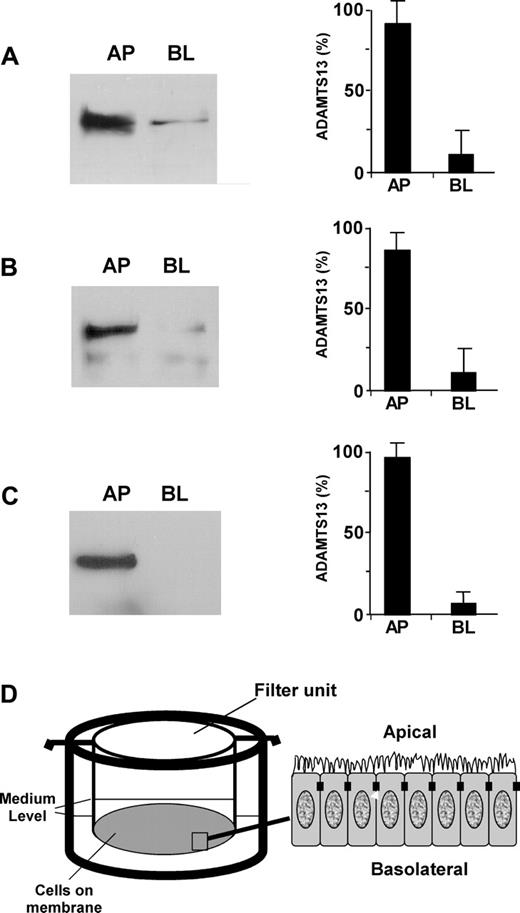
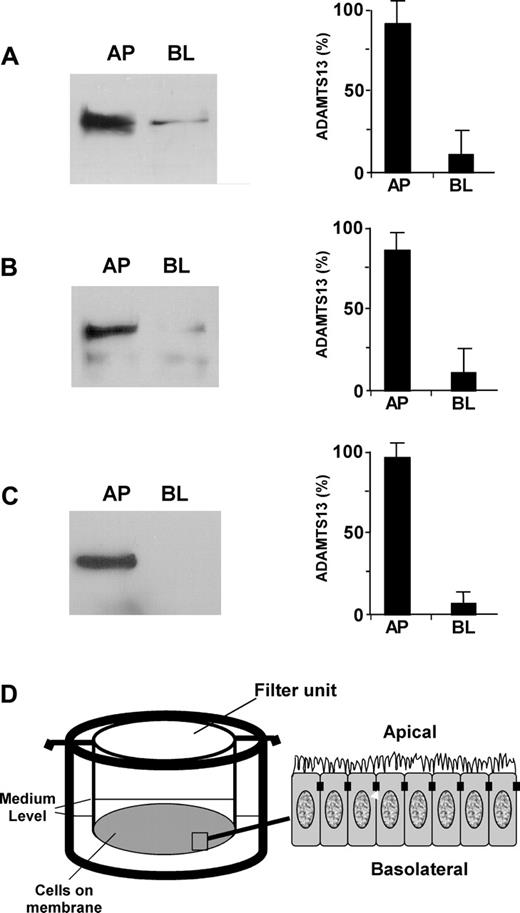
To determine whether ADAMTS13 produced in endothelial cells in fact gets into circulation and to understand how mutations in the ADAMTS13 gene affect ADAMTS13 biosynthesis and secretion, we assessed the polarity of ADAMTS13 in 2 well-characterized polarized cell lines (ECV304 and MDCK) grown on the Transwell filter units (Figure 3D). Approximately 85% of endogenous ADAMTS13 (Figure 3A) and approximately 80% of recombinant ADAMTS13-V5-His (Figure 3B) were sorted into the apical domain of ECV304. The sorting efficiency was even greater in MDCK cells stably expressing recombinant ADAMTS13-V5-His. Approximately 95% of ADAMTS13-V5-His was detected in the apical domain of MDCK cells (Figure 3C). These data suggest that ADAMTS13 may be secreted apically (ie, toward the lumen of the vessels in vivo).
Apical sorting of ADAMTS13 depends on the CUB domains
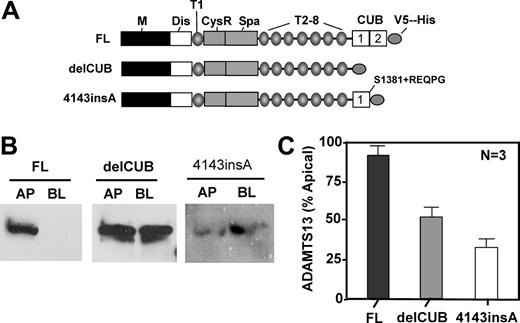
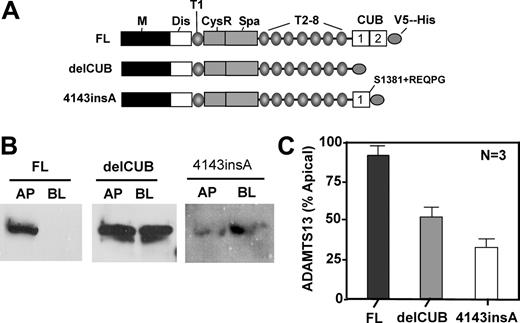
To determine the role of the CUB domains of ADAMTS13 in biosynthesis and secretion, we constructed ADAMTS13 mutants either by deleting the CUB domains (delCUB) or by inserting a nucleotide “A” between the nucleotides 4143 and 4144 in the second CUB domain (4143-4144insA) based on a naturally occurring mutation found in patients with hereditary TTP (Figure 4A). The latter mutant (4143-4144insA) truncates the second CUB domain and appends amino acid REQPG after S1381 (Figure 4A).31 When stably expressed in MDCK cells, construct delCUB was randomly sorted with 52% to apical domain and 48% to basolateral domain of MDCK cells (Figure 4B-C, delCUB), whereas the polarity of 4143-4144insA was surprisingly reversed with only 34% of the mutant protein being sorted into the apical domain (Figure 4B-C, 4143-4144insA). These data suggest that the CUB domains may be important for apical sorting of ADAMTS13 protease in polarized cells.
The CUB domains contain a signal that directs apical sorting
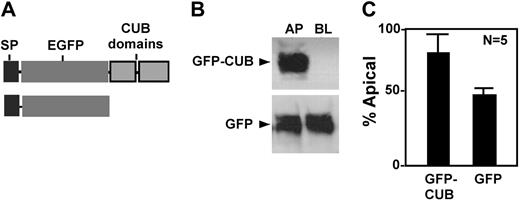
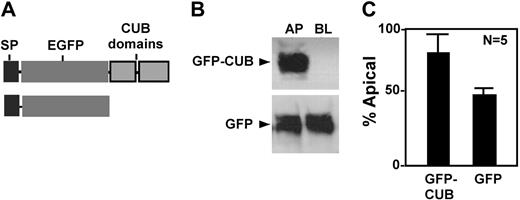
To determine whether the CUB domains contain any information that directs apical sorting, we constructed chimeric proteins consisting of a prothrombin signal peptide (SP) and enhanced green fluorescent protein (GFP) with or without CUB domains (Figure 5A). When stably transfected into MDCK cells, the chimeric protein GFP-CUB was predominantly secreted into the apical domain (Figure 5, GFP-CUB), whereas GFP alone was randomly secreted (Figure 5, GFP). These data confirm that the CUB domains of ADAMTS13 harbor an apical sorting signal that targets a heterogeneous protein into the apical domain of polarized cells.
Localization of ADAMTS13 in human endothelial cells by fluorescent microscopy. HUVECs (A,D,G,J), HAECs (B,E,H,K), and ECV304 (C,F,I,L) grown on chamber slides or cryosections of human umbilical veins (M-N) were fixed with ethanol–acetic acid (9:1). The cells or slides were incubated with rabbit anti–ADAMTS13 IgG (20 μg/mL) and mouse anti–VWF IgG (10 μg/mL), followed by incubation of the cells with Cy3-conjugated anti–rabbit IgG and Cy2-conjugated anti–mouse IgG (1:100). The nuclei were stained with 4′, 6′-diamidino-2-phenylindole (DAPI) in the mounting medium. The digital images (40×) were taken under a Nikon inverse fluorescent microscope. ADAMTS13 staining in red is shown in panels A-C. VWF staining in green is shown in panels D-F. Overlapping images for ADAMTS13, VWF, and DAPI staining were shown in panels J-L and N. The yellow staining (white arrows) indicates the areas of ADAMTS13 colocalized with VWF. Cells (not shown) or tissue sections (M) incubated with only secondary antibody were negative.
Localization of ADAMTS13 in human endothelial cells by fluorescent microscopy. HUVECs (A,D,G,J), HAECs (B,E,H,K), and ECV304 (C,F,I,L) grown on chamber slides or cryosections of human umbilical veins (M-N) were fixed with ethanol–acetic acid (9:1). The cells or slides were incubated with rabbit anti–ADAMTS13 IgG (20 μg/mL) and mouse anti–VWF IgG (10 μg/mL), followed by incubation of the cells with Cy3-conjugated anti–rabbit IgG and Cy2-conjugated anti–mouse IgG (1:100). The nuclei were stained with 4′, 6′-diamidino-2-phenylindole (DAPI) in the mounting medium. The digital images (40×) were taken under a Nikon inverse fluorescent microscope. ADAMTS13 staining in red is shown in panels A-C. VWF staining in green is shown in panels D-F. Overlapping images for ADAMTS13, VWF, and DAPI staining were shown in panels J-L and N. The yellow staining (white arrows) indicates the areas of ADAMTS13 colocalized with VWF. Cells (not shown) or tissue sections (M) incubated with only secondary antibody were negative.
ADAMTS13 is associated with rafts via the CUB domains
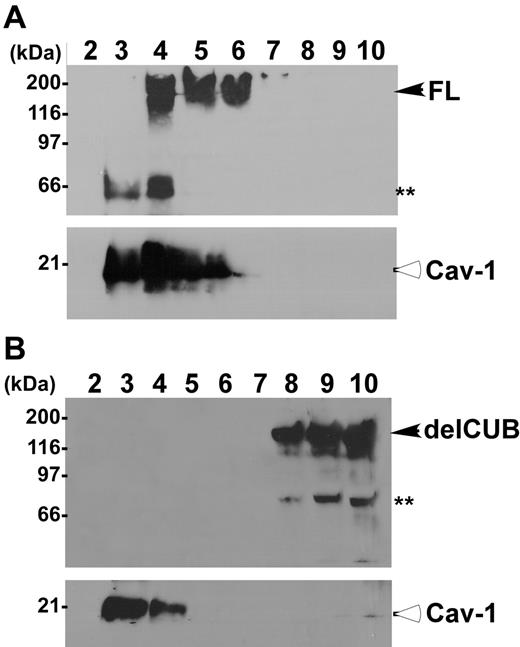
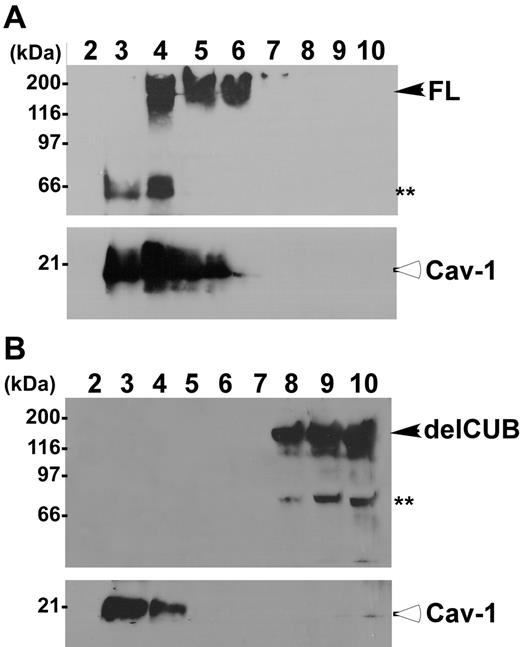
To determine the platform that ADAMTS13 uses for its apical sorting, we determined the association of ADAMTS13 and mutants with rafts. Rafts are operationally defined as the membrane microdomains that are resistant to 1% Triton X-100 at 4°C. They contain high concentrations of glycosphingolipids and cholesterol, and therefore float at the top fractions in sucrose gradients.40-44 The full-length ADAMTS13 was predominantly recovered in the top fractions of sucrose gradients (Figure 6, lanes 4-6) in which caveolin-1 (Cav-1), a marker specific for lipid rafts, was also present at a high concentration (Figure 6). Construct delCUB was almost exclusively recovered in the bottom fractions of the sucrose gradients (Figure 6, lanes 8-10). These data suggest that apical sorting of ADAMTS13 may depend on its association with lipid rafts through the CUB domains.
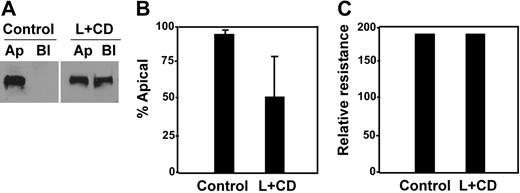
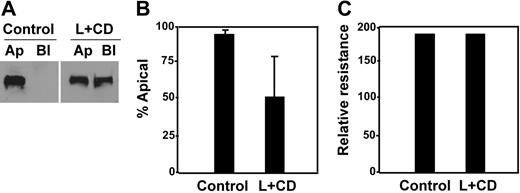
This hypothesis was further supported by a cholesterol-depletion experiment, in which MDCK cells expressing full-length ADAMTS13-V5-His were treated with lovastatin and methyl-β-cyclodextrin.44,45 Upon cholesterol depletion, only approximately 56% of ADAMTS13-V5-His was detected in the apical domain of the cells (Figure 7A-B, L + CD), in contrast to more than 95% of ADAMTS13-V5-His sorted into the apical domain in untreated cells (Figure 7A-B, control). Of note, although the total amount of ADAMTS13 secreted into the conditioned medium was reduced by about 50% (data not shown), such a treatment with a combination of lovastatin and methyl-β-cyclodextrin did not appear to affect the cell viability and monolayer integrity. The cell viability was determined by trypan blue staining (data not shown), whereas the monolayer integrity was determined by almost identical transepithelial resistance prior to (control) and after (L + CD) the drug treatment (Figure 7C).
Discussion
The results presented here demonstrate that endothelial cells produce ADAMTS13 in culture and in vivo. First, the cDNA fragments corresponding to the signal peptide, the metalloprotease domain, the spacer domain, and the second CUB domain of ADAMTS13 were amplified by RT-PCR (Figure 1A) and sequenced to confirm accuracy. Second, full-length ADAMTS13 was detectable in the cell lysates and the conditioned media of HUVECs, HAECs, and ECV304 by Western blotting (Figure 1B) and by metabolic labeling with 35S-methionine/cysteine (Figure 2D). The molecular weight of ADAMTS13 detected in cell lysate was approximately 10 to 15 kDa smaller than in conditioned medium (Figure 1B,D). The difference in molecular masses between the intracellular and the secreted proteins may result from the complexity of the N-glycosylation, additions of possible O-glycosylation, or other types of posttranslational modifications, such as C-mannosylation (WXXW) in the thrombospondin type 1 repeats.10,36 Third, ADAMTS13 in the conditioned media of HUVECs, HAECs, and ECV304 cells cleaved FRETS-VWF73 (Figure 1C), plasma VWF, and GST-VWF73-H (data not shown). The cleavage of FRETS-VWF73 by the cell lysates and conditioned media was completely inhibited by 10 mM EDTA or rabbit anti–ADAMTS13 IgG (Figure 1C), suggesting the specific cleavage of the substrate by ADAMTS13, not other proteases in the cell lysate and conditioned medium.
ADAMTS13 is sorted apically in ECV304 and MDCK cells. ADAMTS13 proteases secreted into the apical (AP) and basolateral (BL) domains of untransfected ECV304 (A), stably transfected ECV304 (B), and stably transfected MDCK (C) cells with pcDNA3.1-ADAMTS13-V5-His that were grown on 6-well Transwell filter units (D) were determined by Western blotting with rabbit anti–ADAMTS13 IgG (2 μg/mL) or mouse anti–V5 IgG (2 μg/mL) and SuperSignal ECL reagents. The relative amount of ADAMTS13 in the apical and basolateral domains was determined by densitometry with NIH ImageJ software. The entries represent means ± SD from 3 independent experiments.
ADAMTS13 is sorted apically in ECV304 and MDCK cells. ADAMTS13 proteases secreted into the apical (AP) and basolateral (BL) domains of untransfected ECV304 (A), stably transfected ECV304 (B), and stably transfected MDCK (C) cells with pcDNA3.1-ADAMTS13-V5-His that were grown on 6-well Transwell filter units (D) were determined by Western blotting with rabbit anti–ADAMTS13 IgG (2 μg/mL) or mouse anti–V5 IgG (2 μg/mL) and SuperSignal ECL reagents. The relative amount of ADAMTS13 in the apical and basolateral domains was determined by densitometry with NIH ImageJ software. The entries represent means ± SD from 3 independent experiments.
ADAMTS13 and VWF appear to be partially colocalized to the endoplasmic reticulum, Golgi apparatus, and certain secretory granules, consistent with the localization of the secretory proteins23 (Figure 2). However, little, if any, ADAMTS13 is detected in the Weibel-Palade bodies of HUVECs and HAECs, in which VWF is strongly positive (Figure 2). The localization of ADAMTS13 and VWF was further verified by immunofluorescent staining with monoclonal antibody against ADAMTS13-disintegrin domain (from Dr David Motto, University of Michigan, Ann Arbor, MI) and visualization of the staining pattern under confocal microscope in HUVECs and HAECs (data not shown). The data suggest that ADAMTS13 and VWF are targeted into the distinct compartments in endothelial cells after they exit from the endoplasmic reticulum, Golgi apparatus, and trans-Golgi network.
However, the colocalization of ADAMTS13 with VWF to the endoplasmic reticulum may be an initial step to regulate the sizes of VWF multimers. This hypothesis is supported by the observation published previously, demonstrating the cleavage of pre-pro-VWF by newly synthesized ADAMTS13 in Hela cells in which both ADAMTS13 and VWF cDNAs were cotransfected. The cleavage of pre-pro-VWF appears to occur within the endoplasmic reticulum and requires the spacer domain of ADAMTS13 protease.36
Not only did the endothelial cells in culture synthesize and secrete active ADAMTS13, but also the endothelial cells in situ may produce the protease. Immunohistochemistry showed that ADAMTS13 antigen was detectable in the endothelial cells on the cryosections of human umbilical veins that coexpressed VWF. However, only a small fraction of the endothelial cells was positive for both ADAMTS13 and VWF antigens (Figure 2N). The low positive rate of ADAMTS13 and VWF antigens indicates that ADAMTS13 and VWF expression in the endothelial cells may be tightly regulated in vivo. Together, our data clearly demonstrated that ADAMTS13 is synthesized and secreted in vascular endothelial cells in culture and most likely in vivo.
Deletion of the CUB domains or a mutation in the second CUB domain of ADAMTS13 abolishes or reverses apical sorting. MDCK cells were stably transfected with full-length ADAMTS13 (FL), CUB domain–deleted ADAMTS13 (delCUB), and a mutant ADAMTS13 (4143-4144insA) that deletes the second CUB domain and appends amino acid REQPG after S1381 as shown in panel A. The cells were grown on the Transwell filter units, and the conditioned media were collected from apical and basolateral domains. ADAMTS13 and mutants secreted were determined in the equal volume of the concentrated conditioned media by Western blotting with anti–V5 IgG (panel B). The relative amount of protein secreted into the apical domain of MDCK cells was quantified by densitometry of the signals on X-ray film with ImageJ software and is shown in panel C. The entries here represent the means ± SD from 3 independent experiments. M indicates metalloprotease domain; Dis, disintegrin domain; T1 to T8, first to eighth thrombospondin type 1 repeat; CysR, cysteine-rich domain; Spa, spacer domain; and CUB, complement C1r/C1s, urinary EGF, and bone morphogenetic protein domain.
Deletion of the CUB domains or a mutation in the second CUB domain of ADAMTS13 abolishes or reverses apical sorting. MDCK cells were stably transfected with full-length ADAMTS13 (FL), CUB domain–deleted ADAMTS13 (delCUB), and a mutant ADAMTS13 (4143-4144insA) that deletes the second CUB domain and appends amino acid REQPG after S1381 as shown in panel A. The cells were grown on the Transwell filter units, and the conditioned media were collected from apical and basolateral domains. ADAMTS13 and mutants secreted were determined in the equal volume of the concentrated conditioned media by Western blotting with anti–V5 IgG (panel B). The relative amount of protein secreted into the apical domain of MDCK cells was quantified by densitometry of the signals on X-ray film with ImageJ software and is shown in panel C. The entries here represent the means ± SD from 3 independent experiments. M indicates metalloprotease domain; Dis, disintegrin domain; T1 to T8, first to eighth thrombospondin type 1 repeat; CysR, cysteine-rich domain; Spa, spacer domain; and CUB, complement C1r/C1s, urinary EGF, and bone morphogenetic protein domain.
Apical sorting of chimeric GFP-CUB domains in MDCK cells. The conditioned media were collected from MDCK cells stably expressing chimeric GFP-CUB-V5-His domains (GFP-CUB) and GFP alone (GFP). The constructs are shown in panel A. The proteins secreted into the apical and basolateral domains were determined by mouse anti–V5 IgG for V5-His tagged construct and by mouse anti–GFP IgG (B). The signals on the luminogram were scanned and quantified by NIH ImageJ software. The relative amount (means ± SD, n = 5) of each protein in the conditioned medium collected from the apical domain of the cells was expressed in panel C.
Apical sorting of chimeric GFP-CUB domains in MDCK cells. The conditioned media were collected from MDCK cells stably expressing chimeric GFP-CUB-V5-His domains (GFP-CUB) and GFP alone (GFP). The constructs are shown in panel A. The proteins secreted into the apical and basolateral domains were determined by mouse anti–V5 IgG for V5-His tagged construct and by mouse anti–GFP IgG (B). The signals on the luminogram were scanned and quantified by NIH ImageJ software. The relative amount (means ± SD, n = 5) of each protein in the conditioned medium collected from the apical domain of the cells was expressed in panel C.
Although the polarity of ADAMTS13 was not directly determined in primary endothelial cells due to their inability to form a tight monolayer on Transwell filter units (Figure 3D), the data from both ECV304 (Figure 3A and 3B)34 and MDCK (Figure 3C) indicate that ADAMTS13 protease may be sorted (or secreted) apically (ie, toward the lumen of the vessels or other tubular structures in vivo).
The apical sorting of ADAMTS13 appeared to depend on the CUB domains, as the removal of CUB domains abolished apical sorting of ADAMTS13 in MDCK cells (Figure 4, delCUB). In addition, the mutant (4143-4144insA) that deleted the second CUB domain was preferentially sorted into the basolateral domain of MDCK cells (Figure 4, 4143-4144insA). The loss of polarity in construct delCUB or the reversal of polarity in construct 4143-4144insA was not due to protein misfolding after certain domains were deleted, because the secreted delCUB and 4143-4144insA mutants were proteolytically as active as the wild-type ADAMTS13 after the protein concentration of each protease was equally adjusted.23,26,31
The signal in the CUB domains of ADAMTS13 was further confirmed by its ability to target a heterogeneous and randomly secreted GFP reporter into the apical domain of MDCK cells (Figure 5). An apical sorting signal is also present in the CUB domains of cubilin, an intrinsic factor/vitamin B12 receptor. Cubilin contains 27 CUB domains and is localized to the apical pole of the epithelial cells in kidney and small intestine.46,47 Most of the CUB domains in cubilin are preferentially sorted into the apical domain of MDCK cells in a carbohydrate-dependent manner.48 However, the carbohydrates are not sufficient to target ADAMTS13 into the apical domain, as the delCUB is fully glycosylated, but sorted randomly. The exact sorting signal within the CUB domains has yet to be determined.
ADAMTS13, but not the mutant without CUB domains, is associated with rafts. MDCK cells stably expressing full-length ADAMTS13-V5-His (A, FL) and the CUB domain–deleted ADAMTS13 mutant (B, delCUB) were lysed with 1% Triton X-100 in MEB buffer, pH 6.5, at 4°C. The cell lysate was subjected to 5% to 40% of sucrose gradient centrifugation as described. Then, 1.2-mL fractions were collected from the top to the bottom. The ADAMTS13 in each faction was determined by Western blotting. Closed arrows indicate intact full-length ADAMTS13 and delCUB mutant. The double asterisks indicate proteolytically degraded ADAMTS13 products. Open arrows indicate caveolin-1 protein (Cav-1), a 21-kDa protein specific for rafts.
ADAMTS13, but not the mutant without CUB domains, is associated with rafts. MDCK cells stably expressing full-length ADAMTS13-V5-His (A, FL) and the CUB domain–deleted ADAMTS13 mutant (B, delCUB) were lysed with 1% Triton X-100 in MEB buffer, pH 6.5, at 4°C. The cell lysate was subjected to 5% to 40% of sucrose gradient centrifugation as described. Then, 1.2-mL fractions were collected from the top to the bottom. The ADAMTS13 in each faction was determined by Western blotting. Closed arrows indicate intact full-length ADAMTS13 and delCUB mutant. The double asterisks indicate proteolytically degraded ADAMTS13 products. Open arrows indicate caveolin-1 protein (Cav-1), a 21-kDa protein specific for rafts.
Full-length ADAMTS13 was found to be associated with lipid rafts in transfected MDCK cells, comigrating into the top fractions of sucrose gradients (Figure 6A), and such raft association appeared to be correlated with the polarity of ADAMTS13 protease. For example, the construct delCUB that was secreted randomly (Figure 4) was not recovered in the fractions containing lipid rafts (Figure 6B). Furthermore, depletion of intracellular cholesterol by treatment of MDCK cells with low dose of lovastatin and methyl-β-cyclodextrin that disrupts formation of the lipid rafts44,49 abolished apical sorting of ADAMTS13 (Figure 7).
Although the exact mechanism by which a soluble ADAMTS13 is associated with lipid rafts is not known, one could hypothesize that such association between the CUB domains of ADAMTS13 and rafts may be indirect, perhaps through the interactions of the CUB domains with other membrane-bound sorting receptors such as v-SNARE,50 VIP17/MAL,51 lectinlike molecule VIP36,41,52 and annexin XIIIb53,54 in the trans-Golgi network (TGN), on the vesicular carriers and/or at the apical cell surface.41,53 Removal of the CUB domains or depletion of intracellular cholesterol would break up the interactions among ADAMTS13, sorting receptors, and rafts, therefore reducing the efficacy of apical sorting. However, these interactions appear to be transient or may occur only intracellularly, as the ADAMTS13 protease was not detected in an association with plasma membrane in transfected COS7 cells by surface biotinylation.23
We conclude that ADAMTS13 is synthesized in human vascular endothelial cells in culture and most likely in vivo as well; ADAMTS13 protease is predominantly sorted apically in polarized cells; the apical sorting depends on the CUB domains and their possibly indirect association with lipid rafts; the mutations in the CUB domains may cause a “traffic jam,” thereby resulting in congenital deficiency of ADAMTS13 activity. The data suggest that the apically secreted ADAMTS13 in the vascular endothelia may contribute significantly to the plasma ADAMTS13 proteases. The data also imply a novel cargo-selective mechanism for apical sorting of a soluble ADAMTS protease in polarized cells.
Depletion of intracellular cholesterol abolished apical sorting of ADAMTS13. MDCK cells expressing full-length ADAMTS13 were treated without (control) or with a combination of lovastatin (L, 40 μg/mL) and methyl-β-cyclodextrin (CD, 250 μg/mL) for 48 hours. The conditioned media on both apical and basolateral domains were collected and analyzed by Western blotting with anti–V5 IgG and SuperSignal ECL system (A). The total volume of the conditioned medium loaded into the gel from the treated group (L + CD) was twice as much as that from the control. The percentage of ADAMTS13 detected in the apical domain of MDCK cells was quantified by densitometry with NIH ImageJ software (B). The entries represent mean ± SD (n = 3) in panel B. The integrity of the tight junction of MDCK cells prior to (control) and after drug treatment (L + CD) is shown in panel C.
Depletion of intracellular cholesterol abolished apical sorting of ADAMTS13. MDCK cells expressing full-length ADAMTS13 were treated without (control) or with a combination of lovastatin (L, 40 μg/mL) and methyl-β-cyclodextrin (CD, 250 μg/mL) for 48 hours. The conditioned media on both apical and basolateral domains were collected and analyzed by Western blotting with anti–V5 IgG and SuperSignal ECL system (A). The total volume of the conditioned medium loaded into the gel from the treated group (L + CD) was twice as much as that from the control. The percentage of ADAMTS13 detected in the apical domain of MDCK cells was quantified by densitometry with NIH ImageJ software (B). The entries represent mean ± SD (n = 3) in panel B. The integrity of the tight junction of MDCK cells prior to (control) and after drug treatment (L + CD) is shown in panel C.
Prepublished online as Blood First Edition Paper, April 4, 2006; DOI 10.1182/blood-2006-02-002139.
Supported by grants from the National Institutes of Health (HL079027) and the American Heart Association–Delaware and Pennsylvania (AHA0465532U).
Presented in abstract form at the 47th annual meeting of the American Society of Hematology, Atlanta, GA, December 10-13, 2005.55
D.S. and X.W.Z. performed experiments and analyzed data; M.N. established certain stable cell lines; and X.L.Z. designed research, analyzed the data, and wrote the article.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Dr Fritz Scheiflinger at Baxter Biotech, Vienna, Austria for providing us with rabbit anti–ADAMTS13 IgG; Drs David Motto and David Ginsburg at the University of Michigan, Ann Arbor, for mouse anti–ADAMTS13 IgG; Dr Peter Davies at University of Pennsylvania, Philadelphia, for HAECs; Dr Douglas Cines for providing us with fresh umbilical cords for isolation of HUVECs; and Dr Harry Ischiropoulos at the Department of Pediatrics, The Children's Hospital of Philadelphia, Pennsylvania for ECV304 cell line.