Abstract
Numerous megakaryocyte-specific genes contain signature Ets-binding sites in their regulatory regions. Fli-1 (friend leukemia integration 1), an Ets transcription factor, is required for the normal maturation of megakaryocytes and controls the expression of multiple megakaryocyte-specific genes. However, in Fli-1–/– mice, early megakaryopoiesis persists, and the expression of the early megakaryocyte-specific genes, αIIb and cMpl, is maintained, consistent with functional compensation by a related Ets factor(s). Here we identify the Ets protein GABPα (GA-binding protein α) as a regulator of early megakaryocyte-specific genes. Notably, GABPα preferentially occupies Ets elements of early megakaryocyte-specific genes in vitro and in vivo, whereas Fli-1 binds both early and late megakaryocyte-specific genes. Moreover, the ratio of GABPα/Fli-1 expression declines throughout megakaryocyte maturation. Consistent with this expression pattern, primary fetal liver–derived megakaryocytes from Fli-1–deficient murine embryos exhibit reduced expression of genes associated with late stages of maturation (glycoprotein [GP] Ibα, GPIX, and platelet factor 4 [PF4]), whereas GABPα-deficient megakaryocytes were mostly impaired in the expression of early megakaryocyte-specific genes (αIIb and cMpl). Finally, mechanistic experiments revealed that GABPα, like Fli-1, can impart transcriptional synergy between the hematopoietic transcription factor GATA-1 and its cofactor FOG-1 (friend of GATA-1). In concert, these data reveal disparate, but overlapping, functions of Ets transcription factors at distinct stages of megakaryocyte maturation.
Introduction
Megakaryocytes and erythroid cells derive from a bipotent megakaryocyte-erythrocyte progenitor.1,2 The relatedness of these 2 lineages is reflected in a highly overlapping set of expressed hematopoietic transcription factors, including GATA-1 and FOG-1 (friend of GATA-1),3 that are important for the normal development of both lineages.4-6 How megakaryocytes acquire a unique phenotype that is dramatically different from erythroid cells is poorly understood. A fruitful approach to study this problem has been the analysis of cis regulatory elements that mediate megakaryocyte-specific gene expression. Thus, numerous regulatory regions of megakaryocyte-specific genes were found to contain GATA sites in close proximity to Ets elements.7 GATA elements are bound by GATA-1 or GATA-2 that are coexpressed in megakaryocytes and that function in an overlapping manner during megakaryopoiesis.4,8,9 Both GATA factors interact with the cofactor FOG-1, which in turn can activate or repress the activity of GATA proteins depending on cell and promoter context.
Among the proteins that can bind Ets elements in megakaryocytic-specific gene promoters is Fli-1 (friend leukemia integration 1), a member of the pointed domain-containing subfamily of Ets proteins.10 Fli-1 plays a critical role in megakaryopoiesis. Forced expression of Fli-1 in the human erythroid leukemia cell line K562 stimulates the expression of megakaryocyte-specific genes.11 Moreover, Fli-1 associates with the αIIb, cMpl, and GPIX genes in vivo and can activate their promoters in transient transfection assays.12-16 Fli-1 knockout (Fli-1–/–) mice die at embryonic (E) day E11.5 due to intracranial hemorrhaging from defective vasculature.17,18 Cell cultures from day E11.5 Fli-1–/– fetal livers in the presence of thrombopoietin (TPO) reportedly contain increased numbers of megakaryocyte colony-forming units compared to wild type (WT) cultures. Fli-1–/– megakaryocytes display immature morphology,17,19 and expression levels of the early megakaryocyte-specific genes, αIIb and cMpl, appear normal to moderately decreased in Fli-1–/– embryos,17,19 whereas levels of the late gene GPIX are dramatically reduced.17 These findings indicate that Fli-1 is required for late-stage maturation of megakaryocytes and imply that other Ets proteins might substitute for Fli-1 during early megakaryopoiesis.
While Fli-1 likely functions in multiple ways,20 our previous work indicates that one mechanism of Fli-1 action involves the regulation of transcriptional synergy between GATA-1 and FOG-1 at the αIIb proximal promoter, which contains a GATA-binding site in close proximity to an Ets-binding site.12 The synergistic activity is lost if the Ets element is deleted. Forced recruitment of Fli-1 to the mutant promoter restores GATA-1/FOG-1 synergy. However, transcriptional synergy between GATA-1 and FOG-1 at the αIIb promoter is observed in transiently transfected fibroblasts, which lack Fli-1, indicating that one or more widely expressed Ets proteins can substitute for the loss of Fli-1.12 Indeed, electrophoretic mobility shift assays (EMSAs) with a probe spanning the Ets binding site of the αIIb proximal promoter and megakaryocyte-derived nuclear extracts reveals multiple specific protein/DNA complexes, only one of which contains Fli-1.12 The most abundant of these complexes also is observed when nuclear extracts from nonmegakaryocytic cell lines are used, suggesting that a widely expressed protein is capable of binding to the same Ets element.
In this study, we have identified this widely expressed, Ets-binding factor as the heterodimeric GA-binding protein (GABP) complex and show that it functions as a regulator of early megakaryocyte-specific gene expression. The GABP complex is formed by 2 components, GABPα, which mediates DNA-binding, and GABPβ, which is required for transcriptional activation and nuclear localization of GABPα (for review see Rosmarin et al21 ). Despite its broad expression pattern, GABPα is known to regulate select hematopoietic-specific genes in the myeloid and lymphoid lineages.22-25 We show that GABPα preferentially binds to the promoters of early megakaryocytic genes and that this contrasts with Fli-1, which binds both early and late megakaryocytic genes. Accordingly, loss of GABPα in gene-targeted mice (GABPαtp/tp) predominantly affects expression of early megakaryocyte-specific genes, whereas loss of Fli-1 predominantly affects the expression of late megakaryocyte-specific genes. Thus, these studies identify a new and unanticipated role for the widely expressed Ets protein GABPα during megakaryopoiesis and reveal developmental stage-specific functions among distinct Ets proteins during this process.
Materials and methods
Cell lines
NIH3T3 cells and COS cells were obtained from American Type Culture Collection (ATCC; Manassas, VA) and were grown in Dulbecco modified Eagle medium (DMEM; Invitrogen, Carlsbad, CA). The murine megakaryocytic line Y1026 was obtained from Dr Katya Ravid (Boston University School of Medicine) and maintained in F-12 nutrient mixture (Invitrogen). All media contained 100 U/mL penicillin, 100 U/mL streptomycin, 2 mM l-glutamine, and 10% fetal bovine serum (HyClone, Logan, UT).
Murine lines and fetal liver–derived megakaryocytes
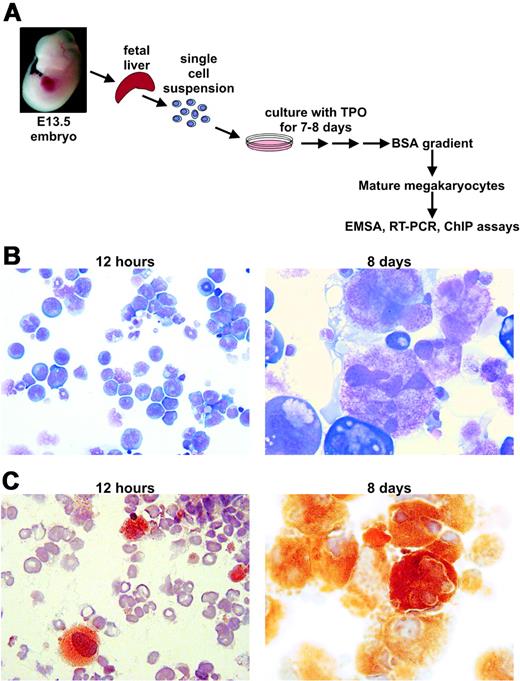
C57BL/6J (The Jackson Laboratory, Bar Harbor, ME), Fli-1+/–18,19 and GABPα+/tp27 mice were maintained. The latter 2 were used for timed matings to generate Fli-1–/– and GABPαtp/tp and littermate WT control embryos. Fetal liver–derived megakaryocytes were obtained on E11-13.5 and cultured in DMEM with TPO (R&D Systems, Minneapolis, MN, 100 ng/mL) for 12 hours or for 8 days to allow differentiation into morphologically mature megakaryocytes.28 May-Grunwald/Giemsa (Sigma Aldrich [SA], St Louis, MO) and acetylcholine esterase (AchE) stainings (SA) were performed either after 12 hours or 8 days of culture as described.29 Light microscopy images were obtained by using a Zeiss Axioskop 2 microscope, Zeiss Axiocam camera, and Zeiss AxioVision 3.1 software (Carl Zeiss Microimaging, Thornwood, NY). The objective lenses used include a Plan-Neofluar 10×/0.50 lens and a Zeiss Ph2 Plan-Neofluar 50×/0.75 lens. The imaging medium was air for all objective lenses. WT, Fli-1–/–, and GABPαtp/tp genotypes were determined by polymerase chain reaction (PCR) on embryonic tissues.18,19,27 All animal studies were done with prior institutional Institutional Animal Care and Use Committees' approval.
EMSA
EMSA was carried out as previously described.12 The sequences of the probes that span the known functional proximal Ets element (underlined) are αIIb: 5′-TAAGCTGAAACTTCCGGTGGTGGGAAC-3′12 ; αIIb Ets mutant: 5′-TAAGCTGA-AACAACCGGTGGTGGGAAC-3′; cMpl: 5′-TCTGACAGGAACCTGAGGGGCTG-3′30 ; GPIX: 5′-GCTATTTTCACCATTTCCTTCCTCCTGTCAGGCA-3′14,31,32 ; and PF4: 5′-GTATCCTGGGTTTCCGGACTGGGCAG-3′.33,34 Probes (0.2 ng) were end-labeled with [γ-32P]-ATP (Amersham, Arlington Heights, IL),12 and approximately 105 cpm were incubated with 15 μg of nuclear extracts. For supershift studies, the following antibodies were used: rabbit polyclonal anti–Fli-1 (c-19, Santa Cruz Biotechnology [SCB], Santa Cruz, CA)12 ; rabbit polyclonal anti-GABPα (H-180, SCB), rabbit polyclonal anti-GABPα, and rabbit anti-GABPβ1 serum,27 mouse monoclonal anti-HA (F-7, SCB) and control rabbit IgG (SA). Each was added to the nuclear extracts for 25 minutes at room temperature prior to the addition of the probe.
Fluorescence-activated cell sorting (FACS)
Immature (CD41+/GPIbα–) megakaryocytes were enriched by FACS. Briefly, E13.5 fetal-liver cells of C57BL/6J WT mice were cultured in TPO-containing medium for 12 hours and stained with the following antibodies (1:100 dilution) for 20 minutes at room temperature: fluorescein isothiocyanate (FITC)–conjugated rat monoclonal anti–mouse CD41 (Becton Dickinson [BD], San Jose, CA), phycoerythrin-conjugated rat monoclonal anti–mouse CD42b (GPIbα; Emfret, Eibelstadt, Germany). An anti–mouse CD16/CD32 (FcR III/II receptor) (BD) was added to each sample to avoid nonspecific binding antibodies to the cells. The CD41+/GPIbα– cells were collected using a BD FACSVantage SE cell sorter. Mature (CD41+/GPIbα+) megakaryocytes were collected after 8 days of culture of C57BL/6J WT fetal liver cells using a bovine serum albumin (BSA) gradient centrifugation as described.35
Flow cytometry
E11 fetal liver–derived cells from gene-targeted mice were filtered through a 100-μm cell strainer (BD), stained with antibodies as described in the previous section, and analyzed on a FACSort flow cytometer (BD). Between 15 000 and 40 000 counts were obtained. Dead cells excluded according to the forward and side scatters. The percentages of immature megakaryocytes (CD41+/GPIbα–) and mature megakaryocytes (CD41+/GPIbα+) were calculated based on quadrate statistical analysis using CellQuest Professional Software (BD).
BSA gradient centrifugation
Mature megakaryocytes were enriched by BSA gradient centrifugation following culture of C57BL/6J WT E13.5 fetal liver cells in TPO-containing medium for 8 days.35 Briefly, cells were collected by centrifugation at 3.5g (400 rpm) (Beckman Coulter, Hialeah, FL, Allegra 6) and resuspended in 5 mL phosphate buffered saline (PBS). The BSA step gradient was prepared by placing PBS containing 1.5% albumin on top of PBS with 3% albumin. Cells were loaded on top of the gradient and spun at 2g (150 rpm) for 30 minutes at room temperature (RT). Mature megakaryocytes formed a pellet at the bottom of the tube.
Quantitative RT-PCR
Total cellular RNA was extracted with Trizol reagent (Invitrogen) as previously described.17 Reverse transcription reactions were performed on 3 μg of total RNA using a Superscript II kit (Invitrogen). Results were quantified using real-time PCR with TaqMan probes/primers on an ABI Prism 7900 system. All probes were from the commercially available collection from TaqMan Gene Expression Assays. Serial dilution of known concentration of pure plasmid DNA or PCR products was used to establish the standard curve for each probe/primer. The probe IDs were as follows: αIIb, Mm 00439768_m1; cMpl, Mm00440310_m1; GPIX, Mm007671_g1; GPIbα, Mm0050 677_g1; PF4, Mm00451315_g1; Fli-1, Mm00484410_m1; GABPα, Mm00484598_m1; HPRT, Mm00446968_m1; and GAPDH, Mm99999915_g1. The relative gene expression levels were normalized by either glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or hypoxanthine phosphoribosyltransferase (HPRT) signal to eliminate the differences in total RNA amount and reverse transcription efficiency. Normalization to GAPDH and HPRT yielded similar results (data not shown), but only data relative to GAPDH are shown.
Glutathione S-transferase (GST)–fusion proteins and Western blots
GST-fusion constructs GST-Fli-11-274 and GST-GABPα1-310 were generated by PCR and insertion into the pGEX2T vector (Pharmacia, Uppsala, Sweden). All constructs were sequenced. Proteins were expressed in the Escherichia coli strain BL-21 and purified with glutathione sepharose 4B beads (Invitrogen).
Proteins were size-fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) using NuPAGE Novex Pre-Cast 10% Bis-tris (Invitrogen) gels with MOPS (3-[N-morpholino] propane sulfonic acid) SDS running buffer, transferred to nitrocellulose membrane (Invitrogen), and probed with rabbit polyclonal anti–Fli-1 H60 (SCB, 1:500 dilution) or rabbit polyclonal anti-GABPα H-180 (1:1000 dilution). Bound antibodies were detected with rabbit horseradish peroxidase–labeled secondary antibody and enhanced chemiluminescence (Perkin-Elmer, Shelton, CT). Signal was quantitated using ImageQuant TL software (Amersham Biosciences, Freiburg, Germany). Concentrations of GABPα and Fli-1 in nuclear extract preparations were determined by comparing the signal intensity of known volumes of nuclear extract to the signal intensity of known amounts of the correspondent recombinant GST-protein.
Chromatin immunoprecipitation (ChIP) assays
ChIP assays were performed as described.36 Antibodies used were anti–Fli-1 (c-19), anti-GABPα (H-180), and rabbit isoimmune IgG control. The source of chromatin was C57BL/6J murine E13.5 fetal liver cells after TPO culture for 8 days following a BSA gradient centrifugation. Results were quantified using real-time PCR with SYBR green dye on an ABI Prism 7900 System (Applied Biosystems, Weiterstadt, Germany) as described.37 A standard curve was generated for each primer pair by serial dilution of an unprecipitated (“input”) sample. All PCR signals from immunoprecipitated samples were referenced to their respective input standard curve to account for potential variations among samples and PCR primer amplification efficiencies. Primers were designed with Primer Express software (Applied Biosystems). αIIb sense: 5′-GCCATGAGCTCCAGTCTGATAA-3′, antisense: 5′-AGCTCTTTCCCTTTCCCTGAA-3′; cMpl sense: 5′-CTGCCAACAGAAGGCTCATG-3′, antisense: 5′CTGTCAGATACAGCCCCACGT-3′; GPIX sense: 5′-GCCTCCTGGCCCTGACA-3′, antisense: 5′-TGTGGCTGCTGCCTGACA-3′; GPIbα sense: 5′-TGGTGGCTAGTAGCTGCAAAGTC-3′, antisense: 5′-TTATCAGCTCTCTGCACAGCATTC-3′; PF4 sense: 5′-GCTGCTGGCCTGCACTTAAG-3′, antisense: 5′-GCCACTGGACCCAAAGATAAAG-3′; and negative control upstream αIIb region sense: 5′-AAATAGATGTCAAGTTGGCATAAACCT-3′, antisense: 5′-TGCCAGCGTTCAAGTACAAAA-3′.
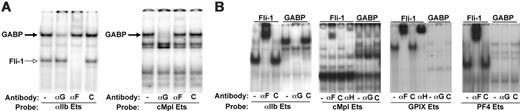
GABPα is the major Ets transcription factor that binds to the proximal αIIb promoter by EMSA. (A) EMSA using nuclear extract from either Y10 cells or NIH3T3 cells. Indicated above the gel is whether a 32P-labeled 27 bp probe from –53 to –27 bp upstream of the murine αIIb gene transcription start site12 (WT) or a similar probe mutated within the core Ets binding motif was included. Indicated below the gel are the antibodies included. αF = anti–Fli-1 antibody. C = isoimmune control antibodies. The fold excess of the cold competitor probe is also indicated. The source of nuclear extracts is indicated at the bottom. The position of migration of Fli-1 is indicated by a thin open arrow. (B) EMSA using WT probe and Y10 nuclear extract. αG1 and αG2 = 2 anti-GABPα antibodies. αGb = anti-GABPβ.C = isoimmune control antibodies. The positions of migration of GABPα and Fli-1 are indicated by a thick black arrow and a thin open arrow, respectively. (C) Competitive binding of GABPα and Fli-1 to the αIIb proximal promoter in vitro. The probe is same as in panel B, but these studies use nuclear extracts from COS cells or COS cells overexpressing HA-Fli-1. The amounts of GABPα and Fli-1 added in each lane are indicated. The position of migration of GABPα and Fli-1 are indicated as in panel B. All gels are representative studies of at least 3 similar studies with similar outcomes.
GABPα is the major Ets transcription factor that binds to the proximal αIIb promoter by EMSA. (A) EMSA using nuclear extract from either Y10 cells or NIH3T3 cells. Indicated above the gel is whether a 32P-labeled 27 bp probe from –53 to –27 bp upstream of the murine αIIb gene transcription start site12 (WT) or a similar probe mutated within the core Ets binding motif was included. Indicated below the gel are the antibodies included. αF = anti–Fli-1 antibody. C = isoimmune control antibodies. The fold excess of the cold competitor probe is also indicated. The source of nuclear extracts is indicated at the bottom. The position of migration of Fli-1 is indicated by a thin open arrow. (B) EMSA using WT probe and Y10 nuclear extract. αG1 and αG2 = 2 anti-GABPα antibodies. αGb = anti-GABPβ.C = isoimmune control antibodies. The positions of migration of GABPα and Fli-1 are indicated by a thick black arrow and a thin open arrow, respectively. (C) Competitive binding of GABPα and Fli-1 to the αIIb proximal promoter in vitro. The probe is same as in panel B, but these studies use nuclear extracts from COS cells or COS cells overexpressing HA-Fli-1. The amounts of GABPα and Fli-1 added in each lane are indicated. The position of migration of GABPα and Fli-1 are indicated as in panel B. All gels are representative studies of at least 3 similar studies with similar outcomes.
Transient expression assays and plasmid constructs
NIH3T3 cells were transfected with Fugene 6 (Roche, Milan, Italy) according to the manufacturer using a Fugene 6: total DNA ratio at 3:1. Total amount of transfected DNA was kept constant in all samples. After 40 hours, luciferase activity was determined using a luciferase assay kit (Promega, Madison, WI) and a BD Monolight 3010C Luminometer (BD). The 100-bp mouse αIIb promoter luciferase reporter construct containing a GAL4 binding site in place of the proximal Ets site has been described.12,38 Expression vectors for Fli-11-274 PU-11-160 fused to the GAL4 DNA binding domain (GAL4 DBD), and pXM-GATA-1 and pMT2-FOG-1 have been described.12,39-41 The GAL4-GABPα1-310 construct was generated by PCR from reverse-transcribed total RNA from Y10 cells. The resulting cDNA was inserted into pCMX-GAL4.40
Results
GABPα binds to the Ets element at the αIIb proximal promoter in vitro
To identify additional proteins that bind to the αIIb Ets element, EMSAs were performed using nuclear extracts of the megakaryocytic cell line Y10 and the fibroblast cell line NIH3T3, and a probe spanning the functional Ets binding site within the proximal promoter of the murine αIIb gene (Figure 1). As observed previously, multiple specific DNA/protein complexes formed on the αIIb Ets element that were competed away by excess unlabeled WT cold probe, but not by a probe with a mutated Ets site (Figure 1A). No specific protein/DNA complexes formed when the mutant probed was used (Figure 1A, right). The fastest migrating specific band seen in Y10 cell was absent in nonmegakaryocytic cells. This band disappeared upon addition of an anti–Fli-1 antibody identifying it as Fli-1 (Figure 1A, open arrow). To identify the other protein complexes, we tested a panel of antibodies raised against the related pointed-domain Ets proteins Ets-1, Ets-2, Erg-1/2/3, TEL, and the GABP. Two independently derived antibodies against GABPα, but not a control antibody, virtually eliminated the most prominent EMSA complex, indicating that GABPα binds to the αIIb Ets element in vitro (Figure 1B, filled arrow and data not shown). Since GABPα interacts with GABPβ, we examined whether GABPβ also is present in this complex, and indeed, this antibody markedly diminished the GABPα-containing complex, indicating that both GABP subunits associate with the αIIb Ets element (Figure 1B).
Schematic for use of murine fetal liver–derived primary megakaryocytes. (A) Illustration of the procedure for isolating mature megakaryocytes from murine fetal liver cells. (B) May-Grunwald/Giemsa stain of the initial cell mixture 12 hours after isolation and dispersion into culture media, and after 8 days of growth in media (original magnification of both images = 500-fold). (C) AchE stain of the initial cell mixture 12 hours after isolation and dispersion into culture media, and after 8 days of growth in media (original magnification of both images = 500-fold).
Schematic for use of murine fetal liver–derived primary megakaryocytes. (A) Illustration of the procedure for isolating mature megakaryocytes from murine fetal liver cells. (B) May-Grunwald/Giemsa stain of the initial cell mixture 12 hours after isolation and dispersion into culture media, and after 8 days of growth in media (original magnification of both images = 500-fold). (C) AchE stain of the initial cell mixture 12 hours after isolation and dispersion into culture media, and after 8 days of growth in media (original magnification of both images = 500-fold).
To compare DNA binding affinities of GABP and Fli-1 in vitro, competitive binding assays were carried out using nuclear extracts from untransfected COS cells that express high levels of endogenous GABPα or from COS cells overexpressing HA-tagged Fli-1. In the presence of a fixed amount of GABPα (3 ng), addition of increasing amounts of nuclear extracts of Fli-1 (0-180 ng) resulted in increased amounts of Fli-1/DNA complexes and a corresponding decrease of GABP-bound DNA (Figure 1C, left). Conversely, beginning with a fixed amount of Fli-1 (90 ng), addition of increasing amounts of GABPα (0-9 ng) resulted in increased amounts of GABP-bound DNA and a corresponding decrease in Fli-1/DNA complexes (Figure 1C, right). We conclude that GABPα and Fli-1 cross-compete for binding to the αIIb Ets binding site in vitro. Based on our measurements of Fli-1 and GABPα concentrations by quantitative Western blots (data not shown), we estimate that GABPα has approximately 30-fold higher binding affinity than Fli-1 for binding to the αIIb proximal Ets element.
Expression of Fli-1 and GABPα in developing megakaryocytes
To investigate the relative expression levels of Fli-1 and GABPα during megakaryocyte maturation, we isolated early, immature, and late, mature megakaryocytes from E13.5 murine fetal liver cells. Fetal liver cells were cultured for 12 hours in the presence of TPO (Figure 2A) to allow nonhematopoietic cells to adhere to the dish. Of the nonadherent cells, 5% to 10% were morphologically recognizable megakaryocytes as judged by May-Grunwald/Giemsa staining and AchE staining (Figure 2B,C, left). Immature CD41+/GPIbα– megakaryocytes were selected from this pool of cells by FACS using antibodies against CD41 and GPIbα. Mature CD41+/GPIbα+ megakaryocytes were obtained after 8 days of culture following purification by BSA gradient centrifugation (Figure 2A). More than 80% of these cells were morphologically mature megakaryocytes, displaying large-sized, multilobular nuclei and granular cytoplasm, and more than 90% were positive for AchE (Figure 2B,C, right).
We determined steady-state mRNA levels of Fli-1 and GABPα by real-time RT-PCR in both immature and mature megakaryocytes and normalized mRNA levels to those of GAPDH. Moreover, signals were normalized to standard curves from PCR reactions with known amounts of cDNA templates of Fli-1 and GABPα, thus allowing quantitative comparisons between PCR signals. We found that Fli-1 mRNA levels were twice that of GABPα in the immature population isolated (Figure 3A). The amounts of Fli-1 mRNA increased approximately 6-fold with respect to GAPDH as cells matured, whereas GABPα levels increased only 2-fold (Figure 3A). Hence, the Fli-1/GABPα ratio increased by 3-fold throughout maturation. Moreover, absolute Fli-1 mRNA levels were approximately 6-fold higher than GABPα levels in mature megakaryocytes (Figure 3A). We also measured the protein levels of Fli-1 and GABPα by quantitative Western blot analysis of nuclear extracts in mature megakaryocytes (Figure 3B). As internal standard, known amounts of recombinant GST-Fli-11-274 or GST-GABPα1-310 proteins were simultaneously analyzed. We found that Fli-1 protein was present at an approximately 6-fold molar excess over GABPα, consistent with the levels of mRNA transcripts. For lack of sufficient amounts of CD41+/GPIbα– cells obtainable by FACS, we could not carry out similar studies on immature megakaryocytes. Thus, these data indicate that Fli-1 is significantly more abundant in late stages of megakaryocyte maturation, while GAPBα and Fli-1 are expressed at more similar levels at early stages.
Relative expression of Fli-1 and GABPα in murine fetal liver–derived megakaryocytes. (A) Steady-state RNA levels measured by quantitative real-time RT-PCR of Fli-1 and GABPα both relative to GAPDH in CD41+/GPIbα–immature megakaryocytes (□) and CD41+/GPIbα+ mature megakaryocytes (▪). (B) Relative protein levels of Fli-1 and GABPα in mature megakaryocytes. On the left, Western blots of known amounts of nuclear extracts were compared to known concentration of the appropriate recombinant proteins to determine the relative ratio of GABPα to Fli-1 proteins in nuclear extracts from fetal liver–derived mature megakaryocytes. Quantitative analysis by QuantImage analysis is shown on the right. For all studies in panels A and B, the mean ± one SD of 3 experiments, each in triplicate, is shown.
Relative expression of Fli-1 and GABPα in murine fetal liver–derived megakaryocytes. (A) Steady-state RNA levels measured by quantitative real-time RT-PCR of Fli-1 and GABPα both relative to GAPDH in CD41+/GPIbα–immature megakaryocytes (□) and CD41+/GPIbα+ mature megakaryocytes (▪). (B) Relative protein levels of Fli-1 and GABPα in mature megakaryocytes. On the left, Western blots of known amounts of nuclear extracts were compared to known concentration of the appropriate recombinant proteins to determine the relative ratio of GABPα to Fli-1 proteins in nuclear extracts from fetal liver–derived mature megakaryocytes. Quantitative analysis by QuantImage analysis is shown on the right. For all studies in panels A and B, the mean ± one SD of 3 experiments, each in triplicate, is shown.
Preference of GABPα and Fli-1 binding to the proximal promoters of early versus late megakaryocytic genes by EMSA. (A) EMSA gels using nuclear extracts from primary megakaryocytes are shown. On the left, the probe is the same as in Figure 1 and on the right, the probe involves the functional Ets site in the proximal promoter region of the murine cMpl gene.30 (B) EMSA using nuclear extract from COS cells or COS cells overexpressing HA-Fli-1 are shown. Equal amounts of extracts were used in each set of gels, allowing direct comparisons of relative binding intensity. αH = anti-HA tag antibody. Supershift of the HA-Fli-1 was done either using αF alone (αIIb and PF4) or with αH (cMpl and GPIX). All gels are representative studies of at least 3 similar studies with similar outcomes.
Preference of GABPα and Fli-1 binding to the proximal promoters of early versus late megakaryocytic genes by EMSA. (A) EMSA gels using nuclear extracts from primary megakaryocytes are shown. On the left, the probe is the same as in Figure 1 and on the right, the probe involves the functional Ets site in the proximal promoter region of the murine cMpl gene.30 (B) EMSA using nuclear extract from COS cells or COS cells overexpressing HA-Fli-1 are shown. Equal amounts of extracts were used in each set of gels, allowing direct comparisons of relative binding intensity. αH = anti-HA tag antibody. Supershift of the HA-Fli-1 was done either using αF alone (αIIb and PF4) or with αH (cMpl and GPIX). All gels are representative studies of at least 3 similar studies with similar outcomes.
GABPα and Fli-1 binding to the proximal promoters of megakaryocytic genes in vitro
The in vitro occupancy of transcription factors is a function of both their abundance and their affinities to their cognate DNA elements. Therefore, we examined the relative DNA binding activities of Fli-1 and GABPα in primary fetal liver–derived megakaryocytes by EMSA. Using a DNA probe containing the αIIb Ets element, we observed a slight preponderance of GABPα binding when compared to the Fli-1 (Figure 4A, left). To determine whether both Fli-1 and GABPα bind to the relevant Ets elements of other megakaryocyte-specific genes with comparable affinities, we examined previously defined Ets-binding elements from the early megakaryocytic gene cMpl13 and the 2 late megakaryocytic genes GPIX14,31 and PF4.33 When the Ets probe from the cMpl promoter was used, multiple distinct protein complexes were observed, one of which reacted with the anti-GABPα antibody (Figure 4A, right). In contrast, the anti–Fli-1 antibody failed to react with any of the protein complexes associated with the cMpl Ets element. These findings suggest that within the limits of this assay, GABPα binds to this element in vitro more efficiently than does Fli-1.
EMSA assays with Ets element-containing probes from the late megakaryocytic genes yielded complex patterns of protein bands, and analysis was difficult (data not shown). Since Fli-1 and GABPα clearly occupy these genes in vivo as determined by ChIP in primary megakaryocytes (see next section), we set out to compare Fli-1 and GABPα binding to early versus late megakaryocyte-specific promoters using a simplified in vitro binding assay. To this end, EMSA studies were carried out using nuclear extracts from untransfected COS cells (that express endogenous GABPα) or from COS cells overexpressing HA-tagged Fli-1. Both GABPα and Fli-1 bound to probes containing the αIIb Ets element (Figure 4B), consistent with our results from Figure 1. When the probe containing cMpl proximal Ets element was used, a GABPα-containing band was clearly visible that was lost upon inclusion of the anti-GABPα antibody (Figure 4B). A specific Fli-1 band was supershifted by both an anti–Fli-1 antibody and an anti-HA antibody. A different binding pattern was observed when Ets element probes from the late genes, GPIX and PF4, were studied. Thus, Fli-1 strongly bound both probes, whereas GABPα exhibited poor binding (Figure 4B, right 2 panels). Together, these in vitro data suggest that GABPα preferentially binds to the promoters of early megakaryocytic genes, while Fli-1 preferentially binds to the promoters of late megakaryocytic genes.
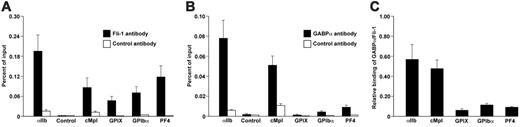
Fli-1 and GABPα bind to the proximal promoters of megakaryocyte-specific genes in vivo. Panels A and B show ChIP assays using fetal liver–derived mature megakaryocytes and examining the proximal promoter regions containing known functional Ets binding sites.13 (A) ChIP with Fli-1 antibody and isotype-matched control antibody. (B) ChIP with GABPα antibody and isotype-matched control antibody. The control represents a region 3 kb upstream of the murine αIIb transcription start site known not to be of functional importance or containing an Ets binding site.12 The mean ± one SD of 3 experiments, each in triplicate, is shown. (C) Signal ratio of GABPα to Fli-1 at the various megakaryocyte-specific promoters is shown.
Fli-1 and GABPα bind to the proximal promoters of megakaryocyte-specific genes in vivo. Panels A and B show ChIP assays using fetal liver–derived mature megakaryocytes and examining the proximal promoter regions containing known functional Ets binding sites.13 (A) ChIP with Fli-1 antibody and isotype-matched control antibody. (B) ChIP with GABPα antibody and isotype-matched control antibody. The control represents a region 3 kb upstream of the murine αIIb transcription start site known not to be of functional importance or containing an Ets binding site.12 The mean ± one SD of 3 experiments, each in triplicate, is shown. (C) Signal ratio of GABPα to Fli-1 at the various megakaryocyte-specific promoters is shown.
Fli-1 and GABPα occupancy at early and late megakaryocytic genes in vivo
To examine occupancy of Fli-1 and GABPα at the proximal promoters of early and late megakaryocytic genes in vivo, we performed ChIP assays on murine fetal liver–derived mature megakaryocytes. Chromatin immunoprecipitated with Fli-1 antibody was enriched for the proximal promoters of αIIb, cMpl, GPIX, GPIbα, and PF4 (Figure 5A). As a control, a more distal region approximately 3 kb upstream from the αIIb transcription start site, which is known to lack functionally important Ets elements,12 was not enriched (Figure 5A). In contrast, chromatin immunoprecipitated with GABPα antibody was enriched in the proximal promoters of the early megakaryocytic genes, αIIb and cMpl (Figure 5B), but little enrichment was found at the proximal promoter regions of the late megakaryocytic genes (Figure 5B). Again, as negative control, the upstream αIIb region was not enriched (Figure 5B). To illustrate the relative occupancy by GABPα and Fli-1 at megakaryocytic promoters, we plotted the ChIP results by dividing the GABPα signal by that of Fli-1. The binding intensity of GABPα to Fli-1 at the proximal promoters of 2 early megakaryocytic genes is 4-8 fold higher than that at the promoters of the 3 late megakaryocytic genes studied (Figure 5C). These data show that while both Fli-1 and GABPα can associate with both early and late megakaryocyte-specific genes in vivo, GABPα binds predominantly to the early genes, while Fli-1 preferentially associates with late megakaryocyte-specific genes. For lack of sufficient available cells, parallel ChIP assays using immature megakaryocytes could not be done.
Analysis of Fli-1–/– and GABPαtp/tp megakaryocytes
To compare the in vivo requirements for Fli-1 and GABPα in megakaryopoiesis, we analyzed primary megakaryocyte precursors in Fli-1–/– and GABPαtp/tp fetal livers. GABPαtp/tp mice were generated by a “gene trap” strategy.27 GABPα is expressed at approximately 10% of WT levels in these animals,27 which permits their survival until E12.5. GABPα–/– embryos die prior to implantation,42 precluding analysis of fetal liver hematopoiesis. Mice bearing 2 targeted alleles of Fli-1 (Fli-1–/–) do not express any Fli-1 protein, resulting in fatal hemorrhaging at around E11.5.18 Thus, we performed comparative studies on both types of animals at E11.
We measured by flow cytometry the number of CD41+ cells in total fetal liver cell populations following 12 hours in culture. CD41 is a surface antigen present on the αIIb/βIII integrin. We found that both Fli-1–/– and GABPαtp/tp E11 fetal livers contained fewer CD41+ cells compared to their WT littermates (Figure 6A, top left). However, when we considered the ratio of immature (GPIbα–) to mature (GPIbα+) cells within the CD41+ population, distinct phenotypes emerged: Fli-1–/– CD41+ cells contained increased proportions of immature megakaryocytes and a marked decrease in mature megakaryocytes relative to their WT littermates (Figure 6A, top and right). In contrast, GABPαtp/tp CD41+ cells showed a decrease in the fraction of immature megakaryocytes and an increase in the proportion of mature megakaryocytes relative to WT littermates (Figure 6A, bottom and right). These experiments are consistent with distinct roles for Fli-1 and GABPα throughout megakaryocyte maturation.
Analysis of megakaryocytes and megakaryocytic gene expression in the Fli-1–/– and GABPαtp/tp mice. (A) Total fetal liver cells from E11 at 12 hours culture as in Figure 2 were used to study the relative number of immature and mature megakaryocytes in fetal hematopoietic tissue. These cells were stained with FITC-conjugated anti-CD41 and phycoerythrin (PE)–conjugated anti-GPIbα antibodies and subjected to flow cytometry. Top left shows the analysis of 9 Fli-1–/– fetal liver samples and 9 WT littermates. Bottom left shows the analysis of 6 GABPαtp/tp fetal liver samples and 6 WT littermates. At the right of the figure, the relative levels of mature to immature megakaryocytes for each phenotype compared to their WT siblings are shown. * = P < .005; ** = P < .001; and *** = P < .0001 relative to WT control. The mean ± one SD is shown for each study. (B) Megakaryocyte-specific gene expression patterns in Fli-1–/– and GABPαtp/tp mice fetuses were studied using RNA extracted from E11 embryos (left 2 panels) or E12.5 fetal liver cells (right panel) and subjected to real-time RT-PCR for the messages indicated. The number of fetuses studied in each group is indicated. For all studies, * = P < .005; ** = P < .001; *** = P < .0001; and **** = P < .000 01 relative to WT control. The mean ± one SD is shown for each study.
Analysis of megakaryocytes and megakaryocytic gene expression in the Fli-1–/– and GABPαtp/tp mice. (A) Total fetal liver cells from E11 at 12 hours culture as in Figure 2 were used to study the relative number of immature and mature megakaryocytes in fetal hematopoietic tissue. These cells were stained with FITC-conjugated anti-CD41 and phycoerythrin (PE)–conjugated anti-GPIbα antibodies and subjected to flow cytometry. Top left shows the analysis of 9 Fli-1–/– fetal liver samples and 9 WT littermates. Bottom left shows the analysis of 6 GABPαtp/tp fetal liver samples and 6 WT littermates. At the right of the figure, the relative levels of mature to immature megakaryocytes for each phenotype compared to their WT siblings are shown. * = P < .005; ** = P < .001; and *** = P < .0001 relative to WT control. The mean ± one SD is shown for each study. (B) Megakaryocyte-specific gene expression patterns in Fli-1–/– and GABPαtp/tp mice fetuses were studied using RNA extracted from E11 embryos (left 2 panels) or E12.5 fetal liver cells (right panel) and subjected to real-time RT-PCR for the messages indicated. The number of fetuses studied in each group is indicated. For all studies, * = P < .005; ** = P < .001; *** = P < .0001; and **** = P < .000 01 relative to WT control. The mean ± one SD is shown for each study.
Gene expression patterns in Fli-1–/– and GABPαtp/tp mice
The experiments in the preceding section suggest that deficiencies of Fli-1 and GABPα affect late and early stages of megakaryocyte maturation, respectively. However, staging by flow cytometry of maturation relied on the expression of GPIbα, a presumed Fli-1 target.43 To obtain a more comprehensive view of the changes in gene expression patterns, we examined the steady-state mRNA levels of 2 early genes, αIIb and cMpl, and 3 late genes, GPIX, GPIbα, and PF4, by real-time RT-PCR using whole E11 Fli-1–/– and GABPαtp/tp embryos, and also E12.5 GABPαtp/tp fetal livers.
In the Fli-1–/– embryos, the expression of all 3 late genes was substantially decreased, while the early genes showed a comparatively mild reduction in expression (Figure 6B, left). In contrast, in GABPαtp/tp embryos (Figure 6B, center) and GABPαtp/tp fetal livers (Figure 6B, right), expression of αIIb and cMpl was reduced up to approximately 25% of WT littermate, while expression of GPIX, GPIbα, and PF4 was maintained at closer to normal levels. Loss of GABPα does not significantly affect expression of Fli-1 (Figure 6B, middle and right), suggesting that Fli-1 is not a downstream target of GABPα and does not account for the observed changes in gene expression in the GABPαtp/tp embryos.
GABPα mediates GATA-1/FOG-1 synergy at the αIIb promoter
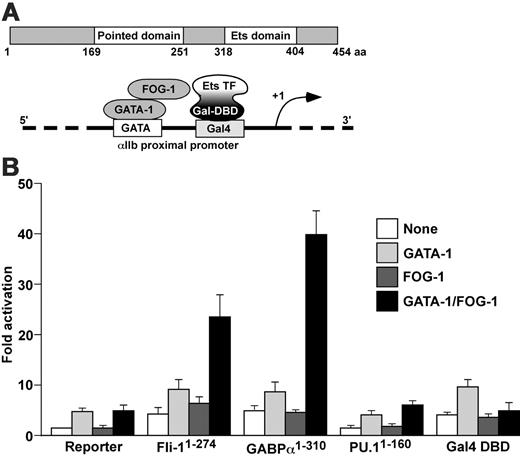
If GABPα is capable of functionally substituting for Fli-1, it is expected that the mechanisms by which these 2 proteins function are similar. Previously, we demonstrated that synergistic activation of the αIIb promoter by GATA-1 and FOG-1 required an intact Ets element.12 Mutation of the Ets site abrogated the ability of FOG-1 to augment the activity of GATA-1. However, when GAL4-Fli-11-274 was recruited to the mutant αIIb via a newly introduced GAL4 DNA binding domain, GATA-1/FOG-1 synergy was restored.12 Using this assay, we examined whether GABPα can regulate the activity of the GATA-1/FOG-1 transcription factor pair in the same manner.
We generated a GAL4-fusion construct (GAL4-GABPα1-310) that contained the N-terminal 310 amino acids of GABPα, lacking the DNA-binding Ets domain, fused to the DNA-binding domain of GAL4 (Figure 7A). Combinations of GATA-1, FOG-1, and GAL4-GABPα1-310 were transiently expressed in NIH3T3 cells together with a reporter gene driven by the αIIb promoter in which the Ets element was replaced with a GAL4 binding site12 (Figure 7A). Thus, potential effects of endogenous Ets factors on αIIb promoter activity were circumvented. Expression of GATA-1 alone moderately activated the reporter gene (Figure 7B). Coexpression of FOG-1 failed to further enhance GATA-1 activity, consistent with our previous observations.12 In contrast, when GAL4-GABPα1-310 was present, FOG-1 substantially increased reporter gene activity in a GATA-1–dependent manner (Figure 7B). The effects of GABPα were specific, as neither GAL4 fused to the Ets protein PU.1 (GAL4-PU.11-160), nor the GAL4 DNA-binding domain itself conferred transcriptional synergy between GATA-1 and FOG-1 (Figure 7B, right). Of note, the synergy between GATA-1 and FOG-1 was more pronounced with GAL4-GABPα1-310 than with GAL4-Fli-11-274. These results demonstrate that GABPα and Fli-1 share at least one mechanism by which they activate a megakaryocyte-specific promoter.
GABPα as well as Fli-1 mediates GATA-1/FOG-1 synergy at the αIIb promoter. (A) Schematic representations of the preserved domains of GABPα (top) and of the vector system used in the assay (bottom). The reporter construct contained 100 bp of αIIb promoter region in which a GAL4-binding site was substituted for the Ets-binding site. GAL4 fusion constructs of 3 different Ets transcription factors, Fli-1, GABPα, and PU.1, were coexpressed with GATA-1 and FOG-1 as indicated in panel B. (B) Transient expression studies done in NIH3T3 cells with the modified 100-bp αIIb promoter reporter construct ± GATA-1 ± FOG-1 in the presence of the indicated recombinant protein at the bottom. The mean ± one SD of 3 experiments, each in triplicate, is shown.
GABPα as well as Fli-1 mediates GATA-1/FOG-1 synergy at the αIIb promoter. (A) Schematic representations of the preserved domains of GABPα (top) and of the vector system used in the assay (bottom). The reporter construct contained 100 bp of αIIb promoter region in which a GAL4-binding site was substituted for the Ets-binding site. GAL4 fusion constructs of 3 different Ets transcription factors, Fli-1, GABPα, and PU.1, were coexpressed with GATA-1 and FOG-1 as indicated in panel B. (B) Transient expression studies done in NIH3T3 cells with the modified 100-bp αIIb promoter reporter construct ± GATA-1 ± FOG-1 in the presence of the indicated recombinant protein at the bottom. The mean ± one SD of 3 experiments, each in triplicate, is shown.
Discussion
Nuclear factors belonging to the same family typically provide overlapping yet distinct functions during gene expression. A prominent example in the hematopoietic system is the GATA family of transcription factors.44 Here we show that during megakaryocyte development, the Ets family transcription factor GABPα performs an important role predominantly during early stages of megakaryocyte maturation by regulating the expression of αIIb and cMpl. This role contrasts with that of the related Ets factor Fli-1 that is important for the expression of late-stage, megakaryocytic genes. ChIP analysis of primary fetal liver–derived megakaryocytes showed that Fli-1 occupies promoters of genes associated with both early and late stages of megakaryocyte maturation, while GABPα associates preferentially with early genes. Our work further suggests that developmental stage specificity by these 2 factors is accomplished in at least 2 ways. First, expression levels of GABPα and Fli-1 change throughout megakaryocyte maturation such that Fli-1 becomes the more abundant protein in mature megakaryocytes. Second, comparison of the relative binding avidities between GABPα and Fli-1 in vitro revealed that while both proteins can compete for the same binding site, GABPα binds with higher avidity to the Ets sites of the αIIb and cMpl promoters, while Fli-1 prefers those in the late expressed genes, GPIX and PF4. Our ChIP studies confirm these binding site preferences in vivo. The molecular basis that underlies this discrimination between Ets sites in vivo remains an open question. We did not observe an obvious consensus among the Ets elements of early versus late megakaryocytic genes by alignments of the core Ets elements and their immediate flanking sequences (data not shown). In vivo, specificity is likely determined by a combination of primary nucleotide sequences flanking the core Ets elements and the broader context provided by neighboring protein binding sites.
Comparative analysis of gene expression patterns in genetargeted, primary megakaryocytes show that loss of Fli-1 impacts predominantly late-expressed genes, while deficiency of GABPα impairs the expression of early genes. These results are consistent with the EMSA and ChIP experiments. However, these data also demonstrate that stage and gene selectivity are not absolute, and suggest that GABPα and Fli-1 might partly compensate for each other. This is supported by several observations: first, loss of either GABPα or Fli-1 results in a partial, but not complete, loss of target gene expression. Second, ChIP experiments show that Fli-1 does occupy early megakaryocyte-specific genes, at least in mature megakaryocytes, and conversely, that GABPα can be detected at late megakaryocyte-specific genes, albeit at low levels. Third, forced expression of Fli-1 in K562 cells induces the expression of the αIIb gene.11 Fourth, functional experiments show that GABPα and Fli-1 share at least one mechanism of action, which involves regulating the activity of GATA-1/FOG-1 complex.
This overlap in function may explain how GABPα-deficient cells traverse early stages of maturation. Additionally, the presence of approximately 10% residual GABPα in the GABPαtp/tp cells also may have allowed progenitor cells to develop into megakaryocytes. Nevertheless, the diminished ratio of immature to mature megakaryocytes observed in GABPα-deficient animals suggests that the GABP complex might be required to properly balance the proliferation and differentiation of megakaryocytes. It is possible that the GABP complex promotes cell cycle progression and/or cell viability to drive expansion of the immature cell population. Alternatively, the GABP complex might prevent premature differentiation. Certainly, the identification of additional GABPα target genes will provide further insights into the mechanisms by which it regulates megakaryocyte proliferation and maturation.
GABPα was first identified as a regulator of viral genes and nuclear respiratory factors.21 Recently, it has been shown to be important for the expression of specific genes in the myeloid and lymphoid lineages23,45 ; however, to our knowledge, this report is the first demonstration that GABPα is important for the normal development of the megakaryocytic lineage. The broad expression pattern of both GABPα and GABPβ raises the question of how GABP can mediate megakaryocyte-specific gene expression. More specifically, why do GATA-1 and FOG-1 not inappropriately activate megakaryocyte-restricted genes in the erythroid lineage? Insights into this question came from our previous work showing that Fli-1 can regulate the activity of the GATA-1/FOG-1 complex.12 Fli-1 expression is extinguished in maturing erythroid cells, thus providing a mechanism by which lineage selectivity is accomplished. To this end, we queried a database of erythroid-expressed genes and found that, like Fli-1, GABPα expression declines rapidly during erythroid maturation and is virtually extinguished at 30 hours after erythroid differentiation.46 Although GABPβ expression persists during erythroid maturation,46 it alone is unable to regulate gene expression, as the GABPα subunit is required for its recruitment to target genes.21
The expression of αIIb and cMpl is not restricted to megakaryocytes. αIIb has been described on early hematopoietic progenitor cells,47,48 and the cMpl/TPO cytokine axis is important for early progenitor development.49,50 Indeed, the absence of functional cMpl in human patients results not only in severe thrombocytopenia, but eventual total bone marrow failure.51 How the expression of αIIb and cMpl is established in early hematopoietic cells (HSCs) is presently unknown, but might involve combinations of GABPα, the early-expressed GATA factor GATA-2, and FOG-1. Our own studies of CD41+ cells from E11 and E12.5 fetal livers likely excluded most early HSCs since they are rare cells in fetal livers,47,48,52 and their level of αIIb expression is low.53,54
There are likely additional biologically relevant Ets transcription factors in megakaryocyte-specific gene expression. For example, TEL is another widely expressed pointed domain Ets transcription factor that is involved in megakaryopoiesis.55 While TEL is essential for the establishment of all hematopoietic lineages in the bone marrow,56 a lineage-specific knockout of TEL in mice established a specific role of TEL in megakaryopoiesis.55 Based on the similar phenotypes observed in these TEL knockout megakaryocytes and in the Fli-1–/– megakaryocytes, we speculate that like Fli-1, TEL may function mainly in the late stages of megakaryocyte maturation.17,55
The mechanism by which GABPα and Fli-1 bestow transcriptional synergy on GATA-1 and FOG-1 is unknown. The presence of a pointed domain in both Fli-1 and GABPα suggests that this domain might contribute to this synergy. GABPα can interact via its pointed domain with the transcriptional cofactor and histone acetyltransferase p300.24,57 Thus, it is tempting to speculate that the pointed domain via p300 binding increases GATA-1/FOG-1 activity, perhaps by antagonizing the effects of the FOG-1–associated histone deacetylase–containing repressor complex NuRD.58 However, the pointed domain by itself is not sufficient to mediate transcriptional synergy by GATA-1 and FOG-1 in transiently transfected cells,12 suggesting that additional domains/mechanisms contribute to this activity.
In summary, megakaryopoiesis is controlled through the cooperative actions of tissue-specific and widely expressed transcription factors. Previously, it has been shown that GATA-1 and FOG-1 are central for the regulated expression of megakaryocyte-specific genes and that Fli-1 controls the activity of this complex. We now show that the GABP complex can carry out a similar function. However, the GABP complex appears to control gene expression preferentially during early megakaryopoiesis, while Fli-1 carries out this role predominantly during late stages of megakaryocyte maturation. Developmental stage-specific functions by the GABP complex and Fli-1 are likely a product of binding-site specificities and expression levels. Further definition of the mechanism by which these activators function along with the identification of additional factor-specific target genes will elucidate the transcriptional regulatory network that balances megakaryocyte progenitor proliferation and maturation.
Prepublished online as Blood First Edition Paper, June 6, 2006; DOI 10.1182/blood-2006-04-019760.
Supported by National Institutes of Health (NIH) grants P01 HL40387 (M.P., G.A.B.), P01 CA78582 (G.S., D.K.W.), DK58044 (G.A.B.), and DK54937 (G.A.B.). H.-H.X. and W.J.L. were supported by the Intramural Research Program, National Heart, Lung, and Blood Institute (NHLBI, NIH). L.P., D.K.W., W.J.L., G.A.B., and M.P. designed research; L.P., H.-H.X., G.S., X.W., and Y.W. performed research; K.W. and W.J.L. contributed vital new reagents or analytic tools; and L.P., G.A.B., and M.P. had a major role in writing the paper.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank James Klein at the Medical University of South Carolina for genotyping Fli-1–/– murine embryos, the flow cytometry core facility at the Medical University of South Carolina for Fli-1–/– fetal liver flow cytometry analysis, Dr Katya Ravid at Boston University School of Medicine for the Y10 cells, and Dr Michael Atchinson at the University of Pennsylvania for the PU.11-160 construct.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal