Abstract
Overexpression of CKS1B, a gene mapping within a minimally amplified region between 153 to 154 Mb of chromosome 1q21, is linked to a poor prognosis in multiple myeloma (MM). CKS1B binds to and activates cyclin-dependent kinases and also interacts with SKP2 to promote the ubiquitination and proteasomal degradation of p27Kip1. Overexpression of CKS1B or SKP2 contributes to increased p27Kip1 turnover, cell proliferation, and a poor prognosis in many tumor types. Using 4 MM cell lines harboring MAF-, FGFR3/MMSET-, or CCND1-activating translocations, we show that lentiviral delivery of shRNA directed against CKS1B resulted in ablation of CKS1B mRNA and protein with concomitant stabilization of p27Kip1, cell cycle arrest, and apoptosis. Although shRNA-mediated knockdown of SKP2 and forced expression of a nondegradable form of p27Kip1 (p27T187A) led to cell cycle arrest, apoptosis was modest. Of importance, while knockdown of SKP2 or overexpression of p27T187A induced cell cycle arrest in KMS28PE, an MM cell line with biallelic deletion of CDKN1B/p27Kip1, CKS1B ablation induced strong apoptosis. These data suggest that CKS1B influences myeloma cell growth and survival through SKP2- and p27Kip1-dependent and -independent mechanisms and that therapeutic strategies aimed at abolishing CKS1B function may hold promise for the treatment of high-risk disease for which effective therapies are currently lacking.
Introduction
The molecular lesions causing myeloma initiation and progression are not completely understood. We have previously shown that overexpression of genes mapping to 1q are highly correlated with increased risk of early death in patients diagnosed with multiple myeloma (MM).1 Gains of the q arm of chromosome 1 are one of the most common genetic abnormalities in MM,2 and we and others have shown that tandem duplications and jumping segmental duplications of the chromosome 1q band, resulting from decondensation of pericentromeric heterochromatin, are frequently associated with disease progression.3–5 Using a combination of array-based comparative genomic hybridization (aCGH) and microarray data on cells isolated from newly diagnosed disease, we recently produced a high-resolution map of recurrent gains and losses of DNA in MM.6 Using unsupervised clustering and nonnegative matrix factorization of the aCGH data, we found that hyperdiploid disease could be segregated into 2 groups, one exhibiting trisomies of the odd-number chromosomes and another also exhibiting gains of chromosomes 1q and 7, deletion of chromosome 13, and the absence of trisomy 11.6 This method also identified 2 nonhyperdiploid subtypes: one characterized by high-level amplification of 1q21 and 1q22 and deletions of chromosome 1p and chromosome 13, and another characterized by an absence of chromosome 1 abnormalities but harboring deletions of chromosomes 8 and 13.6 Using interphase fluorescence in situ hybridization (FISH) analysis, we recently showed that while monoclonal gammopathy of undetermined significance (MGUS) lacked evidence of 1q21 gains, 1q21 gains in smoldering MM conferred a higher risk for conversion.2 We also showed that gains of 1q21 were associated with inferior survival of symptomatic MM and that the incidence of this abnormality increased in relapsing disease.2
Given that the q21 band of chromosome 1 represents an amplification hot spot, we hypothesized that genes within this region may confer an aggressive clinical phenotype. Indeed, 2 genes, PSMD4 and CKS1B, overexpressed in a 70-gene high-risk signature,1 map to a minimal common region (MCR) of gain/amplification at chromosome 1q21.1-1q22 with borders at 142.60 Mb and 153.27 Mb.6 These observations suggest that CKS1B amplification and overexpression may contribute to disease progression.
The eukaryotic cell cycle is controlled by cyclin-dependent kinases (Cdks), which are opposed by cyclin-dependent kinase inhibitors (Cdkis).7 Emerging evidence suggests that deregulation of the G1/S transition of the cell cycle is a critical step in plasma cell tumorigenesis, as activation of 1 of the 3 D-type cyclins is a universal event in MM,8 and deletion of chromosome 13, presumably leading to haploinsufficiency of RB1, is also a significant negative prognostic variable.9,10 Loss of p16INK4a expression by deletion or promoter hypermethylation is frequent in MM,11,12 and germ-line loss of Cdkn2a/p16Ink4a predisposes BALB/c mice to plasmacytoma development.13 Mice lacking p18INK4c exhibit severe defects in plasma cell differentiation,14 and homozygous deletion of CDKN4c/p18INK4c is seen in a majority of MM cell lines.15 Finally, we have recently shown that the CDKN1B locus on chromosome 12p can be affected by biallelic microdeletions that result in loss of gene expression in myeloma cell lines.6
CKS1B is a member of the Cks/Suc1 family of small proteins (9-18 kDa) that bind the catalytic subunit of cyclin-dependent protein kinases and regulate their function.16 Homologues from different species share extensive sequence conservation, and the 2 human homologues can functionally substitute for Cks1 in budding yeast. CKS1B has also been identified as an essential accessory protein for the Skp1-Cul1-F-box protein-SKP2 (SCFSkp2) ubiquitin ligase.17
Reduced protein levels of the Cdki, p27Kip1, which regulates Cdk2–cyclin E activity and the late restriction point of the G1 to S transition of the cell cycle, is associated with a poor prognosis in many cancers18 including MM.19 The absence of inactivating mutations in the CDKN1B/p27Kip1 gene has raised speculation that hyperactivation of CKS1B, representing the rate-limiting component of the SCFSkp2-Cks1 ubiquitin ligase,20,21 may lead to inappropriate degradation of p27Kip1.18 Consistent with this hypothesis, elevated expression of CKS1B has been linked to a poor prognosis in oral,22 gastric,23 breast,24 and colon carcinomas.25
In the present study, we show that ablation of CKS1B in 3 independent MM cell lines resulted in stabilization of p27Kip1, cell cycle arrest, and apoptosis. Of importance, while knocking down SKP2 or forced expression of a nondegradable form of p27Kip1 induced cell cycle arrest, these experimental manipulations induced little or no apoptosis in myeloma cell lines. Moreover, knocking out CKS1B in a MM cell line deficient for p27Kip1 also induced cell cycle arrest and apoptosis. Together these data suggest that CKS1B may influence myeloma cell growth and survival through SKP2- and p27Kip1-dependent and -independent mechanisms and that strategies targeting CKS1B function might be exploited therapeutically.
Materials and methods
Patients
Purified plasma cells were obtained from healthy subjects and patients with MM.26 The institutional review board of the University of Arkansas for Medical Sciences approved these research studies, and all subjects provided written informed consent approving use of their samples for research purposes.
Gene expression profiling (GEP) and microarray analysis
Western blotting
Cytosolic and nuclear fractions were isolated using the Nuclear/Cytosol Fractionation Kit (BioVision Research Products, Mountain View, CA). Equal amounts of lysates were separated by electrophoresis on 4% to 12% sodium dodecyl sulfate–polyacrylamide gels, and Western blotting was carried out using the Western Breeze Chemiluminescent Immunodetection protocol, as described (Invitrogen, Frederick, MD). The following primary antibodies were used: anti–cullin 1A, anti-CKS1B, anti-SKP2, and anti–phospo-thr-187-p27Kip1 (Zymed Laboratories, South San Francisco, CA); anti–cleaved caspase-3, anti-PARP, anti–β-tubulin, and anti–Histone 1 (Upstate Biotechnology, Charlottesville, VA); and anti–total p27Kip1 (BD Transduction Laboratories, San Jose, CA).
Expression of CKS1B and SKP2 shRNA and nondegradable p27Kip1
Two synthetic double-stranded oligonucleotide sequences specific for CKS1B (5′-GATCCCCGGACATAGCCAAGCTGGTCTTCAAGAGAGACCAGCTTGGCTATGTCCTTTTTA-3′22 and 5′-GATCCCCTGGAGGAATCTTGGCGTTCTTCAAGAGAGAACGCCAAGATTCCTCCATTTTTA-3′),28 a synthetic double-stranded oligonucleotide sequence specific for SKP2 (5′-GATCCCCGGGAGTGACAAAGACTTTGTTCAAGAGACAAAGTCTTTGTCACTCC CTTTTTA-3′),29 and a nonsense scrambled oligonucleotide (5′-GATCCCCGACACGCGA CTTGTACCACTTCAAGAGAGTGGTACAAGTCGCGTGTCTTTTTA-3′) were obtained from OligoEngine (Seattle, WA). shRNA double-stranded oligonucleotides were cloned into lentiviral pLVTH vectors (kindly provided by Dr Didier Trono, National Center for Competence in Research, Switzerland). Recombinant lentivirus was produced by transient transfection of 293T cells following a standard protocol.30 Briefly, crude virus was concentrated by ultracentrifugation at 9 000 g for 90 minutes. Viral titers were determined by measuring the amount of HIV-1 p24 antigen by enzyme-linked immunosorbent assay (NEN Life Sciences, Boston, MA). A 99% transduction efficiency of MM cells was achieved with 3000 ng lentiviral p24 particles/106 cells. Plasmids containing a nondegradable form of p27Kip1 (p27T187A), which is resistant to SCFSKP2-Cks1–dependent proteasomal degradation, were a generous gift from Dr Michele Pagano (NYU, New York, NY) and Dr Francois X. Claret (University of Texas, M. D. Anderson Cancer Center, Houston, TX). The amplified mutant CDKN1B cDNA sequence was cloned into the pWPI lentiviral vector (gift from Dr Didier Trono). Production and titration of lentiviral stocks were performed using the protocols outlined above.
Cell cycle and apoptosis analysis
Cells (1 × 106) of each sample were fixed in 75% ethanol at −20°C overnight. The following day, the cells were washed with cold PBS, treated with 100 μg RNase A (Qiagen, Hilden, Germany), and stained with 50 μg propidium iodide (Roche, Mannheim, Germany). Flow cytometric acquisition was performed using a 3-color FACScan flow cytometer and CellQuest software (Becton Dickinson, San Jose, CA). For each sample, 10 000 events were gated. Data analysis was performed using Modfit LT (Verity Software House, Topsham, ME). The apoptotic cell fraction was determined as mean ± standard error of the percent of cells of sub-G0 DNA content of 2 independent experiments.
Caspase inhibition assay
To test whether CKS1B inhibition induced apoptosis through caspase activation, JJN3, OCI-MY5, XG-1, and KMS28PE cells infected with CKS1B shRNA were treated with Z-VAD-fmk (R&D Systems, Minneapolis, MN), a pan-caspase inhibitor used at 100 μM after 12 hours of lentivirus infection. Fresh inhibitor was added every day until the end of the experiments. As positive control, myeloma cells were treated with 10 nM of the proteasome inhibitor bortezomib for 24 hours. All experiments were performed in duplicate.
Cell proliferation assay
JJN3, OCI-MY5, XG-1, and KMS28PE cell lines, infected with CKS1B or SKP2 shRNA or p27T187A cDNA, and their controls, were seeded at a density of 0.5 million cells/mL. Cell number and viability were determined by trypan blue exclusion at various time intervals.
Results
CKS1B mRNA and protein expression are inversely correlated with p27Kip1 protein levels in primary disease
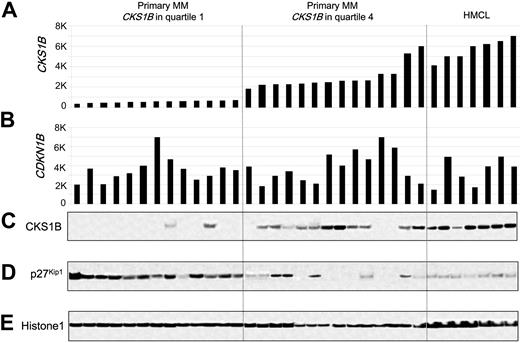
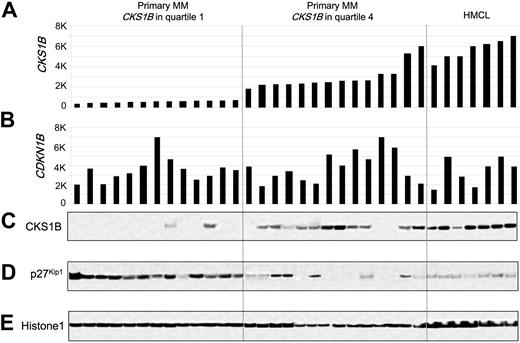
Since CKS1B is known to regulate the ubiquitination and proteasomal degradation of p27Kip1,20,21 we tested the relationship between the expression of CKS1B and p27Kip1 in primary MM. Western blot analysis of nuclear extracts from plasma cells derived from 27 newly diagnosed MM cases and 7 MM cell lines for which microarray data were available revealed a strong, but not absolute, correlation between CKS1B mRNA and protein, with no correlation between CDKN1B mRNA and p27Kip1 protein levels (Figure 1). There was an inverse correlation between CKS1B and p27Kip1 protein levels; however, there were cases that express low or high levels of both p27Kip1 and CKS1B. The variable expression of p27Kip1 seen in Western blot was confirmed by immunohistochemistry (data not shown) and consistent with previous reports.18
CKS1B mRNA and protein expression are inversely correlated with p27Kip1 protein levels in primary disease. (A) CKS1B and (B) CDKN1B (p27Kip1) gene expression signal in 1000-unit increments is plotted on the y-axis. Primary myeloma with CKS1B expression in quartile 1 (n = 13) and quartile 4 (n = 14) and MM cell lines (n = 7) are grouped and plotted from left to right along the x-axis. Each bar represents a sample, and the height indicates the level of gene expression in each sample. (C) CKS1B, (D) p27Kip1, and (E) Histone 1 levels were evaluated by Western blot analysis of nuclear fractions derived from the same aliquots of plasma cells used in panels A-B. Samples are ordered from left to right in the exact same order in all panels.
CKS1B mRNA and protein expression are inversely correlated with p27Kip1 protein levels in primary disease. (A) CKS1B and (B) CDKN1B (p27Kip1) gene expression signal in 1000-unit increments is plotted on the y-axis. Primary myeloma with CKS1B expression in quartile 1 (n = 13) and quartile 4 (n = 14) and MM cell lines (n = 7) are grouped and plotted from left to right along the x-axis. Each bar represents a sample, and the height indicates the level of gene expression in each sample. (C) CKS1B, (D) p27Kip1, and (E) Histone 1 levels were evaluated by Western blot analysis of nuclear fractions derived from the same aliquots of plasma cells used in panels A-B. Samples are ordered from left to right in the exact same order in all panels.
Effects of silencing CKS1B or SKP2 and overexpressing a nondegradable form of p27Kip1 on MM cell growth and survival
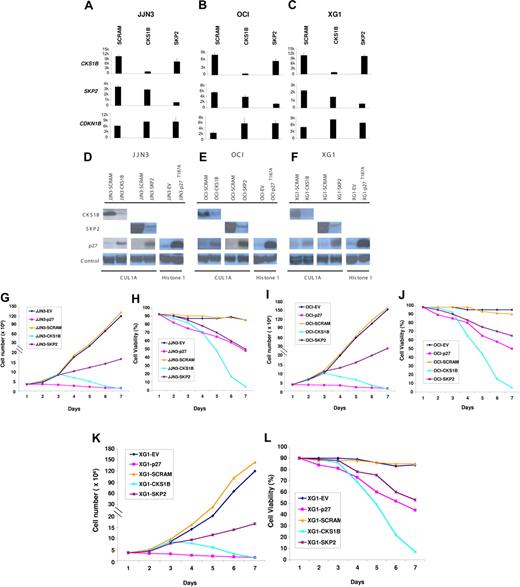
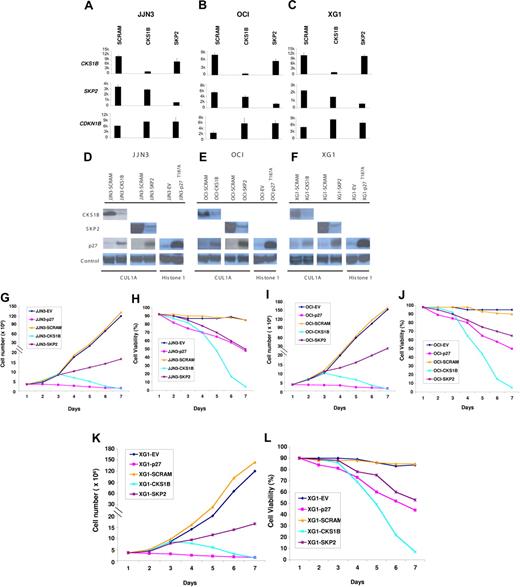
To define a functional role of CKS1B in MM cell growth and survival, we evaluated the effects of a CKS1B shRNA or a nondegradable form of p27Kip1 (p27T187A), resistant to SCFSkp2-Cks1–dependent proteasomal degradation, on the proliferation of the myeloma cell lines JJN3, OCI-MY5, and XG-1. Moreover, to investigate whether CKS1B may have additional functions, which are independent of its role as an accessory protein to SCFSKP2-ubiquitin ligase complex, we also silenced SKP2 and compared the phenotypic effects of silencing each gene.
Separate shRNAs against the CKS1B and SKP2 genes could induce a near-complete ablation of their respective mRNA and protein (Figure 2A-F). Although CKS1B and SKP2 inhibition could similarly increase p27Kip1 mRNA levels, accumulation of p27Kip1 protein was significantly higher in cells expressing SKP2 shRNA (Figure 2A-F). This result could be attributed to the greater effect on p27 protein stabilization induced by silencing SKP2, as determined by measuring p27 half-life in vitro by treatment with cyclohexamide (data not shown). Stable transfection of p27T187A was confirmed by a significant increase of p27Kip1 protein levels on Western blot analysis (Figure 2D-F) compared to the empty vector control (EV). Growth analysis indicated that CKS1B silencing did not inhibit cell proliferation until day 3 after infection (Figure 2G,I,K); nevertheless, on day 5 a dramatic decrease of cell viability could be observed in CKS1B shRNA–expressing cells compared to the cells expressing the scrambled control shRNA (44%-49% vs 86%-93%; P < .05) (Figure 2H,J,L). On day 7, virtually all cells expressing CKS1B shRNA had died, while the controls continued to proliferate. Silencing of SKP2 strongly reduced cell proliferation, but induced only a moderate and gradual decrease in cell viability (Figure 2G-L). Unlike CKS1B and SKP2 shRNA, myeloma cells expressing p27T187A showed a dramatic inhibition in cell proliferation one day after infection. After 3 days, control cells continued to proliferate, while the number of p27T187A cells started to decrease slowly with only 48% to 50% of cells remaining viable on day 7 (Figure 2G-L).
Effects of silencing CKS1B or SKP2 and overexpressing a nondegradable form of p27Kip1 on MM cell growth and survival.CKS1B, SKP2, and CDKN1B (p27Kip1) mRNA expression was evaluated by microarray analysis 5 days after lentiviral infection in (A) JJN3, (B) OCI-MY5, and (C) XG-1 cells expressing a nonspecific scrambled shRNA (SCRAM), CKS1B shRNA, or SKP2 shRNA. All experiments were performed in duplicate, and the results were expressed as the mean ± standard error. Western blot analysis on nuclear fractions of the same aliquots of cells shown in panels A, B, or C confirmed that CKS1B shRNA and SKP2 shRNA efficiently inhibited CKS1B and SKP2 protein expression, respectively, and increased p27T187A levels in (D) JJN3, (E) OCI-MY5, and (F) XG-1. Cullin 1A (Cul1A) expression was used as control for the specificity of CKS1B and SKP2 action. p27T187A overexpression was confirmed in all 3 cell lines 5 days after lentiviral infection, compared to the control cells infected with an empty vector (EV). Histone 1 was used as a loading control. Effects of CKS1B or SKP2 shRNA and p27T187A overexpression on (G,I,K) cell proliferation and (H,J,L) cell viability in (G-H) JJN3, (I-J) OCI-MY5, and (K-L) XG-1. Total number of cells and cell viability were evaluated every day after lentiviral infection by trypan blue exclusion. Error bars represent standard error of the mean for 2 independent experiments.
Effects of silencing CKS1B or SKP2 and overexpressing a nondegradable form of p27Kip1 on MM cell growth and survival.CKS1B, SKP2, and CDKN1B (p27Kip1) mRNA expression was evaluated by microarray analysis 5 days after lentiviral infection in (A) JJN3, (B) OCI-MY5, and (C) XG-1 cells expressing a nonspecific scrambled shRNA (SCRAM), CKS1B shRNA, or SKP2 shRNA. All experiments were performed in duplicate, and the results were expressed as the mean ± standard error. Western blot analysis on nuclear fractions of the same aliquots of cells shown in panels A, B, or C confirmed that CKS1B shRNA and SKP2 shRNA efficiently inhibited CKS1B and SKP2 protein expression, respectively, and increased p27T187A levels in (D) JJN3, (E) OCI-MY5, and (F) XG-1. Cullin 1A (Cul1A) expression was used as control for the specificity of CKS1B and SKP2 action. p27T187A overexpression was confirmed in all 3 cell lines 5 days after lentiviral infection, compared to the control cells infected with an empty vector (EV). Histone 1 was used as a loading control. Effects of CKS1B or SKP2 shRNA and p27T187A overexpression on (G,I,K) cell proliferation and (H,J,L) cell viability in (G-H) JJN3, (I-J) OCI-MY5, and (K-L) XG-1. Total number of cells and cell viability were evaluated every day after lentiviral infection by trypan blue exclusion. Error bars represent standard error of the mean for 2 independent experiments.
Silencing of CKS1B induces apoptosis by activating caspase-3 and PARP
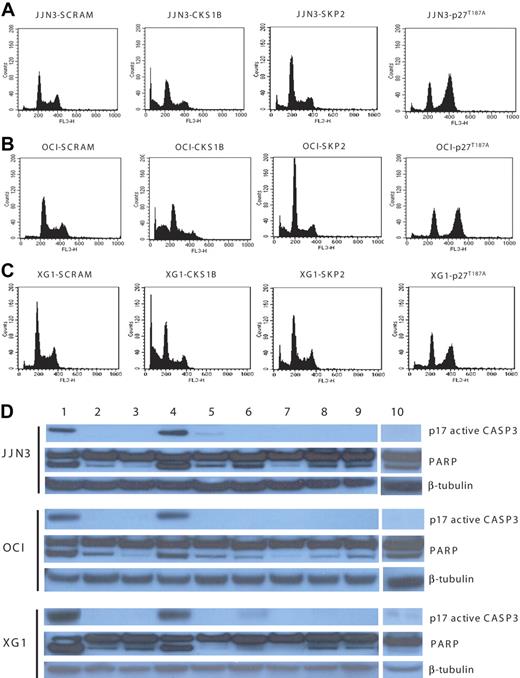
We next investigated whether the distinct effects on MM cell proliferation and cell viability induced by silencing CKS1B and SKP2 and overexpressing p27T187A could be due to their different effects on cell cycle progression and apoptosis.
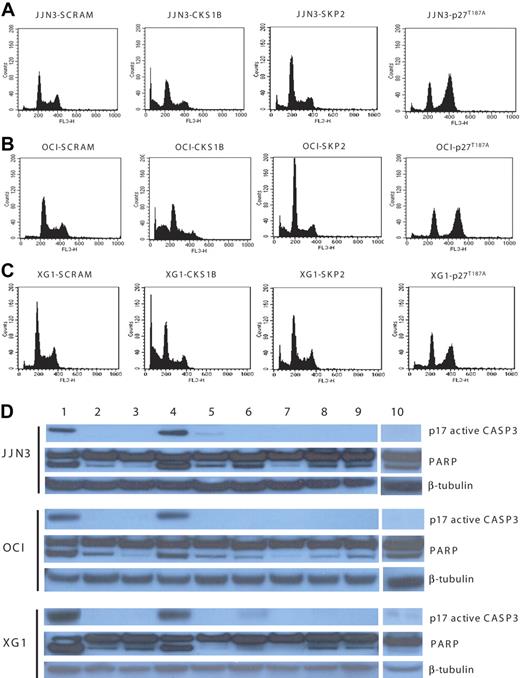
Flow cytometry analysis, performed on day 5, showed that CKS1B shRNA significantly enhanced G0 to G1 phase accumulation (P < .05) and induced strong apoptosis in all 3 myeloma cell lines, compared to controls (P < .05); SKP2 shRNA deregulated the G1-S transition more strongly than CKS1B shRNA, but caused only a moderate increase in the percentage of apoptotic cells. Conversely, the overexpression of p27T187A had no effect on apoptosis, compared to controls, but induced a significant shift to the G2-M phase (P < .05) with a concurrent reduction of cells in S phase (P < .05) (Figure 3A-C; Tables 1,Table 2–3).
Silencing of CKS1B induces apoptosis by activating caspase-3 and PARP. (A) Cell cycle distribution and apoptosis were evaluated by flow cytometry analysis performed 5 days after lentiviral infection in JJN3 cells expressing a scrambled sequence (SCRAM), CKS1B shRNA, SKP2 shRNA, or p27T187A cDNA. Note that silencing of CKS1B induced cell cycle arrest in G0-G1 phase and dramatic increase in the percentage of cells with sub-G0 DNA content (indicative of apoptosis). SKP2 shRNA deregulated the G1-S transition more strongly than CKS1B shRNA, but caused a modest increase of apoptotic cells; the overexpression of p27T187A had no affect on apoptosis, but resulted in a significantly higher percentage of cells in G2-M phase. The same analysis shown in panel A was performed in (B) OCI-MY5 and (C) XG-1. (D) Caspase-3 (active form, p17) and PARP activation was evaluated by Western blot analysis performed in the same aliquots of cells used in panels A-C. The following conditions were analyzed: (1) positive control (myeloma cells treated with bortezomib at 10 nM for 24 hours); (2) positive control + Z-VAD-fmk; (3) negative control (myeloma cells without any treatment); (4) CKS1B shRNA; (5) CKS1B shRNA + Z-VAD-fmk; (6) SCRAM; (7) SCRAM + Z-VAD-fmk; (8) empty vector; (9) p27T187A cDNA; (10) SKP2 shRNA. Note that the activation of caspase-3 and PARP in cells expressing CKS1B shRNA could be abrogated by pretreatment with the pan-caspase inhibitor Z-VAD-fmk, indicating a caspase-dependent mechanism of apoptosis.
Silencing of CKS1B induces apoptosis by activating caspase-3 and PARP. (A) Cell cycle distribution and apoptosis were evaluated by flow cytometry analysis performed 5 days after lentiviral infection in JJN3 cells expressing a scrambled sequence (SCRAM), CKS1B shRNA, SKP2 shRNA, or p27T187A cDNA. Note that silencing of CKS1B induced cell cycle arrest in G0-G1 phase and dramatic increase in the percentage of cells with sub-G0 DNA content (indicative of apoptosis). SKP2 shRNA deregulated the G1-S transition more strongly than CKS1B shRNA, but caused a modest increase of apoptotic cells; the overexpression of p27T187A had no affect on apoptosis, but resulted in a significantly higher percentage of cells in G2-M phase. The same analysis shown in panel A was performed in (B) OCI-MY5 and (C) XG-1. (D) Caspase-3 (active form, p17) and PARP activation was evaluated by Western blot analysis performed in the same aliquots of cells used in panels A-C. The following conditions were analyzed: (1) positive control (myeloma cells treated with bortezomib at 10 nM for 24 hours); (2) positive control + Z-VAD-fmk; (3) negative control (myeloma cells without any treatment); (4) CKS1B shRNA; (5) CKS1B shRNA + Z-VAD-fmk; (6) SCRAM; (7) SCRAM + Z-VAD-fmk; (8) empty vector; (9) p27T187A cDNA; (10) SKP2 shRNA. Note that the activation of caspase-3 and PARP in cells expressing CKS1B shRNA could be abrogated by pretreatment with the pan-caspase inhibitor Z-VAD-fmk, indicating a caspase-dependent mechanism of apoptosis.
Analysis of apoptotic mechanisms showed strong caspase-3 and PARP activation in cells expressing CKS1B shRNA (Figure 3D), without any evidence of caspase-9-activation (data not shown). The caspase-dependent mechanism of apoptosis was further confirmed by pretreating cells expressing CKS1B shRNA with the pan-caspase inhibitor Z-VAD-fmk, which significantly reduced the fraction of cells with sub-G0 DNA content (20.46% ± 1.6% vs 5.7% ± 0.5% in JJN3; 33.3% ± 2.4% vs 8.7% ± 0.9% in OCI-MY5; 31.75% ± 0.8% vs 12.54% ± 0.7% in XG-1; P < .05) and the activation of caspase-3 and PARP (Figure 3D). On the contrary, pretreatment with the specific caspase-9 inhibitor Z-LEHD-fmk did not inhibit CKS1B-induced apoptosis (data not shown). Moreover, cytochrome c release from mitochondria into the cytosol, known to induce caspase-9 activation, could not be observed in CKS1B shRNA–infected cells (data not shown).
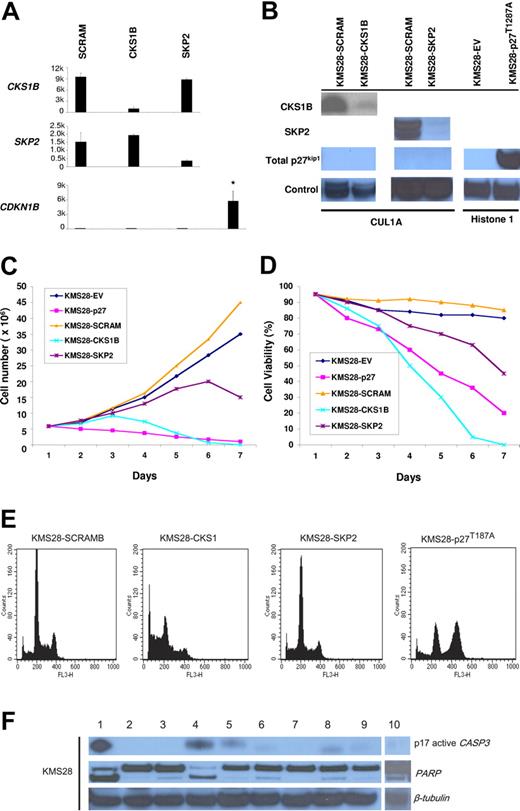
Silencing of CKS1B induces apoptosis in KMS28PE, indicating that CKS1B has a functional role independent of its role in regulating p27Kip1
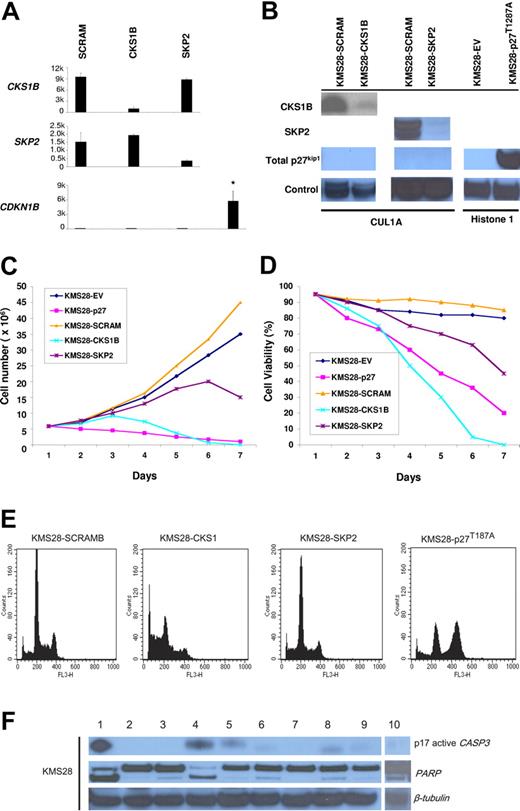
To definitively demonstrate that CKS1B has additional functions in myeloma cells besides regulating p27Kip1 protein stability, we evaluated the effects of introducing CKS1B or SKP2 shRNA on proliferation, cell cycle, and apoptosis of the myeloma cell line KMS28PE, which harbors a biallelic deletion of the CDKN1B locus. As we expected, silencing CKS1B or SKP2, confirmed by microarray (Figure 4A) and Western blot analysis (Figure 4B), could not induce any variation in the expression level of p27Kip1, which remained absent (Figure 4A-B). Growth analysis indicated that silencing CKS1B had dramatic affects on cell viability 4 days after infection, where only 50% of the cells remained alive; on day 6, virtually all KMS28PE-expressing CKS1B shRNA died (Figure 4C-D). Flow cytometry analysis showed a marked increase in the percentage of apoptotic cells (Figure 4E; Table 4), and Western blot results confirmed caspase-3 and PARP activation (Figure 4F). Of interest, no change in cell cycle progression could be observed (Figure 4E; Table 4). Silencing of SKP2 gradually reduced cell proliferation (Figure 4C) and viability (Figure 4D), inducing a moderate G0-G1 cell cycle arrest and a modest increase in the percentage of apoptotic cells (Figure 4E; Table 4). Similarly to the other cell lines, overexpression of p27T187A in KMS28PE (Figure 4B) inhibited cell proliferation (Figure 4C) and induced cell cycle arrest in G2-M phase (Figure 4E; Table 4), without any evidence of apoptosis (Figure 4E-F).
Silencing of CKS1B induces apoptosis independent of p27Kip1 regulation in KMS28PE cell line. (A) CKS1B, SKP2, and CDKN1B (p27Kip1) mRNA expression was evaluated by microarray analysis 5 days after lentiviral infection in KMS28PE cells expressing a nonspecific scrambled shRNA (SCRAM), CKS1B shRNA, or SKP2 shRNA. Note that CDKN1B expression is absent in KMS28PE, compared to the mean expression value of the other cell lines shown in Figure 2A-C. All experiments were performed in duplicate and the results were expressed as the mean ± standard error. (B) Western blot analysis on nuclear fractions of the same aliquots of cells shown in panel A confirmed that CKS1B shRNA and SKP2 shRNA efficiently inhibited CKS1B and SKP2 protein expression, respectively, and did not increase p27Kip1 levels that remained absent. Cullin 1A (Cul1A) expression was used as control for the specificity of CKS1B and SKP2 action. p27T187A overexpression was confirmed in KMS28PE 5 days after lentiviral infection, compared to the control cells infected with an empty vector (EV). Histone 1 was used as a loading control. Effects of CKS1B or SKP2 shRNA and p27T187A overexpression on (C) cell proliferation and (D) cell viability in KMS28PE. (E) Cell cycle distribution and apoptosis were evaluated by flow cytometry analysis performed 5 days after lentiviral infection in KMS28PE cells expressing a scrambled sequence (SCRAM), CKS1B shRNA, SKP2 shRNA, or p27T187A cDNA. Note that silencing of CKS1B induced a dramatic increase in the percentage of cells with sub-G0 DNA content (indicative of apoptosis); SKP2 shRNA caused a modest increase of apoptotic cells, and overexpression of p27T187A had no affect on apoptosis but resulted in a significantly higher percentage of cells in G2-M phase. (F) Caspase-3 (active form, p17) and PARP activation was evaluated by Western blot analysis performed in the same aliquots of cells used in panel E. The following conditions were analyzed: (1) positive control (myeloma cells treated with bortezomib at 10 nM for 24 hours); (2) positive control + Z-VAD-fmk; (3) negative control (myeloma cells without any treatment); (4) CKS1B shRNA; (5) CKS1B shRNA + Z-VAD-fmk; (6) SCRAM; (7) SCRAM + Z-VAD-fmk; (8) empty vector; (9) p27T187A cDNA; (10) SKP2 shRNA. Note that the activation of caspase-3 and PARP in cells expressing CKS1B shRNA could be abrogated by the pretreatment with the pan-caspase inhibitor Z-VAD-fmk, indicating a caspase-dependent mechanism of apoptosis.
Silencing of CKS1B induces apoptosis independent of p27Kip1 regulation in KMS28PE cell line. (A) CKS1B, SKP2, and CDKN1B (p27Kip1) mRNA expression was evaluated by microarray analysis 5 days after lentiviral infection in KMS28PE cells expressing a nonspecific scrambled shRNA (SCRAM), CKS1B shRNA, or SKP2 shRNA. Note that CDKN1B expression is absent in KMS28PE, compared to the mean expression value of the other cell lines shown in Figure 2A-C. All experiments were performed in duplicate and the results were expressed as the mean ± standard error. (B) Western blot analysis on nuclear fractions of the same aliquots of cells shown in panel A confirmed that CKS1B shRNA and SKP2 shRNA efficiently inhibited CKS1B and SKP2 protein expression, respectively, and did not increase p27Kip1 levels that remained absent. Cullin 1A (Cul1A) expression was used as control for the specificity of CKS1B and SKP2 action. p27T187A overexpression was confirmed in KMS28PE 5 days after lentiviral infection, compared to the control cells infected with an empty vector (EV). Histone 1 was used as a loading control. Effects of CKS1B or SKP2 shRNA and p27T187A overexpression on (C) cell proliferation and (D) cell viability in KMS28PE. (E) Cell cycle distribution and apoptosis were evaluated by flow cytometry analysis performed 5 days after lentiviral infection in KMS28PE cells expressing a scrambled sequence (SCRAM), CKS1B shRNA, SKP2 shRNA, or p27T187A cDNA. Note that silencing of CKS1B induced a dramatic increase in the percentage of cells with sub-G0 DNA content (indicative of apoptosis); SKP2 shRNA caused a modest increase of apoptotic cells, and overexpression of p27T187A had no affect on apoptosis but resulted in a significantly higher percentage of cells in G2-M phase. (F) Caspase-3 (active form, p17) and PARP activation was evaluated by Western blot analysis performed in the same aliquots of cells used in panel E. The following conditions were analyzed: (1) positive control (myeloma cells treated with bortezomib at 10 nM for 24 hours); (2) positive control + Z-VAD-fmk; (3) negative control (myeloma cells without any treatment); (4) CKS1B shRNA; (5) CKS1B shRNA + Z-VAD-fmk; (6) SCRAM; (7) SCRAM + Z-VAD-fmk; (8) empty vector; (9) p27T187A cDNA; (10) SKP2 shRNA. Note that the activation of caspase-3 and PARP in cells expressing CKS1B shRNA could be abrogated by the pretreatment with the pan-caspase inhibitor Z-VAD-fmk, indicating a caspase-dependent mechanism of apoptosis.
Discussion
Although sharing virtually all the genetic hallmarks of myeloma, the putative precursor condition, monoclonal gammopathy of undetermined significance (MGUS) rarely converts to overt myeloma,31 and the mechanisms linked to this conversion are currently unknown. MGUS lacks evidence of gains/amplifications of 1q21, but clonal plasma cells from patients with smoldering myeloma harboring this abnormality have a higher risk of converting to overt myeloma than those lacking it.2 Moreover, we recently found that the incidence of, and percentage of cells with, gains/amplification of 1q21 also increase from diagnosis to relapse in patients with symptomatic MM, suggesting that this is a genetic lesion associated with disease progression. We recently used high-resolution aCGH to fine map an amplicon at 1q21 that encompasses the CKS1B locus,6 and in previous studies we also demonstrated that gain of 1q212 and overexpression of CKS1B1 are linked to an inferior survival in patients treated with high-dose therapy and stem cell transplantation. More recent reports have confirmed that both 1q21 gain and increased CKS1B were significantly correlated with reduced survival in patients with MM.32,33
In the current study, we sought to understand the functional role of CKS1B in myeloma growth and survival. As expected, based on its role in regulating p27Kip1 protein turnover, both CKS1B mRNA and protein expression were inversely correlated with the protein levels of its target gene p27Kip1. Silencing of CKS1B mRNA in MM cells led to reduced CKS1B mRNA and protein, accumulation of p27Kip1, and profound growth inhibition. A similar phenotype was observed with 2 independent shRNAs in multiple MM cell lines harboring IGH-mediated translocations, leading to the activation MAF, FGFR3/MMSET, and CCND1. That 2 separate CKS1B shRNAs led to a reduction of both mRNA and protein and a common apoptotic phenotype suggests that the current results are not due to spurious off-target effects. Moreover, the consistency of the phenotypic effects across multiple cell lines would indicate that these effects are not dependent on a specific molecular subtype of the disease. Given that CKS1B and SKP2 function to regulate the SCF ubiquitin ligase, we expected that knocking down either gene might lead to a similar phenotype. While shRNAs to SKP2 led to a dramatic reduction in SKP2 mRNA and protein and a concomitant increase in p27Kip1 protein, it had only modest apoptotic effect. In addition, the overexpression of a nondegradable form of p27Kip1, while inducing a cell cycle arrest, did not induce apoptosis. When CKS1B expression was only partially reduced (> 3000 on Affymetrix microarrays), the apoptotic phenotype as well as p27Kip stabilization were greatly attenuated (data not shown). It is interesting to note that the same CKS1B shRNA used in this study has been previously used to knock down CKS1B in oral carcinomas.22 In these cells, loss of CKS1B led to increased p27Kip1 and growth inhibition but did not induce apoptosis. It is possible that like in myeloma a more pronounced depletion would have induced apoptosis. However, it is possible that myeloma cells growing in suspension are uniquely sensitive to loss of CKS1B.
Taken together, these data suggest that CKS1B has SKP2-independent functions in MM. Perhaps this function is related to its ability to directly bind to and regulate CDK function especially CDK1/cyclin B1.16 It has also been reported that CKS1B has a kinase-independent function in transcription through its ability of recruiting the proteasome to the promoter regions of a subgroup of genes in Saccharomyces cerevisiae.34
Although we have not yet definitively defined the mechanism by which CKS1B loss induces apoptosis in MM, CKS1B ablation induced a mitochondrial-independent programmed cell death involving activation of caspase-3 and PARP that could be rescued by treatment with inhibitors of caspase activation. It is noteworthy that apoptosis could also be inhibited by treatment with IGF-1 but not by IL-6 (data not shown). Knocking down CKS1B levels in myeloma cell lines to those seen in low-risk primary disease (Figure 1) leads to an inability of these cells to survive when grown in suspension. Remarkably, when these same cells are cocultured on osteoclasts, the cells are also resistant to apoptosis (data not shown). Osteoclasts have a powerful survival effect on primary myeloma cells, and this effect is dependent on cell-cell contact.35 It is possible that IGF-1 and cell-cell contact both rescue CKS1B shRNA–mediated apoptosis through activation by AKT. Additional experimental data will be required to confirm this hypothesis.
CKS1B levels are typically high in aggressive primary disease, increase during disease progression, and are uniformly high in myeloma cells lines. Primary disease with low levels of CKS1B is typically associated with a more indolent clinical course and can be characterized as being dependent on microenvironmental cues for survival. In this regard, myeloma cell lines in which CKS1B loss-of-function can be induced by CKS1B shRNA appear to acquire a more benign phenotype, one that is typical of primary disease (ie, rarely survive more than a few days in culture) in that cells cannot survive without support from either IGF-1 or adhesion to osteoclasts. Thus, it is possible that increased CKS1B expression may be a central switch involved in the acquisition of an ability to grow and survive independent of the bone marrow microenvironment. Definitive experimental evidence to support this hypothesis will, at the least, require demonstration that transfection of primary disease with vectors overexpressing CKS1B can impart survival that is not evident in control cells.
Although there are likely to be many genes whose altered expression contributes to disease progression, our data would suggest that increased expression of CKS1B, in part through gains of its genomic locus, leads to increased degradation of p27Kip1 and other unknown events related to cell survival, which may play an important role in tumor cell proliferation, survival, and perhaps drug resistance.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health grants CA55819 (J.D.S., F.Z., B.B.) and CA97513 (J.D.S.) and by the Fund to Cure Myeloma and Peninsula Community Foundation.
National Institutes of Health
Authorship
Contribution: F.Z. designed and performed research, analyzed data, and wrote the paper; S.C. performed research, provided vital reagents, and wrote the paper; J.P.S. and X.W. performed research; B.C. provided critical reagents; M.K. aided in experimental design; B.B. provided critical assessment of the work; J.D.S. designed research, analyzed data, and wrote the paper. F.Z., S.C., and J.D.S. contributed equally to this work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: John D. Shaughnessy Jr, Donna D. and Donald M. Lambert Laboratory of Myeloma Genetics at the Myeloma Institute for Research and Therapy, University of Arkansas for Medical Sciences, 4301 W. Markham St, Slot 776, Little Rock, AR 72205; e-mail: shaughnessyjohn@uams.edu; or Fenghuang Zhan, Donna D. and Donald M. Lambert Laboratory of Myeloma Genetics at the Myeloma Institute for Research and Therapy, University of Arkansas for Medical Sciences, 4301 W Markham St, Slot 776, Little Rock, AR 72205; e-mail: zhanfenghuang@uams.edu.