Abstract
Human telomerase hTERC RNA serves as a template for the catalytic hTERT protein to synthesize telomere repeats at chromosome ends. We have recently shown that some patients with bone marrow failure syndromes are heterozygous carriers for hTERC or hTERT mutations. These sequence variations usually lead to a compromised telomerase function by haploinsufficiency. Here, we provide functional characterization of an additional 8 distinct hTERT sequence variants and 5 hTERC variants that have recently been identified in patients with dyskeratosis congenita (DC) or aplastic anemia (AA). Among the mutations, 2 are novel telomerase variants that were identified in our cohort of patients. Whereas most of the sequence variants modulate telomerase function by haploinsufficiency, 2 hTERC variants with sequence changes located within the template region appear to act in a dominant-negative fashion. Inherited telomerase gene mutations, therefore, operate by various mechanisms to shorten telomere lengths, leading to limited marrow stem cell reserve and renewal capacity in patients with hematologic disorders.
Introduction
Telomerase is a specialized reverse transcriptase (RT) that adds long, repetitive stretches of simple telomeric DNA sequence (ie, TTAGGG in the vertebrates) onto chromosomal termini.1 This cellular RT protein (TERT) copies a short stretch of nucleotides located within the template region of an integral RNA component (TERC) into telomeric DNA repeats.1 Vertebrate TERCs are believed to adopt a complex, folded secondary structure2 as depicted for human TERC (hTERC) in Figure 1A. We have recently conducted extensive site-directed mutagenesis analysis of hTERC to show that much of the structure folded as predicted.3 As is true of many biologically active RNA molecules, most of the internally base-paired regions of hTERC can be extensively mutated without loss of function, provided that the normal base-pairing pattern is preserved.3 However, at certain locations, especially of the single-stranded template region that is copied into telomeric DNA, specific RNA base sequences have been shown to be required for biologic activity.4
Telomerase catalytic proteins (TERTs) from evolutionary distant organisms share a conserved structural organization that can be divided into 3 functional domains (Figure 1C).5 The telomerase-specific domains exist at both the N and C termini of TERTs that are not present in any of the viral RTs.6 The N-terminal region is required to participate in enzymatic function,7,8 in assembly of the protein with its integral hTERC RNA component,7,9 and in the homodimerization of the protein (ie, hTERT protein-protein interaction),7,9,10 whereas the C-terminal domain is required for telomerase-specific activity other than its catalytic function11,12 as well as in the telomeric nucleotide addition processivity process.13-15 The functional RT domain with the universally conserved RT motifs is almost centrally located in the protein primary sequence (Figure 1C). The fact that mutations of key residues that are known to affect its conventional RT catalytic activity also negatively influence telomerase activity strongly argues that telomerase RT domain is the catalytic domain of the enzyme complex.13,15-18
hTERT variant sequences and hTERC variants.
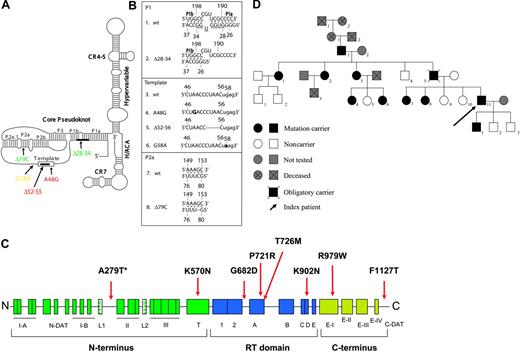
(A) Schematic depiction of the predicted secondary structure of hTERC as proposed by Chen et al.2 The 11-base template sequence (rectangle) and other structural features are indicated, including the core pseudoknot, CR4-CR5, box H/ACA, and CR7 domains, and the hypervariable paired region. Naturally occurring sequence variations of hTERC are indicated in green for those identified in patients with AA, in red for those found in patients with DC, and in yellow for the G58A variant identified in both healthy individuals and patients. Thick black lines indicate nucleotide deletions. (B) Primary sequences located at the sites of the natural hTERC sequence variations. Nucleotide changes are indicated in boldface, and deletions are dashed or underlined. (C) Linear representation of the hTERT protein with the conserved regions (boxed) located throughout the N-terminal, RT, and C-terminal domains indicated. The disease-associated hTERT sequence variations are indicated in black, whereas the A279T variant that has also been identified in healthy individuals are shown with an asterisk. Note the 2 “DAT” domains (telomerase function that can dissociate from its enzymatic activity) located in both the N- and C-termini of the gene (ie, the N-DAT and C-DAT motifs). (D) Pedigree of a patient with the novel hTERT K570N mutation. The proband (V-11) is indicated with an arrow.
hTERT variant sequences and hTERC variants.
(A) Schematic depiction of the predicted secondary structure of hTERC as proposed by Chen et al.2 The 11-base template sequence (rectangle) and other structural features are indicated, including the core pseudoknot, CR4-CR5, box H/ACA, and CR7 domains, and the hypervariable paired region. Naturally occurring sequence variations of hTERC are indicated in green for those identified in patients with AA, in red for those found in patients with DC, and in yellow for the G58A variant identified in both healthy individuals and patients. Thick black lines indicate nucleotide deletions. (B) Primary sequences located at the sites of the natural hTERC sequence variations. Nucleotide changes are indicated in boldface, and deletions are dashed or underlined. (C) Linear representation of the hTERT protein with the conserved regions (boxed) located throughout the N-terminal, RT, and C-terminal domains indicated. The disease-associated hTERT sequence variations are indicated in black, whereas the A279T variant that has also been identified in healthy individuals are shown with an asterisk. Note the 2 “DAT” domains (telomerase function that can dissociate from its enzymatic activity) located in both the N- and C-termini of the gene (ie, the N-DAT and C-DAT motifs). (D) Pedigree of a patient with the novel hTERT K570N mutation. The proband (V-11) is indicated with an arrow.
Inherited mutations in both hTERC RNA and hTERT protein underlie rare bone marrow failure syndromes, autosomal dominant dyskeratosis congenita (DC) and acquired aplastic anemia (AA).19-21 DC is characterized by abnormal skin pigmentation, nail dystrophy, and oral leukoplakia and is often complicated by life-threatening bone marrow failure and immunodeficiency.22 Lymphocytes from patients show decreased hTERC expression, decreased telomerase activity, and significantly reduced telomere lengths as compared to those from age-matched healthy individuals, owing to an inherited mutation in the TERC gene.23,24 Surveys of patients with apparently acquired AA have revealed an assortment of sequence variations not only in hTERC RNA3,25-28 but also in the hTERT protein.20,21,29 We and others have tested many of the reported disease-associated hTERC or hTERT alleles for their ability to support telomerase biologic activity and found them at least partially defective.3,20,24,30,31 Because most patients are heterozygous carriers for the hTERC or hTERT allele, all of the sequence variants tested to date reduce wild-type telomerase enzymatic function by haploinsufficiency. However, because telomerase sequence variants widely modulate enzymatic activity, some of the sequence changes may inhibit the wild-type copy in a dominant-negative fashion. Therefore, in this study, we systematically examined the functional properties of 9 additional disease-associated variants of TERC and TERT that have recently been reported, and a novel hTERC as well as a novel hTERT variant that was recently identified in 2 separate patients in our cohort. Whereas most of the variants moderate telomerase enzymatic function by haploinsufficiency, as has been previously observed, we describe for the first time 2 hTERC sequence changes that appear to inhibit wild-type telomerase enzymatic activity in a dominant-negative fashion. Our data, therefore, suggest that natural sequence variations in telomerase gene components can operate by different mechanisms to cause enzymatic dysfunction that leads to telomere shortening and ultimately a reduced replicative potential of marrow stem cells.
Patients, materials, and methods
Cloning of novel hTERC Δ28-34 and hTERT K570N variants
Peripheral blood leukocytes were collected from patients with acquired AA and some of their relatives, all of whom had given informed consent according to protocols approved by the Institutional Review Board of the National Heart, Lung, and Blood Institute. The TERC and TERT genes were amplified as described previously.20,26 The disease-associated mutations were introduced into the appropriate expression vectors (ie, pcDNA3-hTERC, pcI-3HA-hTERT, or pcI-3FLAG-hTERT) as previously described.3
Telomere length measurements
In vivo reconstitution of telomerase activity
Wild-type or mutant pcDNA3-hTERC DNAs (2 μg) were transfected into VA13+hTERT cells (at approximately 70% confluency) in 6-well polystyrene dishes using SuperFect transfection reagent (Qiagen, Valencia, CA) according to the manufacturer's instructions. Similarly, wild-type or mutant pcI-3HA-hTERT DNAs (2 μg) were transfected into VA13+hTERC cells. Whereas VA13+hTERC cells were engineered to constitutively express a wild-type version of the hTERC RNA, the VA13+hTERT cells would constitutively express wild-type hTERT protein.20,24 In the cases where both the wild-type and mutated versions of the gene were coexpressed in cells, they (each at 1 μg) were cotransfected. Transfection efficiency was monitored by scoring for green fluorescent protein expression under confocal microscopy in parallel transfection reactions supplemented with the reporter vector pEGFP-N1 (Stratagene, La Jolla, CA). Approximately 48 hours after transfection, cells were scraped from the dish in the presence of 1 mL cold phosphate-buffered saline. Cellular extracts were then prepared in 1 × CHAPS lysis buffer as suggested by the manufacturer (Chemicon International, Temecula, CA). Telomerase activity of the cellular extract from 2 × 104 cells was assayed using the TRAPeze telomerase detection kit following the manufacturer's directions (Chemicon International), except that polymerase chain reaction (PCR) was performed as follows: 95°C for 2 minutes, 25 cycles of 94°C for 10 seconds, 50°C for 30 seconds, 72°C for 30 seconds, and 72°C for 5 minutes. Products were analyzed on a 12% native polyacrylamide gel and examined by phosphor imaging (Molecular Dynamics, Piscataway, NJ). Cellular lysates were prepared from 2 × 104 primary lymphocytes collected from a patient with the hTERT K570N variant and a healthy age-matched person and subjected to telomere repeats amplification protocol (TRAP) analysis as outlined in this section.
In vitro reconstitutions of telomerase activity using rabbit reticulocyte lysates
In vitro telomerase reconstitution assay was carried out as described.4 Briefly, the TNT quick-coupled transcription-translation system (Promega, Madison, WI) was used to synthesize hTERT protein from the pcI-3FLAG-hTERT vector in the presence of 100 ng and 10 ng of the various in vitro transcribed and gel-purified hTERC RNAs. One microliter of the reconstituted lysates was used in the TRAP assay with the Cx-ext reverse primer34 and the standard TS forward primer35 as described.4
Northern blotting analysis
Wild-type or mutant pcDNA3-hTERC vectors (6 μg) were transfected into VA13+hTERT cells (at approximately 70% confluency) in 100-mm polystyrene dishes using SuperFect transfection reagent as described (see “In vivo reconstitution of telomerase activity”). Approximately 48 hours after transfection, TRIzol reagent was used to extract total cellular RNA as suggested by the manufacturer (Invitrogen, Carlsbad, CA). Similarly, total cellular RNA was extracted from primary cells (at ∼106 cells) of the hTERT K570N carrier and a healthy individual. Northern blot analysis was performed essentially as described.36
Immunoprecipitation–Northern blotting analysis
FLAG-tagged hTERT protein was expressed in vitro from the pcI-3FLAG-hTERT vector using the TNT quick-coupled transcription-translation system (Promega) in the presence of 100 ng in vitro transcribed full-length hTERC RNA of either the wild-type or mutated sequences at 37°C for 2 hours. In the cases where the different hTERT mutants (in pcI-3FLAG-hTERT plasmid backbone) were used, 1 μg of the wild-type full-length hTERC RNA was used in each of the reactions. The resulting telomerase complexes were affinity enriched on anti-FLAG agarose beads (Sigma, St Louis, MO). To detect hTERT-bound telomerase RNAs, Northern blotting was performed on the enriched telomerase preparations as described in our previous report.3
Western blotting analysis
Wild-type or mutant pcI-3HA-hTERT vectors (2 μg) were transfected into VA13+hTERC cells (at approximately 70% confluency) in 6-well polystyrene dishes using SuperFect transfection reagent as described (see “In vivo reconstitution of telomerase activity”). Approximately 48 hours after transfection, cellular extracts were prepared in SDS lysis buffer. Western blot analysis was performed using the commercially available anti-HA (Covance, Madison, WI) and anti–β-actin (Santa Cruz Biotechnology, Santa Cruz, CA) antibodies as suggested by the manufacturers.
Results
This study focused on 8 distinct hTERT variant sequences and 5 hTERC variants that had recently been identified in patients with DC or AA (Figure 1).21,28,29 These sequence changes and the associated clinical and laboratory findings are summarized briefly in Table 1. Except for the telomerase sequence polymorphisms that were observed in healthy controls as well as in patients with AA (ie, hTERC G58A and hTERT A279T),20,21,37 the remaining 11 nucleotide sequence changes were each detected only in affected individuals and their relatives (Figure 1; Table 1).21,29,38 The latter group includes a novel hTERC Δ28-34 (a deletion of nucleotides 28-34) and hTERT K570N that we recently discovered in 2 unrelated patients, who had otherwise typical AA. These 2 novel mutations were not identified among 528 healthy persons of different ethnic backgrounds analyzed in our previous study.20
Clinical presentation and family history of patients with novel telomerase mutations
The patient with the novel hTERC Δ28-34 deletion is a 28-year-old man with a 13-year history of AA and early hair graying. He first presented to us as he was undergoing G-CSF therapy (300 μg every other day) and experienced pancytopenia (hemoglobin, 101 g/L [10.1 g/dL]; absolute neutrophil count, 1.4 × 109/L [1400/μL]; and platelet count, 44 × 109/L [44 000/μL]) and severe marrow aplasia (< 10% cellularity) but with a normal karyotype. He failed to respond to 2 courses of immunosuppression and growth factors. However, he later responded to androgens (400 mg/wk, maintaining safe levels of neutrophil counts without G-CSF administration, and reducing transfusion requirements. Family history of hematologic dyscrasias was also evident. His father had been previously diagnosed with chronic idiopathic thrombocytopenic purpura and died of pulmonary fibrosis and fungal sepsis. Necropsy results revealed both liver cirrhosis and pulmonary fibrosis consistent with α1-antitrypsin deficiency; however, no specific test was performed to confirm the diagnosis. The bone marrow was hypoplastic, at the time attributed to cimetidine administration. His brother also was diagnosed with hypoplastic anemia. Eight years prior to his presentation, his paternal uncle was diagnosed with myelodysplastic syndrome rapidly evolving to death. No information on mutational status of the family members was available.
The patient with the novel hTERT K570N is a 26-year-old man with a 10-year history of severe AA (5% bone marrow cellularity) and is unresponsive to immunosuppressive therapy. A family history of blood diseases was also evident (Figure 1D). His father (IV-5) had myelodysplastic syndrome evolving to acute myeloid leukemia. His paternal aunt (IV-2) has had AA for decades and is effectively responsive to androgen therapy. His paternal great-grandmother (I-2) also died of a blood disorder. Genetic analysis showed that the mutation is present in his paternal but not maternal relatives, making his father an obligatory carrier of the K570N mutation (Figure 1D). Other family members carrying the mutation also presented with macrocytosis but with the absence of cytopenias (Table 2). Telomere shortening in both neutrophils and lymphocytes strongly correlated with the mutational status of members of the family (Table 2). Although liver disease also seems to be associated with this family (paternal aunt [IV-1], and not shown on the pedigree are a fourth cousin, 3 times removed from the index patient, and a third cousin, twice removed from the index case), the mutation could only be associated with the aunt (Table 2, IV-1), but not with the descendants of these 2 cousins. No evidence of nail dystrophy, leukoplakia, or skin hyperpigmentation was observed in any of the hTERT K570N carriers. Although the index patient and some of his relatives showed early hair graying, this characteristic did not track with mutational status, suggesting that telomerase insufficiency might not be associated with this phenotype in this family. All of the patients whose telomerase variant alleles were the subject of this study were heterozygous carriers.
Two hTERT disease-associated sequence changes located within the conserved catalytic domain abolish telomerase activity
Because half of the number of hTERT natural sequence variants analyzed here (4 of 8) localized to the catalytic RT domain (Figure 1C), we first determined whether any of them could modulate telomerase enzymatic function in a telomerase reconstitution assay. This assay makes use of a human lung fibroblast (VA13+hTERC) cell line,20 which normally expresses only the wild-type copy of the telomerase hTERC RNA, and hence lacks telomerase enzymatic function. However, upon transfection of these cells with a vector that expresses a wild-type copy of the hTERT catalytic protein, telomerase enzymatic activity can be reconstituted and detected as PCR products of 6-bp telomeric DNA ladders using the TRAP protocol.
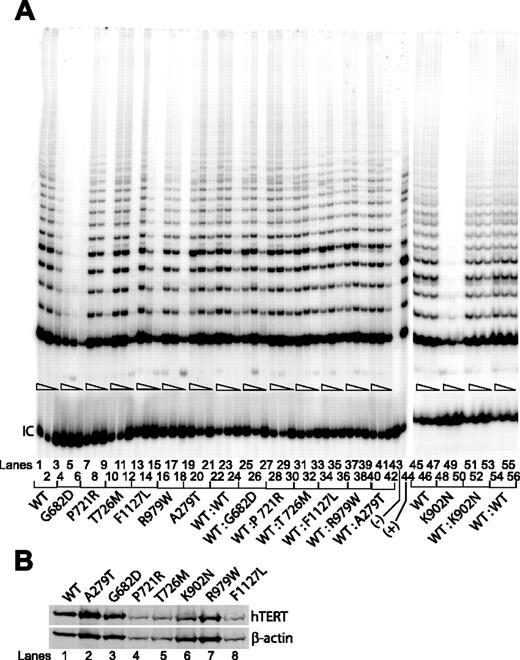
We tested lysates prepared from cells that were transfected with the individual hTERT variants, 4 of which involved residues located within the RT domain (G682D, P721R, T726M, K902N), a novel K570N variant located within the telomerase-specific so-called T domain, and 2 others (R979W and F1127L) located within the telomerase C-terminal extension domain (Figure 1C). Except for K902N,38 the functional significance of these other variants has yet to be demonstrated. We found that only 2 of the sequence changes located within the RT domain (G682D and K902N) abolished telomerase function (Figure 2A lanes 4-6 and 48-50), each yielding less than 1% of the catalytic activity achieved by the wild-type hTERT (lanes 1-3 and 45-47). The other RTs (ie, P721R and T726M) showed as much telomerase activities (lanes 7-12) as did those with either the wild-type hTERT (lanes 1-3) or the known A279T polymorphic sequence20 (lanes 19-21). Similarly, both of the remaining variants located within the C-terminal domain of hTERT (ie, R979W and F1127L) showed little to no reduction in telomerase enzymatic activities (lanes 13-18). We also tested a novel hTERT K570N found recently in an AA patient in our cohort (Table 1). The average telomere length in peripheral lymphocytes collected from this patient was shorter than that of age- and sex-matched individuals (3.1 kb versus 7.5 kb), suggesting that this sequence change was involved in the disease pathogenesis. Indeed, when reconstituted in VA13+hTERC cells, this hTERT K570N mutant drastically reduced telomerase enzymatic activity (Figure 3A, lanes 4-6). None of the hTERT sequence changes tested here altered its protein expression level or stability in Western blotting analysis in transfected cells (Figures 2B and 3B). These data, summarized in Table 1, suggest that only 3 of the disease-associated hTERT protein variants tested in this study, all of which are located within the known functional domains of telomerase (ie, the RT and the telomerase-specific T motif; Figure 1C), drastically reduced levels of telomerase enzymatic function under our assay conditions.
Telomerase enzymatic activities as determined in VA13+hTERC cells for naturally occurring hTERT variants.
(A) Representative TRAP gels showing the relative telomerase enzymatic activities obtained from the substitution mutations and the wild-type hTERT constructs either individually transfected (lanes 1-21, 45-50) or cotransfected (lanes 22-42, 51-56). Serial 5-fold dilutions of the transfected cell lysates (indicated by triangles) were assayed for each sample to ensure linearity of the PCR-based TRAP assay. Lane 43 shows a negative control composed of wild-type cell lysate denatured at 95°C for 5 minutes prior to assaying. Lane 44 shows PCR products amplified from the non-hTERC control TSR8 DNA template supplied in the TRAP kit. IC indicates PCR products amplified from an unrelated DNA template, which is included as an internal control for PCR amplification efficiency in each reaction. (B) Western blot of wild-type and various natural hTERT variants isolated from transfected cell lysates and probed with an anti-HA probe against the N-terminal HA tag at each of the hTERT constructs.
Telomerase enzymatic activities as determined in VA13+hTERC cells for naturally occurring hTERT variants.
(A) Representative TRAP gels showing the relative telomerase enzymatic activities obtained from the substitution mutations and the wild-type hTERT constructs either individually transfected (lanes 1-21, 45-50) or cotransfected (lanes 22-42, 51-56). Serial 5-fold dilutions of the transfected cell lysates (indicated by triangles) were assayed for each sample to ensure linearity of the PCR-based TRAP assay. Lane 43 shows a negative control composed of wild-type cell lysate denatured at 95°C for 5 minutes prior to assaying. Lane 44 shows PCR products amplified from the non-hTERC control TSR8 DNA template supplied in the TRAP kit. IC indicates PCR products amplified from an unrelated DNA template, which is included as an internal control for PCR amplification efficiency in each reaction. (B) Western blot of wild-type and various natural hTERT variants isolated from transfected cell lysates and probed with an anti-HA probe against the N-terminal HA tag at each of the hTERT constructs.
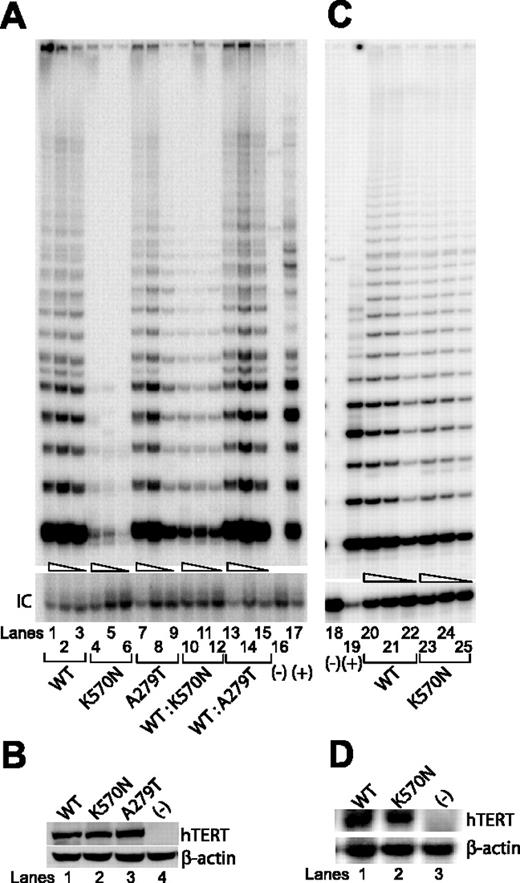
Telomerase enzymatic activities of additional naturally occurring hTERT variants as determined in VA13+hTERC cells or in primary cells.
(A) Representative TRAP gels showing the relative telomerase enzymatic activities obtained from the substitution mutations and the wild-type hTERT constructs either individually transfected (lanes 1-9) or cotransfected (lanes 10-15). Serial 5-fold dilutions of the transfected cell lysates (indicated by triangles) were assayed for each sample to ensure linearity of the assay. Lanes 16 and 18 show negative controls composed of cell lysate denatured at 95°C for 5 minutes prior to assaying. Lanes 17 and 19 show PCR products amplified from the non-hTERC control TSR8 DNA template supplied in the TRAP kit. IC indicates PCR products amplified from an unrelated DNA template, which is included as an internal control for PCR amplification efficiency in each reaction. (B) Western blot of wild-type and various natural hTERT variants isolated from transfected cell lysates and probed with an anti-HA probe against the N-terminal HA tag at each of the hTERT constructs. (C) Telomerase enzymatic assay of cell lysates prepared from about 106 primary lymphocytes collected from a patient with the hTERT K570N variant (lanes 23-25) or a healthy age-matched individual (lanes 20-22). (D) Northern blot analysis of hTERT gene expression in lymphocytes of a patient with the hTERT K570N variant (lane 2), a healthy age-matched individual (lane 1), or in the VA13+hTERC cells (lane 3) that is known to lack hTERT gene expression.
Telomerase enzymatic activities of additional naturally occurring hTERT variants as determined in VA13+hTERC cells or in primary cells.
(A) Representative TRAP gels showing the relative telomerase enzymatic activities obtained from the substitution mutations and the wild-type hTERT constructs either individually transfected (lanes 1-9) or cotransfected (lanes 10-15). Serial 5-fold dilutions of the transfected cell lysates (indicated by triangles) were assayed for each sample to ensure linearity of the assay. Lanes 16 and 18 show negative controls composed of cell lysate denatured at 95°C for 5 minutes prior to assaying. Lanes 17 and 19 show PCR products amplified from the non-hTERC control TSR8 DNA template supplied in the TRAP kit. IC indicates PCR products amplified from an unrelated DNA template, which is included as an internal control for PCR amplification efficiency in each reaction. (B) Western blot of wild-type and various natural hTERT variants isolated from transfected cell lysates and probed with an anti-HA probe against the N-terminal HA tag at each of the hTERT constructs. (C) Telomerase enzymatic assay of cell lysates prepared from about 106 primary lymphocytes collected from a patient with the hTERT K570N variant (lanes 23-25) or a healthy age-matched individual (lanes 20-22). (D) Northern blot analysis of hTERT gene expression in lymphocytes of a patient with the hTERT K570N variant (lane 2), a healthy age-matched individual (lane 1), or in the VA13+hTERC cells (lane 3) that is known to lack hTERT gene expression.
Because these hTERT variants were identified in individuals who are heterozygous carriers for the gene, an altered allele might modulate normal telomerase function through either a haploinsufficiency or dominant-negative mechanism. To address these possibilities, we performed TRAP assays on cell lysates prepared from VA13+hTERC cells that had been cotransfected with plasmids expressing the wild-type hTERT sequence and the individual disease-associated variants. As shown in Figures 2A and 3A, little to no effects were observed between cells that carried only the wild-type hTERT vector and those that carried both the wild-type and the individual mutated copy (Figures 2A, lanes 22-42, and 3A, lanes 10-15), consistent with the hypothesis that the natural variants functioned in a haploinsufficient manner to modulate wild-type telomerase function.20,24,30 To confirm these findings in patient tissue, we prepared primary T-cell extracts collected from the hTERT K570N carrier and an age-matched healthy individual. TRAP analysis of the patient's extract showed only a slightly reduced telomerase enzymatic function as compared to that of the healthy individual (Figure 3C), despite the fact that the hTERT transcripts were expressed at similar levels in both cell extracts (Figure 3D). Taken together, these observations suggest that haploinsufficiency also may operate in patient's tissue, as having only half the amount of functional hTERT protein, and hence, telomerase enzymatic function led to telomere shortening effect (Table 1).
Mutations of essential template region of hTERC RNA abolish telomerase function
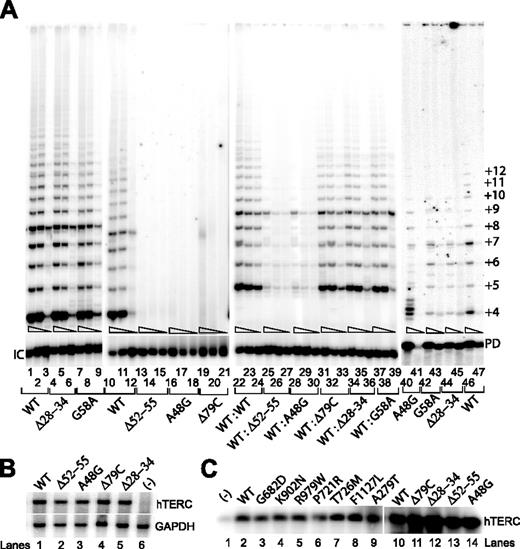
We next examined the properties of 3 separate hTERC RNA sequences that have recently been reported in patients28 (Figure 1; Table 1) and a fourth hTERC variant (ie, Δ28-34) that was identified in a patient with AA in our cohort. The average telomere length in peripheral blood leukocytes collected from the patient with the Δ28-34 mutation was significantly shorter than age- and sex-matched control individuals (4.0 kb versus 7.2 kb; Table 1), suggesting that this deletion in the hTERC sequence might play an important role in telomerase function or telomere maintenance or both. We transfected the individual hTERC variants into VA13+hTERT24 cells to reconstitute telomerase activities. Surprisingly, we found that deletion of nucleotides 28-34 of the hTERC RNA (ie, the Δ28-34 variant) produced similar telomerase enzymatic activity (Figure 4A, lanes 4-6) as compared to cell lysate with either the wild-type hTERC (lanes 1-3) or the known G58A sequence polymorphism (lanes 7-9). On the contrary, other variants, which are located within either the template region (Δ52-55 and A48G) or in the known P2a paired region (Δ79C) effectively abolished telomerase enzymatic function (lanes 13-21), consistent with prior observations that both the template's primary sequence and the secondary structural formation of the P2a stem loop to be absolutely required for optimal telomerase enzymatic function.3,4,39 Our results were not attributable to differences in hTERC synthesis, processing, or stability, because Northern blotting verified that each construct produced comparable steady-state levels of hTERC expression in transfected cells (Figure 4B).
Telomerase enzymatic activity in VA13+hTERT cells expressing various hTERC natural variants.
(A) Representative TRAP gels showing the relative telomerase enzymatic activities obtained from the hTERC variants either individually transfected (lanes 1-21) or cotransfected (lanes 22-39). Serial 5-fold dilutions of the transfected cell lysates (indicated by triangles) were assayed for each sample to ensure linearity of the assay. Lanes 40-47 indicate telomerase enzymatic activity reconstituted in vitro using the rabbit reticulocyte lysate as previously described.4 The smallest telomeric band (+4) represents the initial repeat of 4. Repeats shorter than 4 are not observed on the gel due to the length and design of the Cx-ext reverse primer used in the reaction. Primer dimer (PD) PCR products are also observed due to the partial complementarity of the Cx-ext reverse primer and the TS forward primer used in the assay. (B) Northern blot analysis of naturally occurring hTERC sequence variants expressed in transfected VA13+hTERT cells. Lane 6 shows RNA prepared from cells that were transfected with the pcDNA3.1 vector lacking the hTERC coding sequence. Cellular GAPDH mRNA (lower blot) was assayed in parallel using random-primed probe, which is specific to a specific region of the GAPDH gene. (C) Northern blotting analysis of affinity-enriched telomerase complexes assembled in vitro using full-length wild-type hTERC RNA with the selected hTERT sequences (lanes 2-9), or using the full-length wild-type hTERC or its mutant forms with a wild-type copy of the hTERT protein (lanes 10-14). Telomerase RNA-protein complexes were first assembled in the rabbit reticulocyte lysates. The negative control (lane 1) was a lysate that received no hTERT expression vector.
Telomerase enzymatic activity in VA13+hTERT cells expressing various hTERC natural variants.
(A) Representative TRAP gels showing the relative telomerase enzymatic activities obtained from the hTERC variants either individually transfected (lanes 1-21) or cotransfected (lanes 22-39). Serial 5-fold dilutions of the transfected cell lysates (indicated by triangles) were assayed for each sample to ensure linearity of the assay. Lanes 40-47 indicate telomerase enzymatic activity reconstituted in vitro using the rabbit reticulocyte lysate as previously described.4 The smallest telomeric band (+4) represents the initial repeat of 4. Repeats shorter than 4 are not observed on the gel due to the length and design of the Cx-ext reverse primer used in the reaction. Primer dimer (PD) PCR products are also observed due to the partial complementarity of the Cx-ext reverse primer and the TS forward primer used in the assay. (B) Northern blot analysis of naturally occurring hTERC sequence variants expressed in transfected VA13+hTERT cells. Lane 6 shows RNA prepared from cells that were transfected with the pcDNA3.1 vector lacking the hTERC coding sequence. Cellular GAPDH mRNA (lower blot) was assayed in parallel using random-primed probe, which is specific to a specific region of the GAPDH gene. (C) Northern blotting analysis of affinity-enriched telomerase complexes assembled in vitro using full-length wild-type hTERC RNA with the selected hTERT sequences (lanes 2-9), or using the full-length wild-type hTERC or its mutant forms with a wild-type copy of the hTERT protein (lanes 10-14). Telomerase RNA-protein complexes were first assembled in the rabbit reticulocyte lysates. The negative control (lane 1) was a lysate that received no hTERT expression vector.
Mutations in neither hTERC RNA nor hTERT protein disrupt telomerase RNP assembly process
Because proper assembly of hTERC RNA and hTERT protein in the telomerase holoenzyme complex is required for its enzymatic function,7,9 we next asked whether the natural sequence variations of either the hTERT protein or hTERC RNA could affect this interaction in cells. We compared telomerase ribonucleoprotein (RNP) complexes, assembled in vitro using the rabbit reticulocyte lysates (RRLs) to express a FLAG-tagged hTERT protein, with either the wild-type sequence or mutated sequences in the presence of the full-length wild-type hTERC RNA. Telomerase RNP complexes were immunoprecipitated from the lysates using an anti-FLAG antibody against the FLAG-tagged hTERT protein and were then probed for hTERC by Northern blotting.3 As illustrated in Figure 4C, none of the hTERT mutants tested showed any significant deficiency in binding to hTERC RNA (lanes 1-9). Although the G682D and P721R mutants seemed to show a modestly reduced hTERC binding activity, each of these effects alone could not fully explain the different levels of telomerase TRAP activity shown in Figure 2A. Using a similar assay, we asked whether the disease-associated mutations in the context of the full-length hTERC RNA tested here could also affect their functional interactions with the wild-type FLAG-tagged hTERT protein. Neither of the hTERCs showed any defect in assembly with the catalytic hTERT protein (lanes 10-14). These findings suggest that specific sequence changes of either the hTERT or hTERC tested here did not alter their optimal RNP assembly process.
hTERC template mutations seem to act as dominant negatives against wild-type telomerase enzymatic function
Testing of the disease-associated RNA template mutations (Δ52-55 and A48G variants) indicated that each was expressed at normal concentrations within transfected cells and was capable of forming functional RNP complexes with the catalytic hTERT protein (Figure 4B-C), but was nevertheless functionally defective (Figure 4A, lanes 13-18). When the A48G RNA variant was transcribed in vitro and used to assemble with the hTERT protein expressed in an RRL, irregular telomeric repeats appeared to be synthesized (Figure 4A, lane 40); these unusual repeats, however, were not produced by complexes with either the wild-type hTERC (lanes 46-47) or other hTERC RNA variants (G58A or Δ28-34, lanes 42-45). These data prompted experiments to determine if any of these RNA sequence changes inhibited the normal level of telomerase enzymatic function in a dominant-negative fashion, as had been observed for some genetically engineered RNA template mutations.39 When tested in our assay, in which equal amounts of the wild-type and mutated hTERC copy were cotransfected into VA13+hTERT cells, the 2 disease-associated template-sequence variants (Δ52-55 and A48G) almost completely abolished telomerase enzymatic activity, each constituting less than 1% of TRAP activity as compared to lysates prepared from cells with the wild-type hTERC RNA and each of the nontemplate mutants Δ79C, Δ28-34, or the polymorphic G58A version, or with the wild-type sequence alone (Figure 4A, lanes 22-39). These results suggest that disease-associated mutations in hTERC template domain appear to inhibit wild-type telomerase activity in a dominant-negative fashion.
Discussion
We provide functional evidence that disease-associated alleles of the RNA or protein component of the enzyme telomerase in some patients with bone marrow failure are unable to support normal levels of telomerase function. One of the novel findings in this study is the identification of 2 mutations located within the hTERC RNA template region that may act as dominant negatives against the wild-type copy of telomerase RNP in cells (Figure 4A, lanes 25-30). Such an experimental finding allows us to consider the different means by which disease-associated telomerase mutations inhibit the wild-type enzymatic function. Together with earlier reports,24,37 our data are consistent with the hypothesis that inherited defects in telomerase function and telomere maintenance contribute to the pathogenesis of hematologic disorders in a subset of patients with marrow failure syndromes.
We first considered several novel hTERT sequence variants that have recently been identified in patients with AA or DC.21,28,29 Our telomerase enzymatic assays shown in Figures 2A and 3A suggest that only 3 (ie, G862D, K902N, and K570N) of the 8 naturally occurring hTERT variants tested in this series have defective enzymatic functions. We showed that residue changes located within the functionally conserved RT domain of telomerase (ie, the G682D and K902N mutants) effectively reduced telomerase enzymatic function to less than 1% of that of the wild-type hTERT (Figure 2A). Whereas the lysine residue change at position 902 is located within the conserved motif D of the RT domain and itself is highly conserved among the retroviral and cellular reverse transcriptases,11,16 the G862D residue change is located within the generally nonconserved linker region (Figure 1C). It is noteworthy that we had recently reported 2 other disease-associated mutations (V694M and Y772C), also located within separate linker motifs of the RT domain, to drastically reduce telomerase enzymatic activity,20 suggesting that these seemingly nonconserved residues may be involved in either the structural formation or functional property of telomerase. Similarly, the K570N mutation effectively abolished telomerase enzymatic function, even though it is highly divergent among the telomerases of the different species.7 Using a yeast genetic screening assay, this residue of hTERT has also been shown to be required for proper telomerase function.40 We showed here that 2 other variants (P721R and T726M), both located within the highly conserved motif A of the RT domain, did not show any defect. These observations reiterate the fact that we still do not fully understand the important contribution each residue of hTERT makes to its overall structure or enzymatic function of the telomerase holoenzyme complex.
The remaining R979W and F1127L variants analyzed, both located within the telomerase-specific C-terminal domain of hTERT, each showed TRAP activity that was nearly equivalent to that of the wild-type hTERT (Figure 2A, lanes 13-18). The inconsequential phenotype of the amino acid substitution at position 979 is consistent with published data obtained for 2 engineered variants with amino acid substitutions flanking this residue (ie, the L978A and L980A substitutions).14 Similarly, substitution at position 1127 of hTERT did not result in any significant defect in telomerase enzymatic function. However, when a 6-amino acid substitution was incorporated into this position, this altered version of telomerase yielded only 15% to 50% of telomerase enzymatic activity and failed to immortalize cells,11 a phenotype that has been dubbed “C-DAT” for the C-terminal region that can dissociate telomerase enzymatic function, attributed to the disruption of the yet undefined function of telomerase to maintain telomere lengths even in the presence of the seemingly wild-type level of telomerase enzymatic function.11,41 The observed short telomere in cells of the patient with the F1127L mutation (Table 1) serves as the first case report of the effect of such a unique property of the telomerase protein in a human disease. We are in the process of testing all known disease-associated hTERT variants for such a phenotype in human primary cells.
Because hTERC RNA is an integral unit of the telomerase RNP complex, we had previously undertaken a comprehensive site-directed mutagenesis analysis to show that a high proportion of the seemingly minor point mutations can severely compromise telomerase function by perturbing its secondary structural formation.3 As further illustrated here, a single nucleotide deletion (Δ79C) in a small P2a stem is sufficient to abolish telomerase enzymatic activity (Figures 1A and 4A). On the contrary, most of the nucleotides located near the 5′ end of the hTERC RNA, including sequences that make up the lower strand of the P1a stem (Figure 1A), are dispensable for telomerase enzymatic function,3 consistent with knowledge that the murine version of the telomerase RNA (mTERC) lacks most of these sequences.2 However, the P1b paired region most proximal to the template has been shown by us to be absolutely required for optimal enzymatic activity.3 It has also been shown that stable formation of this P1b helix acts as the template boundary determinant to mediate the precise copying of only nucleotides located within the template region by the catalytic hTERT protein.42,43 The inconsequential effects of the disease-associated Δ28-34 mutation reported in the current study as judged by both the in vitro and in vivo TRAP assays (Figure 4A, lanes 4-6 and 44-45) can be explained by the deletion serendipitously maintaining the formation of the P1b helical structure as predicted and shown in Figure 1B. The apparently normal length of the repeats synthesized by the Δ28-34 mutant (Figure 4A, lanes 44-45) as compared to the A48G template mutation (lanes 40-41) implied that the Δ28-34 mutation did not alter the template boundary. Direct primer extension telomerase assay,43 however, will still need to be carried out to verify the prediction.
All patients whose variant hTERC or hTERT alleles were the subject of this study were heterozygous for the mutant allele, which may be relevant to the observation that most of the disease-associated telomerase variants tested thus far have yielded an effect suggestive of a haploinsufficiency to modulate the wild-type telomerase enzymatic function.24,30,37 For the first time, we show here 2 naturally derived hTERC variants, in which the mutations are located within the template region, to significantly inhibit telomerase function in a dominant-negative fashion (Figure 4A). This may help explain the greater degrees of disease severity and perhaps an earlier onset of the disease experienced by these 2 patients than those who carried other types of hTERC mutations (Table 1 and related reports24,30,37 ). For instance, whereas the 13-year-old boy with the Δ52-55 template mutation and the 16-year-old boy with the A48G template mutation presented with hypoplastic myelodysplasia and nail dystrophy, and nail dystrophy and marrow failure, respectively, 2 other patients in their forties with the Δ79C mutation presented with AA without marrow failure or other physical anomalies.28
The dominant-negative effect of artificially engineered hTERC template mutants has been noted elsewhere.39,44 By using lentiviral vector to deliver these artificial hTERC template mutants into cancer cell lines, it has been demonstrated that these RNAs can quickly kill cancer cells and that the effect depends on a catalytically active telomerase complex to possibly incorporate the mutated telomeric sequence onto the ends of the chromosomes.39,44 Although not yet tested, we suspect a similar detrimental effect of the naturally derived hTERC template mutants (A48G or the Δ52-55) in cells of the patients. However, the effect may not be as evident in blood cells as compared to cancer cell lines used in the previous study,39 because these cancer cells are known to have been endowed with very short telomeres. Recently, Cerone and colleagues have demonstrated that a mutant template hTERC RNA can dictate the synthesis of aberrant telomeric repeats onto chromosome ends of human breast cancer cell lines with long overall telomeres.45 Cells with the mutant repeats appeared to be more sensitive to anticancer drugs etoposide and doxorubicin. Upon drug treatment, these cells showed a significant reduction in proliferative potential and revealed an increase in telomere length heterogeneity, even though the overall length of telomeres was maintained with no signal-free ends or chromosomal fusions, suggesting that the incorporated mutant repeats interfere with telomere structure maintenance and heighten tumor cells to harmful stimuli.45 A similar process may occur in patients when only a single hTERC template is mutated. It is known that some proportion of patients with heterozygote telomerase allele would be at risk of exhausting the regenerative capacity of their marrow stem cells during adulthood, especially when challenged by environmental insults.27,46,47 If the clonal life spans of those stem cells were limited by telomere shortening, as has been observed for the mouse,48 even minor stochastic variations in the initial numbers and replicative histories of marrow stem cells could dictate which heterozygous individuals develop symptomatic marrow failure and the age at which it occurs. Additional variability in disease phenotype could be imparted by the specific telomerase variant that an individual inherits, as some variants have been shown in this and other studies to function in either a haploinsufficiency manner or a dominant-negative fashion to modulate wild-type levels of telomerase activity (Figures 2A, 3A, and 4A).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisment” in accordance with 18 U.S.C. section 1734.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Contribution: Z.-T.X. and A.D.B. equally participated in the functional characterization of the telomerase mutations; R.T.C. was responsible for patient selection, diagnosis, and genotyping; J.W.B. participated in the construction of the expression vectors; J.A.R. participated in the identification of the disease-associated mutations; A.S. was responsible for patient care and sample collections; Y.L. participated in the writing of the manuscript and in the study design; P.M.L. provided data on telomere length measurement and interpretation; N.S.Y. was responsible for patient care; H.L. conceived the idea for the study, performed some of the assays, participated in the analysis and interpretation of the data, and wrote the manuscript. All authors contributed to the final version of the manuscript.
Z.-T.X., A.D.B., and R.T.C. contributed equally to this work.
Acknowledgments
We thank Irma Vulto and Huiying Gao for excellent technical assistance, Olga Nuñez for assistance in patient care, and the patients and their families for their continuing support of and participation in the study.
This work was supported in part by grants from the American Cancer Society (RSG-06-162-01-GMC), the Leukemia and Lymphoma Society of America (3327-04), the AA and MDS International Foundation, the Emory University Research Committee (URC), the Emory Center for AIDS Research CFAR (P30 AI050409), and the Emory Digestive Diseases R&D Center P/F fund (DK64399) (H.L.).
H.L. wishes to dedicate this manuscript to the untimely passing of his former doctoral thesis advisor (Dr Andrew H. Kaplan, University of North Carolina, Chapel Hill).