Abstract
The transcription factor PAX5 is a critical regulator of B-cell commitment and development. Although normally not expressed in myeloid progenitors, PAX5 has recently been shown to be frequently expressed in myeloid malignancies and to suppress expression of myeloid differentiation genes, compatible with an effect on the differentiation or maintenance of myeloid progenitors. However, previous studies in which PAX5 was ectopically expressed in normal myeloid progenitors in vivo and in vitro provided conflicting results as to the effect of PAX5 on myeloid development. Herein, we demonstrate that on ectopic expression of PAX5 in bone marrow multipotent stem/progenitor cells, cells with a biphenotypic B220+GR-1/MAC-1+ phenotype are produced. These remain cytokine-dependent, but unlike control-transduced cells they sustain long-term generation of myeloid progenitors in vitro and remain capable of myeloid differentiation. Notably, PAX5+B220+GR-1/MAC-1+ myeloid progenitors coexpress, at the single-cell level, myeloid genes and otherwise B-cell–specific PAX5 target genes. These findings establish that ectopic expression of PAX5 introduces extensive self-renewal properties in otherwise short-lived myeloid progenitors. Along with the established ectopic expression of PAX5 in acute myeloid leukemia, this motivates a careful investigation of the potential involvement of ectopic PAX5 expression in myeloid and biphenotypic leukemias.
Introduction
The development of distinct mature blood cell lineages from hematopoietic stem cells (HSCs) is a highly ordered process involving a network of transcription factors and extracellular regulators.1 The paired domain transcription factor B-cell–specific activator protein (BSAP/PAX5), encoded by the Pax5 gene, is a critical regulator of B-cell commitment and development,2 and Pax5-deficient mice show a block in early B-cell development.3–5 Further, pro-B cells derived from Pax5-deficient mice reacquire an ability to differentiate into myeloid lineages both in vitro and in vivo,6,7 and more recent data have suggested that Pax5 might in part promote B-cell commitment in multipotent progenitors by repressing the expression of myeloid genes.7–9 These findings, combined with the fact that Pax5 normally is not expressed in the myeloid lineage in mice nor humans,2,10 would be compatible with ectopic expression of Pax5 in myeloid progenitors, resulting in suppression of myeloid programs and, as a consequence, suppression of myelopoiesis or expansion of early myeloid progenitors
The reported expression of PAX5 message in numerous cases of acute myeloid leukemia (AML) and other myeloid malignancies,11–13 including the promyelocytic HL60 and megakaryoblastic MOLM-1 cell lines,14 could potentially be explained by ectopic expression of PAX5 contributing toward expansion of myeloid progenitors or precursors. However, previous studies in which PAX5 has been ectopically expressed in the myeloid lineage have not provided support for such an effect of PAX5 in myeloid progenitors. Rather, overexpression of PAX5 in vivo failed to demonstrate any significant effect on myelopoiesis,15,16 and in seemingly contrast to these data, in vitro studies focusing on the effect of PAX5 on terminal myeloid differentiation in response to all-trans-retinoic acid (ATRA) and myeloid growth factors suggested a suppressive effect of PAX5 on myeloid differentiation and survival.17,18
Herein, we pursued experiments aimed to reconcile these findings by focusing on the ability of ectopically expressed PAX5 to affect the maintenance as well as differentiation of myeloid progenitors in vitro. Notably, when cultured under myeloid conditions, PAX5-expressing cells coexpressed lymphoid PAX5 targets as well as myeloid genes and acquired a biphenotypic B220+GR-1/MAC-1+ phenotype. These myeloid progenitors were sustained long-term in cultures supplemented with myeloid growth factors.
Materials and methods
Hematopoietic growth factors
Recombinant rat (rr) stem cell factor (SCF) and recombinant human (rh) granulocyte colony-stimulating factor (G-CSF) were generously provided by Amgen (Thousand Oaks, CA), rhFMS-like tyrosine kinase 3 ligand (FL), recombinant mouse (rm) interleukin-3 (IL-3), and rm granulocyte-macrophage colony stimulating factor (GM-CSF) by Immunex (Seattle, WA), rhIL-6 by Genetics Institute (Cambridge, MA); rhIL-7 by Sanofi (Labege Cedex, France); rh thrombopoietin (THPO) by Genentech (San Francisco, CA); rh erythropoietin (EPO) by Boehringer Mannheim (Mannheim, Germany); and rh colony-stimulating factor 1 (CSF-1) by Cetus (Emeryville, CA). Unless otherwise indicated, predetermined optimal cytokine concentrations used in this study were as follows: 50 ng/mL rrSCF, 50 ng/mL rhG-SCF, 25 ng/mL rmGM-CSF, 50 ng/mL rhFL, 25 ng/mL rmIL-3, 50 ng/mL rhIL-6, 100 ng/mL rhIL-7, 100 ng/mL rhTHPO, 5 U/mL rhEPO, and 50 ng/mL rhCSF-1.
Antibodies
All antibodies were from PharMingen (Becton Dickinson, San Diego, CA). Antibodies used for cell surface staining were E13-161.7 (SCA-1), 2B8 (c-KIT), RA3-6B2 (B220), 6c3 (BP-1), 1D3 (CD19), R6-60.2 (IgM), A7R34 (IL-7Rα), 3C7 (CD25), RB6/8C5 (GR-1), M1/70 (CD11b, MAC-1), 30-F1 (CD24), S7 (CD43), 145-2C11 (CD3ϵ), H129.19 (CD4), 53-6.7 (CD8α), HL3 (CD11c), B7-2 (CD86), 2.4G2 (CD16/32, FCγR), RAM34 (CD34), AA4.1, 6C3/BP-1 (Ly5.1), TER119; A20 (CD45.1), 104 (CD45.2), streptavidin-PE (phycoerythrin), streptavidin-APC (allophycocyanin), and streptavidin-PE-Cy7 (Caltag, Burlingame, CA) were used to visualize biotin-conjugated primary antibodies.
Isolation and FACS of stem and progenitor cell populations
For fluorescence-activated cell sorting (FACS) 8- to 12-week-old congenic C57BL/6 mice (CD45.1 or CD45.2; Jackson Laboratories, Bar Harbor, ME) were used as bone marrow (BM) donors for purification of lineage (LIN) low and negative, SCA-1– and c-KIT–expressing (LSK) HSCs, performed as previously described.19 LIN−c-KIThiSCA-1−IL-7Rα−FCγRloCD34+ common myeloid progenitors (CMPs), LIN−c-KIThiSCA-1−IL-7Rα−FCγRloCD34− megakaryocyte/erythrocyte lineage-restricted progenitors (MEPs), and LIN−c-KIThiSCA-1−IL-7Rα−FCγRhiCD34+ granulocyte/macrophage lineage-restricted progenitors (GMPs)20,21 were purified by incubating adult BM cells with a cocktail of lineage-specific antibodies including anti-IL-7Rα antibody. Lineage-depleted cells were subsequently stained with FITC-conjugated anti-CD34, PE-conjugated anti-CD16/32 (FCγR), APC-conjugated anti-c-KIT, and biotinylated anti-SCA-1 antibodies. The biotinylated antibody was visualized with streptavidin-PE-Cy7. AA4.1+CD43+CD19+ pro-B cells and double-positive (DP) CD4+CD8+ thymic progenitors were sorted from normal adult BM and thymuses, respectively. Cells with low viability were excluded from the sorting gate by staining with 7-aminoactinomycin D (7AAD; Sigma-Aldrich, St Louis, MO). Stained cells were sorted on a FACSVantage or FACSDiva Cell Sorter (Becton Dickinson).
Retroviral constructs
The human PAX5 cDNA, generously provided by Dr Meinrad Busslinger (Vienna Biocenter, Vienna, Austria), was inserted into a MSCV promoter driven retroviral vector 5′ of an internal ribosome entry site (IRES) followed by enhanced green fluorescence protein (eGFP). The construct was stably integrated in the GP+E86 cell line (a generous gift from Arthur Bank, College of Physicians and Surgeons, Columbia University, New York, NY) and the selected clone produced a virus titer of around 1.14 × 107 transducing units (TU)/mL. The control vector containing yellow fluorescent protein (YFP; from Dr James Hagman, University of Colorado Health Sciences Center, Denver, CO) was introduced into the virus producer 293 GPG cell line to generate media with a virus titer of 1.1 × 107 TU/mL. Another control vector (generous gift from Drs Thomas Relander and Johan Richter, Department of Gene Therapy, Lund, Sweden) contained eGFP upstream of an IRES and a neomycin phosphotransferase (NEO) with virus titer 3.6 × 106 TU/mL.
Transduction and ex vivo culture of transduced progenitors
Hematopoietic cell cultures were performed either in serum-free X-vivo 15 medium (BioWhittaker, Walkersville, MD) supplemented with 1% bovine serum albumin (BSA; StemCell Technologies, Vancouver, BC, Canada), 0.1 mM 2-mercaptoethanol, 2 mM l-glutamine, 100 U/mL penicillin, 100 U/mL streptomycin (hereafter serum-free medium [SFM]) or in Iscove modified Dulbecco medium (IMDM), supplemented with 20% fetal calf serum (FCS), both from BioWhittaker (Verviers, Belgium), 0.1 mM 2-mercaptoethanol, 2 mM l-glutamine, 100 U/mL penicillin, 100 U/mL streptomycin (hereafter serum-containing medium [SCM]). LSK cells were prestimulated in SFM supplemented with SCF, IL-3, IL-6, FL, and either IL-7 or THPO for 48 hours and subsequently transduced on retronectin-coated (Takara Bio, Otsu, Japan) and virus-preloaded non–tissue culture-treated 96-well plates in SFM with the same cytokine combination as indicated for another 48 hours. Subsequently, cells were washed and transferred either into SFM supplemented with SCF, FL, and IL-7 for pro-B cell assay or into SCM supplemented with SCF, GM-CSF, G-CSF, and IL-3 for myeloid progenitor assay. Cells were analyzed for YFP/GFP expression another 72 hours later using FACSCalibur. Transduction efficiency was defined as the percentage of YFP/GFP positive cells 7 days after transduction. For evaluation of cytokine dependence and potential phenotype change, GFP+B220+GR-1/MAC-1+ cells from 24 days of culture in SCM under myeloid conditions were washed and transferred to either SCM or SFM supplemented with only medium or one of the following cytokines: GM-SCF, IL-3, G-CSF, FL, IL-7, EPO, THPO, and CSF-1. Cytokines were resupplied twice a week and cells were split 2 or more times per week to keep cell density below 1 × 106 cells/mL.
Myeloid progenitor assay
For detecting committed myeloid progenitors, a granulocyte-macrophage colony-forming unit (CFU-GM) assay was performed. GFP+ control and PAX5-transduced cells obtained by FACS were plated in methylcellulose (M3434; StemCell Technologies) supplemented with 2 mM l-glutamine, 100 U/mL penicillin, 100 U/mL streptomycin, 0.1 mM 2-mercaptoethanol, 10 ng/mL rrSCF, 25 ng/mL rhFL, 10 ng/mL rmIL-3, 25 ng/mL rhG-CSF, 25 ng/mL rmGM-CSF, and 100 ng/mL rhTHPO in 35-mm Petri dishes. Colonies (>50 cells) in methylcellulose were scored using an Olympus IX70 inverted microscope (Olympus, Hamburg, Germany) equipped with LCPlan FI 40×/0.60 Ph2 and UPlan FI 10×/0.30 Ph1 objectives (Olympus, Tokyo, Japan) after 14 days of incubation at 37°C and 5% CO2. Photographs of colonies in methylcellulose were captured with a Sony DKC-5000 3CCD camera (Sony, Tokyo, Japan), using Infan View software. Images were further processed with Adobe Photoshop software (Adobe Systems, Mountain View, CA).
Cell morphology
Cells (104) from liquid cultures or individual colonies were centrifuged onto glass slides using a Cytospin 3 (Shandon, Labex Instrument, Helsingborg, Sweden), stained for 5 minutes in May-Grünwald stain and 20 minutes in Giemsa stain (both from Histolab, Gothenburg, Sweden), thereafter washed, dried, and finally analyzed for lineage-specific cell type by microscopy (Olympus BX51TF, Olympus, Hamburg, Germany). Photographs of cytospins and colonies in methylcellulose were captured with either Olympus DP70 (Olympus) or Sony DKC-5000 3CCD camera, using an Olympus BX51TF microscope (Olympus, Tokyo, Japan) equipped with a UPlan FI 100×/1.30 oil-immersion objective lens. Photographs of cytospins were captured with an Olympus DP70 camera using DP controller software. Images were further processed with Adobe Photoshop software.
Southern blot analysis
gDNA was extracted by lysis of cells in 250 mM Tris, pH 8.0, 150 mM NaCl, 10 mM EDTA, and 0.1% SDS. The mixture was incubated overnight at 56°C with Proteinase K (Invitrogen, Stockholm, Sweden) and DNA purified by phenol extraction and precipitated by addition of 0.7 volumes of isopropanol (Sigma, Stockholm, Sweden). The precipitated DNA was washed 2 times with 70% ethanol, dried, and redissolved in water. Approximately 10 μg DNA was digested with HindIII (Boehringer Mannheim) and size-fractionated on 0.8% agarose gel. After transfer to a nylon membrane by capillary blotting, the membrane was hybridized with 32P-labeled random primed probe in 6 × SCC, 5 × Denhart solution, 100 μg/mL salmon sperm DNA, and 0.1% SDS at 65°C. Membranes were washed at room temperature in 2 × SSC (0.015M Na citrate, pH 7.0, containing 0.15 M NaCl) supplemented with 0.1% SDS for 15 minutes and 0.1 × SSC with 0.1% SDS for 5 to 10 minutes. The hybridized membrane was subjected to autoradiography.
Linker-mediated polymerase chain reaction
For linker-mediated polymerase chain reaction (LM-PCR), single myeloid colonies (CFU-GMs) were picked from methylcellulose and gDNA was extracted using Qiamp DNA mini kit (Qiagen, Solna, Sweden) according to the manufacturer's instructions. gDNA was digested with TaqI in 20 μL reaction and denatured at 95°C for 10 minutes in the presence of 1 μL of 1 μM solution of primer 1. After cooling on ice, 1 μL of 5 mM solution of dNTP (Roche, Bromma, Sweden) and 5 U Klenow (Roche) enzyme were added to allow the formation of double-stranded blunt end templates. The DNA was then diluted to 50 μL by the addition of distilled water and subsequently precipitated with 5 μL of 3 M NaAc, pH 5.2, and 125 μL 95% ethanol. After freezing, the pellets were collected by centrifugation and washed twice with 70% ethanol. The dry pellets were redissolved in 15 μL water, 2 μL ligation buffer, and 2 μL annealed linker oligonucleotide (10 μM). Subsequently, 5 U T4 DNA ligase (Roche) was added to the reaction and incubated at 20°C overnight. Then, 2 μL of template was used for PCR 1 together with primer 2 and linker sense primer for 30 cycles of amplification (94°C, 45 seconds; 56°C, 45 seconds; 72°C, 60 seconds). PCR product 1 (2 μL) was diluted in 100 μL water and 2 μL diluted template was used for PCR 2 together with primer 3 and linker sense primer for another 30 cycles of amplification (94°C, 45 seconds; 61°C, 45 seconds; 72°C, 60 seconds). The PCR products were transferred to a nylon membrane and the filters were hybridized to T4 PNK-labeled oligonucleotide directed against the LTR (LMHyb). Primer 1: 5′-TTACCACAGATATCCTGTTTGG-3′; primer 2: 5′-CTGCTGTCTCTCTGTTCCTAACC-3′; primer 3: 5′-GATCTGAACTTCTCTATTCTCAG-3′; linker primer sense: 5′-GCGGTGACCCGGGAGATCTGAATTC-3′; linker primer antisense: 5′-GAATTCAGATC-3′; LMHyb: 5′-TTGCAAGGCATGGAAAATACATAACTG-3′.
Protein extracts and EMSA
Nuclear extracts were prepared according to Schreiber et al22 and electrophoretic mobility shift assay (EMSA) performed as previously described.23 Nuclear extracts (5 μg) were incubated with 32P-labeled double-stranded probes (20 000 cpm, 3 fmol) and either anti-PAX5 (sc-1974), anti-CCAAT enhancer binding protein-α (C/EBPα; sc-61 X; both from Santa Cruz Biotechnology, Santa Cruz, CA), or competitor DNA. The samples were separated on 6% polyacrylamide TBE gel, which was dried and subjected to autoradiography.23 CD19-Pax5 sense: 5′-GCAGACACCCATGGTTGAGTGCCCTCCAGG-3′ and CD19-Pax5 antisense: 5′-CCTGGAGGGCACTCAACCATGGGTGTCTGC-3′ oligonucleotides were used.
Cell lines and global gene expression (Affymetrix) analysis are presented in the supplemental methods (available on the Blood website; see the Supplemental Materials link at the top of the online article).
RT-PCR and Q-PCR
Total RNA was prepared using TRIzol (Gibco, Paisley, Scotland). Obtained RNA samples were reverse transcribed using SuperScript II and random hexamers (Invitrogen) according to protocol supplied by the manufacturer and as previously described.19 Quantitative reverse transcription-PCRs (Q-RT-PCRs) were performed by mixing 2 × TaqMan universal PCR master mix, 20 × Assays-on-Demand (primer/MGB-probe mix), RNase-free H2O, and 5 μL cDNA to a final reaction volume of 20 μL. The following TaqMan Assays-on-Demand probes were used: for mb-1 (Cd79a): Mm00432423_m1; Blnk: Mm00456139_m1, Rag2: Mm00501300_m1; Cd19: Mm00515420_m1; Ebf1: Mm01288947_g1; Il7ra: Mm00434295_m1; Csf2ra: Mm00438331_g1; Cebpa: Mm00514283_s1; Cebpe: Mm02030363_s1, Gata1: Mm_00484678_m1. Sequences for Assay-by-Design Q-PCR (Applied Biosystems, San Diego, CA) were as follows: Rag1: forward 5′TGTGGAGCAAGGTAGCTTAGC-3′; reverse 5′-TCATCGGGTGCAGAACTGAAG-3′; MGB-probe 5′-ATGGCTGCCTCCTTG-3′; λ5: forward 5′-GGAACAACAGGCCTAGCTATGG-3′; reverse 5′-CTCCCCGTGGGATGATCTG-3′; MGB-probe 5′-CCGGCAGCTCCTGTTC-3′; Nfe2: forward 5′-GACTTGGCAAGAGATCATGTCCAT-3′; reverse 5′-GGCTCAAAAGATGTCTCACTTGGAA-3′; MGB-probe 5′-CTGAGCTGCAGGGTCT-3′. Normalizing was carried out against Hprt expression levels. For conventional PCR one-twentieth of the RT reaction was amplified using 1 U Taq-polymerase (Life Technologies, Bethesda, MD) in the manufacturer's buffer supplemented with 0.2 mM dNTP in a total volume of 25 μL. Oligonucleotides: Pax5: forward 5′-CTACAGGCTCCGTGACGCAG-3′; reverse 5′-GTCTCGGCCTGTGAAATAGG-3′; 36B4: forward 5′-GAGGAATCAGATGAGGATATGGGA-3′; reverse 5′-AAGCAGGCTGACTTGGTTGC-3′.
Immunoglobulin recombination PCR analysis
gDNA was prepared using TRIzol (Gibco) according to the manufacturer's instructions. IgH D-J rearrangements and germline DNA were amplified in the same reaction by 33 cycles (94°C, 30 seconds, 60°C, 45 seconds, and 72°C, 1 minute) using the DH and J3 primer at 1 μM and the Mu0 (amplifies germline DNA together with J3) primer at 0.1 μM final concentration. The PCR products were blotted and hybridized as described above under “Southern blot analysis.” A J3-specific 32P-labeled oligonucleotide (JH3) was hybridized to the filters at 45°C overnight; thereafter the filters were washed and subjected to autoradiography as above. Oligonucleotides: DH: 5′-GGAATTCG(A/C)TTTTTGT(C/G)AAGGGATCTACTACTGTG-3′; J3: 5′-GTCTAGATTCTCACAAGAGTCCGATAGACCCTGG-3′; Mu0: 5′-CCGCATGCCAAGGCTAGCCTGAAAGATTACC-3′; JH3: 5′-AGACAGTGACCAGAGTCCCTTGG-3′.
Analysis of single cells by RT-PCR
Multiplex single-cell RT-PCR analysis of GFP(PAX5)+B220+GR-1/MAC-1+ cells was performed according to the methods described previously.19,24 Single cells were deposited by a single-cell depositor coupled to a FACSDiva into 96-well PCR plates containing 4 μL lysis buffer followed by multiplex PCR.19,24 Aliquots of second-round PCR products were subjected to gel electrophoresis and visualized by ethidium bromide staining on E-gels (Invitrogen). Oligonucleotides: mb-1: external forward 5′-CCTCCTCTTCTTGTCATACG-3′; reverse 5′-GAACAGTCATCAAGGTTCAGG-3′; internal forward 5′-AAACAATGGCAGGAACCC-3′; reverse 5′-TGATGATGCGGTTCTTGG-3′; Cebpe: external forward 5′-ACTTTCTGACCTCTTTGCC-3′; reverse 5′-GTTTTCAGCCATGTACTCC-3′; internal forward 5′-TCGTTTCCTCACTACCTGC-3′; reverse 5′-CGATGTTGTTACGTTCACG-3′; Hprt: external forward 5′-GGGGGCTATAAGTTCTTTGC-3′; reverse 5′-TCCAACACTTCGAGAGGTCC-3′; internal forward 5′-GTTCTTTGCTGACCTGCTGG-3′; reverse 5′-TGGGGCTGTACTGCTTAACC-3′.
Statistics
Student t test was performed for statistical analyses to assess differences between the groups.
Results
Enforced expression of PAX5 promotes development and proliferation of B-cell progenitors and immortalization of biphenotypic myeloid progenitors
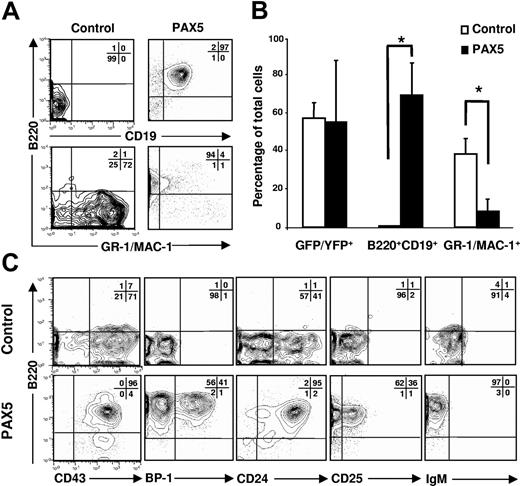
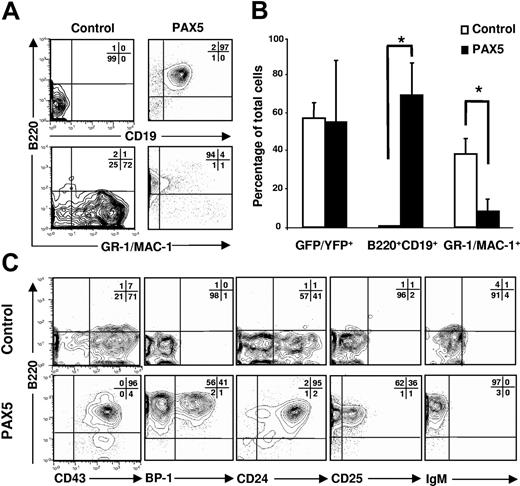
Retroviral overexpression of human PAX5 was targeted to multipotent LSK cells that were subsequently cultured under conditions compatible with B-lymphocyte and myeloid development (see “Materials and methods”). We have previously shown that purified multipotent LSK cells cultured in FL and IL-7 efficiently develop into pro-B cells under serum-free conditions.25 Later we observed that when transduced under conditions containing myeloid cytokines, the ability of LSK cells to generate pro-B cells is severely impaired (K.A. and S.E.W.J., unpublished observation, October 2001). By 14 days of culture in the presence of SCF, FL, and IL-7, control and PAX5-transduced cells had expanded at comparable levels (mean 1016-fold and 1054-fold expansion, respectively). However, whereas control cultures contained primarily myeloid cells and virtually no B220+CD19+ cells, B220+CD19+ cells constituted as much as 97% of PAX5-transduced cells (Figure 1A-B), translating into a 73-fold increase in generation of GFP(PAX5)+B220+CD19+ cells from PAX5-transduced LSK cells. GFP(PAX5)+B220+CD19+ cells coexpressed CD43, BP-1, CD24, and CD25, but not IgM, compatible with representing late pro-B progenitors (Figure 1C). In contrast, control vector-transduced cells failed to efficiently produce B220+ cells, but generated B220− cells expressing CD43 and CD24, and were negative for the B-cell–specific markers BP-1, CD25, and IgM (Figure 1C).
PAX5 promotes development of B220+CD19+ pro-B cells in vitro. LSK cells transduced with either control or PAX5-containing vector were cultured in SFM supplemented with SCF, FL, and IL-7 for 14 days. (A) Expression of B220, CD19, and GR-1/MAC-1 on cells already gated as YFP/GFP+. Representative profiles from 3 independent experiments. (B) Mean percentages (SD) of transduced cells expressing YFP/GFP, B220, CD19, and GR-1/MAC-1 from 3 independent experiments. *P < .05. (C) Typical coexpression profiles of B220 together with CD43, BP-1, CD24, CD25, and IgM on cells produced in control (top panels) and PAX5 (bottom panels) transduced cultures. Representative profiles from 3 independent experiments.
PAX5 promotes development of B220+CD19+ pro-B cells in vitro. LSK cells transduced with either control or PAX5-containing vector were cultured in SFM supplemented with SCF, FL, and IL-7 for 14 days. (A) Expression of B220, CD19, and GR-1/MAC-1 on cells already gated as YFP/GFP+. Representative profiles from 3 independent experiments. (B) Mean percentages (SD) of transduced cells expressing YFP/GFP, B220, CD19, and GR-1/MAC-1 from 3 independent experiments. *P < .05. (C) Typical coexpression profiles of B220 together with CD43, BP-1, CD24, CD25, and IgM on cells produced in control (top panels) and PAX5 (bottom panels) transduced cultures. Representative profiles from 3 independent experiments.
We next investigated how enforced PAX5 expression might affect myeloid differentiation from multipotent LSK cells. Whereas previous in vivo studies have suggested that myeloid development from HSCs is not affected by PAX5,15,16 other in vitro studies have implied that PAX5 under certain growth factor conditions can suppress myeloid development.17,18
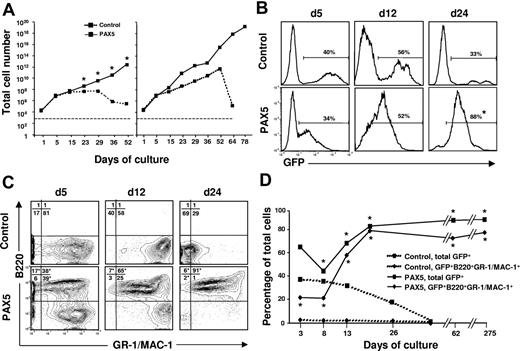
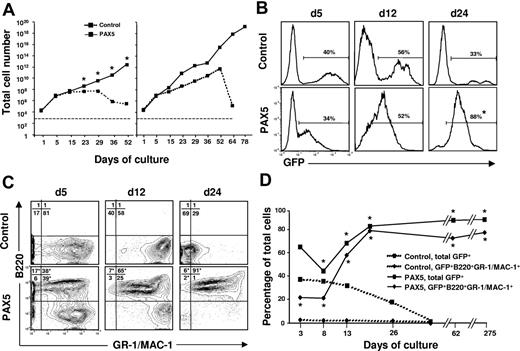
In an attempt to reconcile these findings we cultured control vector and PAX5-transduced LSK cells under conditions promoting myeloid commitment and differentiation. Strikingly, under these conditions, PAX5-overexpressing cells showed enhanced and sustained expansion when compared to control vector-transduced cells (Figure 2A). The finding that control cultures drop off after 25 to 50 days reflects that the normal progenitors, as expected, eventually are exhausted in such cultures (Figure 2A). Although the initial transduction efficiency was comparable, the percentage of GFP+ cells gradually increased in PAX5 but not control-transduced cultures (Figure 2B). Notably, whereas GFP+ control-transduced cells, as expected, were predominantly B220−GR-1/MAC-1+ myeloid cells, PAX5-transduced cultures were dominated by a rapidly expanding biphenotypic GFP(PAX5)+B220+GR-1/MAC-1+ population, which was already detectable at day 5 and by day 24 represented most of the cells (Figure 2C-D). No other B-cell surface antigens, such as IL-7Rα, BP-1, CD19, or IgM, were expressed on biphenotypic GFP(PAX5)+B220+GR-1/MAC-1+ cells nor were erythroid (TER119) or T-cell (CD3) antigens (K.A. and S.E.W.J., unpublished observations, May 2002).
Generation of biphenotypic B220+GR-1/MAC-1+ cells on overexpression of PAX5 in LSK cells. (A) Control and PAX5-transduced LSK cells were cultured under myeloid conditions as described in “Materials and methods” and total (GFP+ and GFP− cells combined) cellular expansion was determined at different time points as indicated. Dashed and solid lines demonstrate expansion of control and PAX5 cultures, respectively. Shown are 2 patterns of growth. Left panel represents mean values of 3 individual transduction experiments (*P < .05), with typical exhaustion of control cultures. Right panel shows the experiment (of totally 15) with the longest sustained control culture. Stapled line indicates starting cell number (2 × 104 cells). (B) GFP expression of control and PAX5-transduced LSK cells cultured under myeloid conditions at different time points. Percentages are mean values from 3 to 6 control and PAX5 experiments. (C) Expression of B220 and GR-1/MAC-1 on GFP+ cells in control (top panels) and PAX5-transduced (bottom panels) cultures. Values in quadrants show mean percentages from 3 experiments for control and PAX5-transduced cultures, respectively. (D) Percentages of total GFP+ control and PAX5-transduced cells coexpressing B220 and GR-1/MAC-1. One of 5 representative experiments is shown. The asterisk indicates statistically significant (P < .05) differences between control and PAX5-transduced cultures.
Generation of biphenotypic B220+GR-1/MAC-1+ cells on overexpression of PAX5 in LSK cells. (A) Control and PAX5-transduced LSK cells were cultured under myeloid conditions as described in “Materials and methods” and total (GFP+ and GFP− cells combined) cellular expansion was determined at different time points as indicated. Dashed and solid lines demonstrate expansion of control and PAX5 cultures, respectively. Shown are 2 patterns of growth. Left panel represents mean values of 3 individual transduction experiments (*P < .05), with typical exhaustion of control cultures. Right panel shows the experiment (of totally 15) with the longest sustained control culture. Stapled line indicates starting cell number (2 × 104 cells). (B) GFP expression of control and PAX5-transduced LSK cells cultured under myeloid conditions at different time points. Percentages are mean values from 3 to 6 control and PAX5 experiments. (C) Expression of B220 and GR-1/MAC-1 on GFP+ cells in control (top panels) and PAX5-transduced (bottom panels) cultures. Values in quadrants show mean percentages from 3 experiments for control and PAX5-transduced cultures, respectively. (D) Percentages of total GFP+ control and PAX5-transduced cells coexpressing B220 and GR-1/MAC-1. One of 5 representative experiments is shown. The asterisk indicates statistically significant (P < .05) differences between control and PAX5-transduced cultures.
Because B220, GR-1 and MAC-1 antigens are expressed not only on myeloid cells, but also on dendritic cells (DCs),26 including plasmacytoid DCs (PDCs), we performed phenotypic analysis of PAX5-transduced cells to investigate expression of several cell surface antigens expressed on DCs. PAX5-expressing B220+GR-1/MAC-1+ cells did not express CD4 and CD8α, which have been shown to be expressed on PDCs, whereas CD11c was expressed on 0.2% and CD86 on 0.8% of GFP(PAX5)+B220+GR-1/MAC-1+ cells, respectively (Figure S1). Moreover, PAX5-transduced cultures expressed high levels of MAC-1 (in contrast to PDCs, which express MAC-1 at low levels), displayed a typical myeloid morphology (Figure 3), lacked expression of Il7r, and were not DJ rearranged, in contrast to what has been reported for PDCs.26
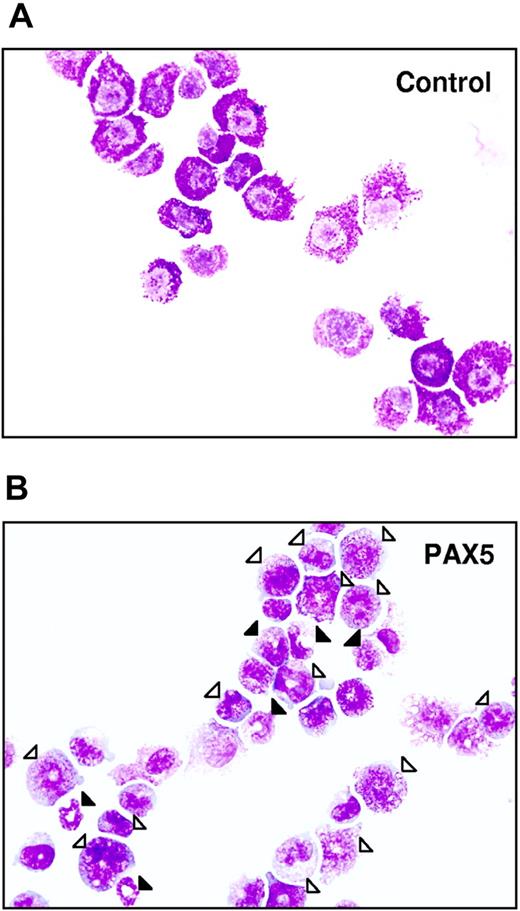
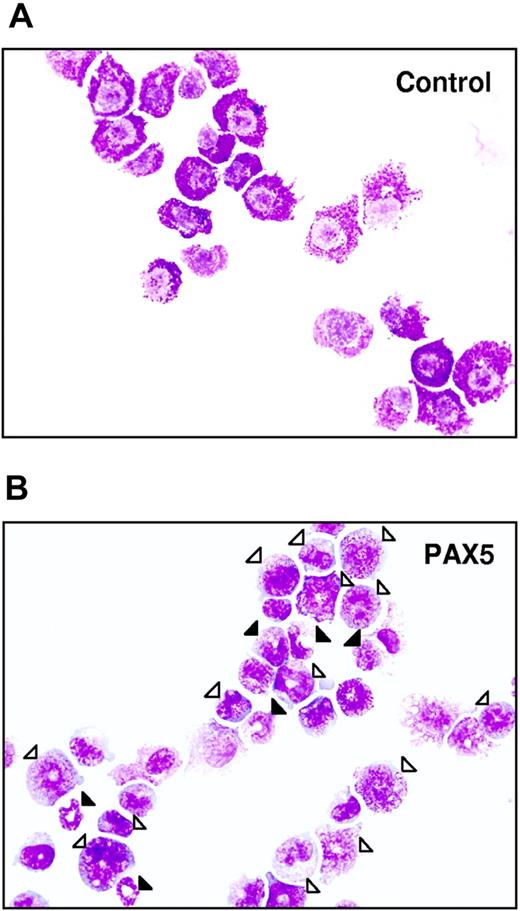
Biphenotypic B220+GR-1/MAC-1+ cells generated from PAX5-transduced LSK cells have an immature myeloid morphology. Typical morphology of May-Grünwald– and Giemsa-stained FACS-purified GFP+ control (A) and PAX5-transduced (B) biphenotypic GFP(PAX5)+B220+GR-1/MAC-1+ cells from liquid culture at 24 days after transduction. Open triangles indicate immature granulocytes (promyelocytes, myelocytes), whereas filled triangles point at more mature forms of granulocytes (metamyelocytes, band and segmented forms). Original magnification × 500.
Biphenotypic B220+GR-1/MAC-1+ cells generated from PAX5-transduced LSK cells have an immature myeloid morphology. Typical morphology of May-Grünwald– and Giemsa-stained FACS-purified GFP+ control (A) and PAX5-transduced (B) biphenotypic GFP(PAX5)+B220+GR-1/MAC-1+ cells from liquid culture at 24 days after transduction. Open triangles indicate immature granulocytes (promyelocytes, myelocytes), whereas filled triangles point at more mature forms of granulocytes (metamyelocytes, band and segmented forms). Original magnification × 500.
Importantly, whereas control-transduced cells ceased to proliferate in long-term culture, cells with a GFP(PAX5)+B220+GR-1/MAC-1+ phenotype could be sustained and expanded for up to 275 days (longest time investigated; Figure 2D). The fact that GFP(PAX5)+B220+GR-1/MAC-1+ cells (and not GFP− cells) expanded enormously in long-term culture indicates that PAX5-transduced cells had a competitive advantage and that this effect is cell intrinsic. Morphologically, the biphenotypic GFP(PAX5)+B220+GR-1/MAC-1+ cell population was highly heterogeneous, consisting of immature as well as mature myeloid cells, predominantly of the granulocytic but also the monocytic lineages, whereas control cultures, as expected, already at day 25 predominantly contained mast cells and macrophages with a few immature and mature granulocytes (Table 1; Figure 3). In contrast to control cultures, PAX5-transduced cultures also contained a low but consistent number of blast cells, compatible with sustaining a population of myeloid progenitors. Importantly, similar proportions of immature and mature granulocytes were observed at later time points when control cultures were no longer able to sustain any myeloid cell production (K.A. and S.E.W.J., unpublished observation, June 2002).
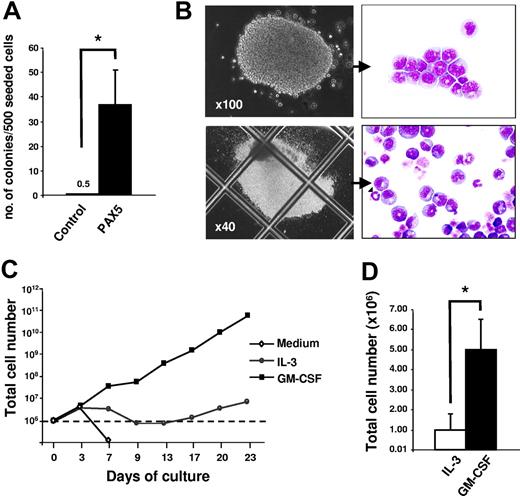
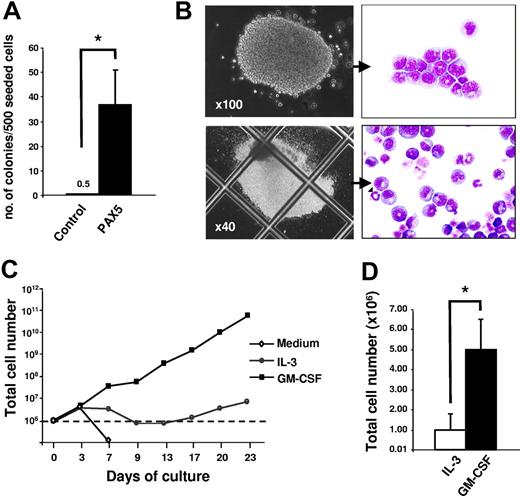
To further confirm that PAX5-transduced cultures are sustained by granulocyte-macrophage restricted progenitors (CFU-GMs), GFP(PAX5)+B220+GR-1/MAC-1+ from PAX5-transduced cultures and GFP+GR-1/MAC-1+ cells from control cultures (obtained by FACS) were plated into methylcellulose (Figure 4A-B). PAX5-overexpressing progenitors generated more and larger myeloid colonies than control-transduced cultures, and these consisted of both immature and mature granulocytes and, to a lesser degree, monocytic cells (Figure 4B). The estimated frequencies of CFU-GMs were 1 in 1000 for control-transduced cells and as much as 1 in 15 for PAX5-transduced cells, respectively (Figure 4A). Neither erythroid burst-forming units (BFU-Es) nor megakaryocyte colonies were detected in both control and PAX5-transduced cultures (K.A. and S.E.W.J., unpublished observations, September 2006). Thus, this finding demonstrates that overexpression of PAX5 leads to enhanced maintenance of GM progenitors in long-term culture.
Biphenotypic B220+GR-1/MAC-1+ cells sustain CFU-GM progenitors at high frequencies and require cytokines for sustained proliferation. (A) At 24 days after transduction, GFP+GR-1/MAC-1+ cells from control-transduced cultures and GFP(PAX5)+B220+GR-1/MAC-1+ from PAX5-transduced cultures were purified by FACS and plated into MethoCult as described in “Materials and methods.” Colony numbers were counted at day 14. Data are mean (SD) values from 3 control and 2 PAX5 experiments with 3 to 5 replicates in each experiment. *P < .05. (B) Appearance of typical myeloid colonies (CFU-GMs) generated in methylcellulose from GFP(PAX5)+B220+GR-1/MAC-1+ cells, with corresponding May-Grünwald and Giemsa morphology of myeloid cells in colonies. (C) At 24 days after transduction, 1 × 106 cells (stapled line) from PAX5-transduced culture were washed and seeded in medium alone or in medium supplemented with either GM-CSF or IL-3. Thereafter, cellular expansion was evaluated at different time points as indicated. One of 2 experiments with similar results is shown. (D) GFP(PAX5)+B220+GR-1/MAC-1+ cells from 24 days of culture were sorted by FACS and 1 × 104 GFP(PAX5)+B220+GR-1/MAC-1+ cells were seeded in SCM supplemented with either IL-3 or GM-CSF. Total number of cells was assessed 2 weeks later. Mean (SD) values of 3 experiments. *P < .05.
Biphenotypic B220+GR-1/MAC-1+ cells sustain CFU-GM progenitors at high frequencies and require cytokines for sustained proliferation. (A) At 24 days after transduction, GFP+GR-1/MAC-1+ cells from control-transduced cultures and GFP(PAX5)+B220+GR-1/MAC-1+ from PAX5-transduced cultures were purified by FACS and plated into MethoCult as described in “Materials and methods.” Colony numbers were counted at day 14. Data are mean (SD) values from 3 control and 2 PAX5 experiments with 3 to 5 replicates in each experiment. *P < .05. (B) Appearance of typical myeloid colonies (CFU-GMs) generated in methylcellulose from GFP(PAX5)+B220+GR-1/MAC-1+ cells, with corresponding May-Grünwald and Giemsa morphology of myeloid cells in colonies. (C) At 24 days after transduction, 1 × 106 cells (stapled line) from PAX5-transduced culture were washed and seeded in medium alone or in medium supplemented with either GM-CSF or IL-3. Thereafter, cellular expansion was evaluated at different time points as indicated. One of 2 experiments with similar results is shown. (D) GFP(PAX5)+B220+GR-1/MAC-1+ cells from 24 days of culture were sorted by FACS and 1 × 104 GFP(PAX5)+B220+GR-1/MAC-1+ cells were seeded in SCM supplemented with either IL-3 or GM-CSF. Total number of cells was assessed 2 weeks later. Mean (SD) values of 3 experiments. *P < .05.
We next investigated whether the sustained growth of GFP(PAX5)+B220+GR-1/MAC-1+ biphenotypic cells is cytokine dependent. Whereas GFP(PAX5)+B220+GR-1/MAC-1+ cells expanded well in the myeloid cytokines, such as GM-CSF, and were also sustained in IL-3, they rapidly lost viability in the absence of cytokines (Figure 4C-D), and neither EPO, THPO, FL, nor IL-7 had any ability to promote viability, proliferation, or differentiation along the erythroid, megakaryocytic, and lymphoid lineages, respectively (K.A. and S.E.W.J., unpublished observations, May 2002).
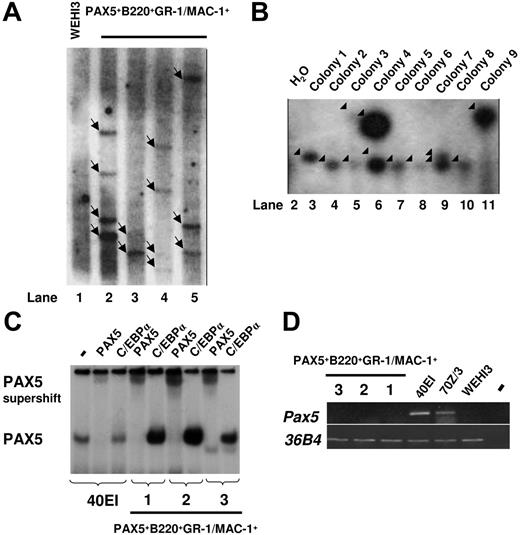
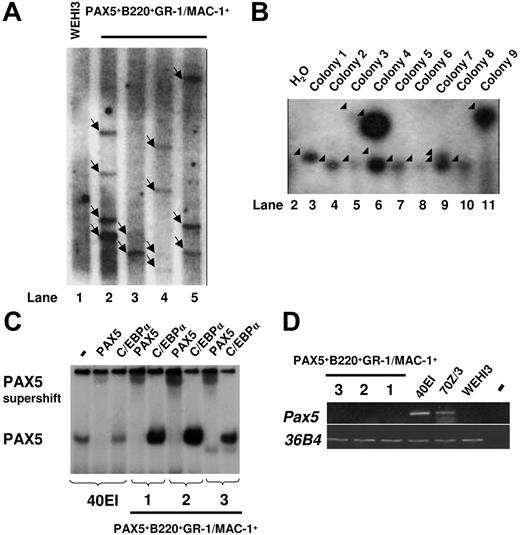
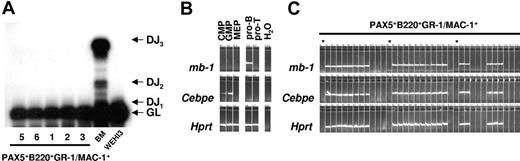
To investigate whether the sustained GM progenitors in PAX5-transduced cultures are monoclonally or oligoclonally derived, we next performed Southern blot analysis of HindIII-digested gDNA using a probe directed against PAX5. We observed multiple different integration sites of the retroviral vector, suggesting that the generated GFP(PAX5)+B220+GR-1/MAC-1+ cell populations were sustained through oligoclonal or polyclonal expansion and that the biphenotype and expansion of GFP(PAX5)+B220+GR-1/MAC-1+ cells was related to the expression of PAX5 rather than to any specific vector integration sites (Figure 5A). To verify the oligoclonal nature of PAX5-transduced cells and exclude that single clones contained the multiple integration sites observed, LM-PCR was performed on 9 individual colonies derived from a single cell line. Several different viral integration patterns could be seen in individual colonies (Figure 5B). Thus, although we cannot exclude the presence of a dominant integration site with the degree of resolution obtained with Southern blot of PCR products, these data support that the GM progenitors sustained by GFP(PAX5)+B220+GR-1/MAC-1+ cells are polyclonally derived.
Biphenotypic B220+GR-1/MAC-1+ cells generated from PAX5-transduced LSK cells represent an oligoclonal population with multiple viral integration sites and high expression of functional human PAX5. (A) Southern blot autoradiogram of 10μg HindIII-digested gDNA from PAX5-transduced cells from 4 different experiments 108 (lane 2), 96 (lane 3), 154 (lane 4), and 106 (lane 5) days after transduction. Lane 1 served as a negative control and contains DNA prepared from the WEHI3 cell line. Samples were hybridized with a probe specific for the retroviral vector. Arrows indicate major integration sites. As indicated by the black line, the gel has been cut to show results from 4 of 5 independently generated PAX5-transduced cultures. One experiment did not give satisfactory resolution and is therefore not shown. Consecutive lane numbers of the original gel are indicated below the panel. (B) Southern blots of LM-PCR products from 9 single CFU-GM colonies generated from FACS-purified GFP(PAX5)+B220+GR-1/MAC-1+ cells 55 days after transduction. The gel has been cut on the right side to delete a control bulk-transduced culture sample due to too high signal at this exposure and on the left side to remove results from another experiment. Consecutive lane numbers for the experiment on the original gel are indicated below the panel. Arrowheads indicate generated PCR products. (C) Autoradiogram of EMSAs using a 32P-labeled PAX5-binding site from the mouse CD19 promoter and nuclear extracts from either a mouse pre-B cell line (40EI) or from the PAX5-transduced myeloid cell cultures established (Exp nos. 1-3). The generated protein DNA complexes were further analyzed by either addition of anti-PAX5 or anti-C/EBPα antibodies as indicated, with the latter used as a negative control. Free probe is not shown. (D) Expression of mouse Pax5. Ethidium bromide-stained agarose gels with PCR products obtained by RT-PCR analysis of biphenotypic B220+GR-1/MAC-1+ cells generated in 3 independent experiments (Exp nos. 1-3) through PAX5 transduction of LSK cells, 2 pre-B cell lines (40EI, 70Z/3) and the myelomonocytic WEHI3 cell line. Water controls were always negative.
Biphenotypic B220+GR-1/MAC-1+ cells generated from PAX5-transduced LSK cells represent an oligoclonal population with multiple viral integration sites and high expression of functional human PAX5. (A) Southern blot autoradiogram of 10μg HindIII-digested gDNA from PAX5-transduced cells from 4 different experiments 108 (lane 2), 96 (lane 3), 154 (lane 4), and 106 (lane 5) days after transduction. Lane 1 served as a negative control and contains DNA prepared from the WEHI3 cell line. Samples were hybridized with a probe specific for the retroviral vector. Arrows indicate major integration sites. As indicated by the black line, the gel has been cut to show results from 4 of 5 independently generated PAX5-transduced cultures. One experiment did not give satisfactory resolution and is therefore not shown. Consecutive lane numbers of the original gel are indicated below the panel. (B) Southern blots of LM-PCR products from 9 single CFU-GM colonies generated from FACS-purified GFP(PAX5)+B220+GR-1/MAC-1+ cells 55 days after transduction. The gel has been cut on the right side to delete a control bulk-transduced culture sample due to too high signal at this exposure and on the left side to remove results from another experiment. Consecutive lane numbers for the experiment on the original gel are indicated below the panel. Arrowheads indicate generated PCR products. (C) Autoradiogram of EMSAs using a 32P-labeled PAX5-binding site from the mouse CD19 promoter and nuclear extracts from either a mouse pre-B cell line (40EI) or from the PAX5-transduced myeloid cell cultures established (Exp nos. 1-3). The generated protein DNA complexes were further analyzed by either addition of anti-PAX5 or anti-C/EBPα antibodies as indicated, with the latter used as a negative control. Free probe is not shown. (D) Expression of mouse Pax5. Ethidium bromide-stained agarose gels with PCR products obtained by RT-PCR analysis of biphenotypic B220+GR-1/MAC-1+ cells generated in 3 independent experiments (Exp nos. 1-3) through PAX5 transduction of LSK cells, 2 pre-B cell lines (40EI, 70Z/3) and the myelomonocytic WEHI3 cell line. Water controls were always negative.
To verify that the expressed PAX5 protein was able to bind DNA, we performed EMSA with nuclear extracts from 3 independently generated GFP(PAX5)+B220+GR-1/MAC-1+ lines (1, 2, and 3) and the PAX5-binding site from the mouse CD19 promoter. One prominent complex was detected using nuclear extracts from the pre-B cell line 40EI and all 3 analyzed GFP(PAX5)+B220+GR-1/MAC-1+ lines (Figure 5C). This complex was disrupted by the addition of anti-PAX5 antibody but not control (anti-C/EBPα) antibody demonstrating that the cells express high levels of functional PAX5 protein.
To resolve if any of the PAX5 protein detected by the EMSA was due to activation of the endogenous Pax5 gene we performed RT-PCR analysis using mouse-specific primers (Figure 5D). Mouse Pax5 transcript was detected in the pre-B cell lines 40EI and 70Z/3, whereas no message was detected either in the myelomonocytic cell line WEHI3 or in any of the 3 GFP(PAX5)+B220+GR-1/MAC-1+ cell populations. This suggests that enforced expression of human PAX5 protein in myeloid progenitor cells otherwise negative for endogenous Pax5 results in development of immortalized B220+GR-1/MAC-1+ biphenotypic cells.
PAX5 expressing biphenotypic B220+GR-1/MAC-1+ cells coexpress otherwise myeloid- and lymphoid-associated genes
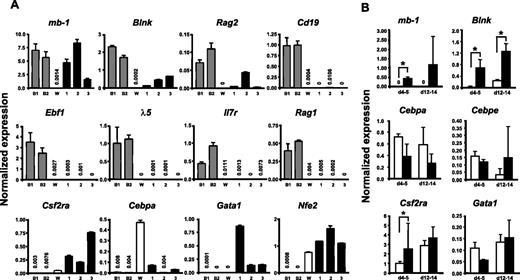
To investigate whether GFP(PAX5)+B220+GR-1/MAC-1+ cells also are biphenotypic with regard to expression of myeloid- and lymphoid-lineage programs, we performed microarray analysis using cells from one of the experiments (Exp no. 1) and compared the RNA expression patterns in these cells to that in Abelson-transformed pre-B cell lines (18-81 and 230-238) and the myelomonocytic cell line WEHI3 (Figure S2; Table S1). This comparison revealed that the GFP(PAX5)+B220+GR-1/MAC-1+ cells had a unique gene expression pattern with significant activity of genes found selectively either in the pre-B cell lines or the myeloid cell line. Using present (P)/absent (A) classification on the pre-B/myeloid cell-associated genes, we identified among the pre-B cell-associated genes, mb-1 and Blnk, which encode functional components of the B-cell receptor, being expressed in the GFP(PAX5)+B220+GR-1/MAC-1+ cells.
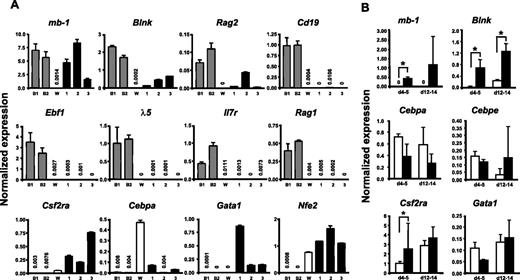
To verify that the long-term cultured GFP(PAX5)+B220+GR-1/MAC-1+ cells expressed a mix of lymphoid and myeloid genes, we performed Q-RT-PCR analysis of a set of lineage-associated genes. This revealed that the GFP(PAX5)+B220+GR-1/MAC-1+ cells expressed significant levels of the B-lymphoid–associated PAX5 targets mb-1, Blnk, and Rag2, whereas Cd19 expression was low or undetectable (Figure 6A). We could only detect very low or no expression of Ebf1, λ-5, Rag1, and Il7r in GFP(PAX5)+B220+GR-1/MAC-1+ cells (Figure 6A). As expected based on their morphology and sustained CFU-GM activity, biphenotypic cells did, however, express the myeloid-associated genes Csf2ra, Cebpa, Nfe2, and Gata1 (Figure 6A).
GFP(PAX5)+B220+GR-1/MAC-1+ cells coexpress genes characteristic for both lymphoid and myeloid cells. (A) Diagrams of TaqMan Q-RT-PCR analysis of gene expression in 2 pre-B cell lines 230-238 (B1) and 18-81 (B2), WEHI3 cell line (W), and 3 GFP(PAX5)+B220+GR-1/MAC-1+ long-term cell lines (Exp no. 1, 190 days; Exp no. 2, 80 days; Exp no. 3, 65 days) independently generated through PAX5 transduction of LSK cells. (B) GFP+ cells were sorted from control (□) and PAX5-transduced (▪) cultures at indicated times and RNA extracted for gene expression analysis. Differences in cDNA input were compensated by normalizing against Hprt expression levels. Mean (SD) values of 3 experiments. *P < .05. All water controls were negative (data not shown).
GFP(PAX5)+B220+GR-1/MAC-1+ cells coexpress genes characteristic for both lymphoid and myeloid cells. (A) Diagrams of TaqMan Q-RT-PCR analysis of gene expression in 2 pre-B cell lines 230-238 (B1) and 18-81 (B2), WEHI3 cell line (W), and 3 GFP(PAX5)+B220+GR-1/MAC-1+ long-term cell lines (Exp no. 1, 190 days; Exp no. 2, 80 days; Exp no. 3, 65 days) independently generated through PAX5 transduction of LSK cells. (B) GFP+ cells were sorted from control (□) and PAX5-transduced (▪) cultures at indicated times and RNA extracted for gene expression analysis. Differences in cDNA input were compensated by normalizing against Hprt expression levels. Mean (SD) values of 3 experiments. *P < .05. All water controls were negative (data not shown).
Although the comparison between the PAX5-transduced cells and the myelomonocytic and pre-B cell lines investigated was useful to establish whether myeloid and B-cell genes were coexpressed in GFP(PAX5)+B220+GR-1/MAC-1+ cells, they were less relevant for establishing whether overexpression of PAX5 would result in altered levels of these genes. Thus, we next compared the expression of mb-1, Blnk, and several key myeloid genes in GFP+ cells purified from control as well as PAX5-transduced cultures at 2 time points (day 4-5 and 12-14; Figure 6B). Importantly, these 2 early time points were chosen to ensure that the control and PAX5-transduced cells were as comparable as possible. These experiments confirmed the specific up-regulation of mb-1 and Blnk by PAX5 and also demonstrated that the investigated myeloid genes Cebpa, Cebpe, Csf2ra, and Gata1 were expressed at similar levels in control and PAX5-transduced cultures (Figure 6B).
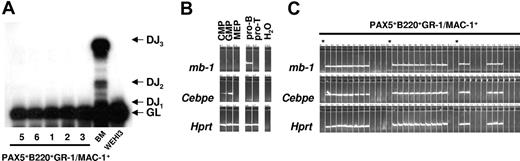
Next we analyzed immunoglobulin heavy (IgH) chain D to J (DJ) recombination in the GFP(PAX5)+B220+GR-1/MAC-1+ cells by PCR. The analysis used a set of 3 primers designed to amplify either the germline gene or any recombination event between D regions and J1-J3. Polyclonal rearrangements were easily detectable in the BM DNA, whereas no rearrangements could be detected in the GFP(PAX5)+B220+GR-1/MAC-1+ cell populations (Figure 7A). These data indicate that GFP(PAX5)+B220+GR-1/MAC-1+ cells do not contain committed B-cell progenitors, but rather myeloid cells with a unique mixed gene expression pattern imposed on them by ectopic expression of PAX5.
GFP(PAX5)+B220+GR-1/MAC-1+ cells coexpress lymphoid and myeloid programs at the single-cell level. (A) Polyclonal IgH chain D to J (DJ) recombination was detected in BM cells but not in the GFP(PAX5)+B220+GR-1/MAC-1+ cell populations from experiments 1, 2, 3, 5, and 6 analyzed by PCR. The analysis used a set of 3 primers designed to amplify either the germline (GL) gene or any recombination event between D regions and J1-J3. (B) Expression of mb-1, Cebpe, and Hprt in cells from populations (sorted by FACS) of common myeloid progenitors (CMPs), granulocyte-macrophage progenitors (GMPs), megakaryocyte-erythrocyte progenitors (MEPs), as well as, pro-B and DP T-cell progenitors (pro-T). (C) Co-expression of mb-1 and Cebpe in single GFP(PAX5)+B220+GR-1/MAC-1+ cells, as evaluated by multiplex single-cell RT-PCR.19 Asterisk indicates water control.
GFP(PAX5)+B220+GR-1/MAC-1+ cells coexpress lymphoid and myeloid programs at the single-cell level. (A) Polyclonal IgH chain D to J (DJ) recombination was detected in BM cells but not in the GFP(PAX5)+B220+GR-1/MAC-1+ cell populations from experiments 1, 2, 3, 5, and 6 analyzed by PCR. The analysis used a set of 3 primers designed to amplify either the germline (GL) gene or any recombination event between D regions and J1-J3. (B) Expression of mb-1, Cebpe, and Hprt in cells from populations (sorted by FACS) of common myeloid progenitors (CMPs), granulocyte-macrophage progenitors (GMPs), megakaryocyte-erythrocyte progenitors (MEPs), as well as, pro-B and DP T-cell progenitors (pro-T). (C) Co-expression of mb-1 and Cebpe in single GFP(PAX5)+B220+GR-1/MAC-1+ cells, as evaluated by multiplex single-cell RT-PCR.19 Asterisk indicates water control.
Even though the detection of both myeloid and B-lymphoid genes as well as coexpression of B220 and GR-1/MAC-1 on the cell surface supports the idea that PAX5 immortalized a myeloid precursor population coexpressing predominantly myeloid but also some lymphoid genes, Q-PCR could not exclude that the lymphoid and myeloid genes might be expressed in different subsets of progenitors. Thus, to get conclusive evidence for coexpression of myeloid and lymphoid genes in the immortalized myeloid progenitors, we further performed single-cell multiplex PCR on GFP(PAX5)+B220+GR-1/MAC-1+ cells.19 Purified myeloid- and lymphoid-restricted progenitors including CMP, GMP, MEP, pro-B cells, and DP T-cell progenitors were used to confirm lineage specificity of the gene expression patterns (Figure 7B). Notably, every GFP(PAX5)+B220+GR-1/MAC-1+ cell that expressed the housekeeping gene Hprt coexpressed the B-lineage–specific PAX5 target gene mb-1 and the myeloid associated gene Cebpe (Figure 7C).10
Discussion
The present studies demonstrate that PAX5 in addition to favoring B-cell commitment and development from multipotent stem/progenitor cells, if ectopically expressed in the myeloid lineage pathway, promotes enhanced maintenance of otherwise short-lived myeloid progenitors with a biphenotypic B220+GR-1/MAC-1+ cell surface phenotype. Notably, biphenotypic GFP(PAX5)+ B220+GR-1/MAC-1+ cells coexpress lymphoid PAX5 target genes mb-1, Blnk, and Rag2, along with myeloid genetic program. Importantly, multiplex single-cell PCR demonstrated that this coexpression did indeed reflect coexpression of B-cell–specific PAX5 target genes and myeloid genes in the same single cells rather than in different subpopulations. In fact, virtually every GFP(PAX5)+B220+GR-1/MAC-1+ cell coexpressed myeloid and PAX5 target genes.
Although GFP(PAX5)+B220+GR-1/MAC-1+ cultures sustained a population of myeloid progenitors in long-term culture, these were, as control-transduced progenitors, able to differentiate into fully mature granulocytes, macrophages, and mast cells, in agreement with previous studies demonstrating that ectopic PAX5 expression is fully compatible with normal myeloid differentiation in vivo.15,16 In fact, also using our vectors, PAX5-transduced LSK cells contributed normally to myelopoiesis in vivo (K.A. and S.E.W.J., unpublished observation, March 2002). Thus, myeloid progenitors expressing PAX5 appear to have extensively enhanced self-renewal potential but remain capable of balanced normal myeloid differentiation. Moreover, PAX5-transduced myeloid progenitors have an oligoclonal origin, as picked clones derived from individual progenitors in a CFU-GM assay showed largely distinct (although also overlapping) integration sites. Thus, these findings indicate that PAX5 promotes sustained polyclonal regeneration of myeloid progenitors, which remain dependent on GM-CSF or IL-3 stimulation. In agreement with this, all GFP(PAX5)+B220+GR-1/MAC-1+ cells expressed high levels of Csf2ra and Il3.
Our findings are in striking contrast to those of Chiang and Monroe who reported that overexpression of PAX5 in myeloid progenitors was accompanied by reduced terminal differentiation and enhanced apoptosis, and reduced GM-CSFRα expression.17,18 Although the reason for this discrepancy remains unclear, it could reflect the different myeloid differentiation conditions used in the 2 studies. In fact, during the first 8 days of cultures when Chiang and Monroe used similar conditions as ours, neither study showed any suppressive effects of PAX5 on myelopoiesis. It was first after 8 days when they switched to culture conditions (including ATRA) forcefully promoting terminal myeloid differentiation that they observed reduced cellular output.17 In contrast, under our conditions, which also included early stem/progenitor factors such as SCF, we observed that ectopic PAX5 expression prohibited the exhaustion of myeloid progenitors normally seen in such cultures. Regardless, our studies establish that ectopic expression of PAX5 introduces extensive self-renewal properties in otherwise short-lived myeloid progenitors. The potential mechanism for this profound effect on myeloid progenitor maintenance remains to be established but is unlikely to exclusively be explained by the reported ability of PAX5 to down-regulate expression of myeloid genes.7–9
Our findings are of particular interest in light of the observation that PAX5, normally being B-cell restricted,2–4,27 is frequently ectopically expressed in myeloid malignancies.11,12 The fact that PAX5 is widely expressed in the myeloid blasts in these cases,11,12 and also in clonally derived myeloid leukemic cell lines,28 virtually excludes that this finding is due to contaminating PAX5+ B cells. Interestingly, considering the B220+GR-1/MAC-1+ biphenotype of the sustained myeloid progenitors expressing PAX5, ectopic PAX5 expression has also been observed in human biphenotypic leukemias.12
It is unlikely that ectopic PAX5 expression in itself can induce myeloid leukemia since GFP(PAX5)+B220+GR-1/MAC-1+ cells generated and sustained in vitro failed to reconstitute normal or leukemic myelopoiesis on transplantation (K.A. and S.E.W.J., unpublished observations, July 2002). Thus, combined with the frequent ectopic expression of PAX5 in myeloid leukemia, our data would rather suggest that ectopic PAX5 expression in some cases might be involved in the progression of AML.
Authorship
Contribution: K.A. designed and performed research, contributed vital new reagents and analytical tools, analyzed data, and wrote the paper; C.R. designed and performed research, contributed vital new reagents, and analyzed data; R.M. designed and performed research, contributed vital new analytical tools, analyzed data, and wrote the paper; C.T.J. performed research; K.B. performed research; S.Z. performed statistical validation of data; Y.S. designed research; C.N. designed research and analyzed data; M.S. designed and performed research, contributed vital new analytical tools, analyzed data, and wrote the paper; and S.E.J. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sten Eirik W. Jacobsen, Hematopoietic Stem Cell Laboratory, Lund Strategic Research Center for Stem Cell Biology and Cell Therapy, Lund University, BMC B10, Klinikgatan 26, 221 84 Lund, Sweden; e-mail: sten.jacobsen@med.lu.se.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This work was supported by grants from the Swedish Cancer Society, the Swedish Research Council, and the Swedish Foundation for Strategic Research. The Lund Stem Cell Center is supported by a Center of Excellence grant from the Swedish Foundation for Strategic Research.
The authors are grateful to Dr Meinrad Busslinger for generously providing PAX5 cDNA, Drs Thomas Relander and Johan Richter for MGIN vector, Dr James Hagman for MIY vector, and Arthur Bank for the GP+E86 cell line. We also thank Ewa Sitnicka, Natalija Buza-Vidas, Jörg Cammenga, Carole Ramirez, Kim Theilgaard-Moench, and Jennifer Antonchuk for valuable feedback on our manuscript; Anna Fossum, Zhi Ma, and David Bryder for essential assistance in cell sorting; and Lilian Wittman, Gunilla Gärdebring, Ingbritt Åstrand-Grundström, Ann Brun, Jens Nygren, Jalal Taneera, Sidinh Luc, and Ramin Tehranchi for expert technical assistance and advice.