Abstract
Vascular endothelial growth factor A (VEGFA) and the type III receptor tyrosine kinase receptors (RTKs) are both required for the differentiation of endothelial cells (vasculogenesis) and for the sprouting of new capillaries (angiogenesis). We have isolated a duplicated zebrafish VegfA locus, termed VegfAb, and a duplicate RTK locus with homology to KDR/FLK1 (named Kdrb). Morpholino-disrupted VegfAb embryos develop a normal circulatory system until approximately 2 to 3 days after fertilization (dpf), when defects in angiogenesis permit blood to extravasate into many tissues. Unlike the VegfAa121 and VegfAa165 isoforms, the VegfAb isoforms VegfAb171 and VegfAb210 are not normally secreted when expressed in mammalian tissue culture cells. The Kdrb locus encodes a 1361–amino acid transmembrane receptor with strong homology to mammalian KDR. Combined knockdown of both RTKs leads to defects in vascular development, suggesting that they cooperate in mediating the vascular effects of VegfA in zebrafish development. Both VegfAa and VegfAb can individually bind and promote phosphorylation of both Flk1 (Kdra) and Kdrb proteins in vitro. Taken together, our data support a model in the zebrafish, in which duplicated VegfA and multiple type III RTKs mediate vascular development.
Introduction
Differentiation of endothelial cells to form the vascular network of arteries and veins requires a critical mitogenic signal, vascular endothelial growth factor A (VEGFA), which is transduced to the nucleus through a number of functionally redundant receptors. These receptors include the type III receptor tyrosine kinases VEGFR1/FLT1, VEGFR2/FLK1 (human KDR), and type I nonkinase transmembrane receptors neuropilin 1 (NRP1) and NRP2,1 which are critical mediators of both vasculogenesis (de novo formation of blood vessels) and angiogenesis (sprouting of vessels from preexisting vessels).2 Mice hemizygous at their single VegfA locus show defective vasculogenesis and die between 11 and 12 days after conception (dpc).3-5 Embryos homozygous for a targeted disruption of Flk1 (Vegfr2) die between 8.5 and 9.5 dpc from defects in both hematopoiesis and vasculogenesis, but disruption of Flt1 (Vegfr1) only leads to perturbed vascular patterning in full-term embryos.6,7 The tight regulation of VEGFA levels and the requirement for its receptors in hematopoiesis and vasculogenesis point to its critical role in the formation of blood vessels during development.
Differential splicing of the single VEGFA locus in humans gives rise to multiple protein isoforms, ranging in size from 121 to 206 amino acids (aa's) (VEGFA121, VEGFA145, VEGFA165, VEGFA189, and VEGF206).8,9 The VEGF isoforms differ mainly by the presence or absence of 2 heparin-binding domains encoded within exon 6 and 7 sequences.10-12 Expression of a single VEGFA isoform rescues VegfA mutant mice through birth, although the vascular networks differ with expression of different isoforms.13-16 VEGFA splicing is regulated during development and in disease, resulting in tissue- and stage-specific isoform ratios, which allow for distinct, context-dependent VEGFA signaling.17
Zebrafish have proven useful for dissecting vascular developmental pathways.18 A single zebrafish vegfA gene has been previously described, encoding the 121- and 165-aa isoforms.19,20 Although VegfA121 is the predominant isoform in early embryos and VegfA165 in adults, other alternate transcripts exist in specific tissues.21 The initial establishment of axial vasculature patterning does not require VegfA, but the sprouting of intersegmental vessels does.22 Together, these results show that VegfA signaling is required in zebrafish angiogenesis.
Many genes, such as the neuropilin genes, are duplicated in the zebrafish.23-26 The 4 zebrafish neuropilin genes, 2 orthologues each of neuropilin-1 (nrp1a and nrp1b) and of neuropilin-2 (nrp2a and nrp2b), are differentially expressed and play an important role in vascular development, possibly by interacting with VegfA.27-30 Previously, we have described a putative orthologue of KDR, flk1, that is expressed in the developing embryonic vasculature.31 A separate and yet unpublished type III RTK with homology to Flt1 (GenBank accession number: AAS92271) has been designated flt1, reflecting the similar structure of these receptors (Stefan Schulte-Merker, personal oral communication, June 2006).
The utility of the zebrafish as a model organism is largely based on the conservation of pathways involved in developmental and human disease processes.32-36 As only 2 VegfA isoforms have been described in the zebrafish and the previously described mutant for zebrafish Flk1 does not phenocopy the murine Flk1 knockout, we searched the zebrafish genome for other VegfA genes and FLK1/KDR homologues.37 By comparison of consensus mammalian VEGFA and KDR sequences to the available zebrafish genome, we indeed identified a duplicated vegfA gene (hereafter referred to as vegfAb) and a putative KDR gene (hereafter referred to as kdrb).
Zebrafish embryos with reduced levels of VegfAb, and separately Kdra and Kdrb, show defects in vascular development. In addition, in vitro overexpression of either the 171- or 210-aa isoforms of VegfAb in tissue culture cells demonstrates that both VegfAb isoforms are poorly secreted, whereas both VegfAa121 and VegfAa165 isoforms are highly secreted. However, both VegfAa and VegfAb can bind Kdra and Kdrb in vitro. These data support a role for multiple VegfA and type III RTKs in zebrafish vascular development.
Materials and methods
Zebrafish husbandry
Zebrafish were maintained and staged as described.38
Cloning, sequencing, RT-PCR, and RACE reactions
Using versions 2 and 3 of the Sanger Center zebrafish genome assembly, we used a standard tblastn algorithm to find the duplicated genes. We designed primers based on search results and isolated the full-length cDNA clones using a combination of oligo dT reverse-transcription–polymerase chain reaction (RT-PCR; BD Clontech, Palo Alto, CA) and rapid amplification of cDNA ends (RACE; BD Clontech). We isolated total RNA for the RT-PCR and RACE reactions from the TU strain using Absolutely RNA (Stratagene, La Jolla, CA) and used it for the RT-PCR and RACE reactions. RT-PCR of the open reading frames required amplification by nested PCR to visualize a product by agarose electrophoresis. Appropriately staged cDNA was first amplified in an MJ thermocycler (MJ Research, Watertown, MA) for 40 cycles (denaturing: 94°C for 20 seconds; annealing: 58°C for 30 seconds; and extension: 72°C for 1 minute) using the 5′ primer CCCTGTGTGAAGAAGCCGCTACT and 3′ primer CAACCACTTCACTTCATTCTCTCG. The resultant reaction was diluted 1/100 and 2 μL used as template for a subsequent PCR reaction using the 5′ primer GCTTTCTGAGACCTTTACCAATGC and 3′ primer TCTCAAGTCAGTTCGGTTCACCTCC for 30 more cycles. The GenBank accession numbers are DQ026828 for vegfAb210, DQ150576 for vegfAb171, and DQ026829 for kdrb.
In situ hybridization and photography
Whole-mount in situ hybridizations were performed as previously described.31,39 Antisense RNA probes corresponded to the complete ORF of each of the genes. Processed embryos were photographed in 90% glycerol on a Leica MZFLIII dissecting microscope (Leica Mikrosysteme Vertrieb, Wetzlar, Germany), PLAN Apo 1.0 objective, 8×-100× magnification. Images were acquired using a Zeiss Axiocam MNRC5 camera and Axiovision software version 4.001(both from Carl Zeiss, Thornwood, NY).
Alkaline phosphatase and hemoglobin staining
O-dianisidine staining for hemoglobin was performed as previously described.31 Alkaline phosphate staining of the vasculature was carried out by 4% paraformaldehyde fixation of the embryos for 2 hours at room temperature, followed by immersion in acetone for 30 minutes at −20°C. The embryos were rinsed twice in 1 × PBST (Gibco-BRL, Carlsbad, CA) and equilibrated in staining buffer (NTMT, 100 mM Tris pH 9.5, 50 mM MgCl2, 100 mM NaCl, 0.1% Tween-20) 3 times for 15 minutes each. The vasculature was visualized by staining in NTMT containing NBT and BCIP for 5 to 20 minutes.
Morpholino injections
A 1-nL volume of a 150-μM to 2-mM morpholino (Gene-Tools, Philomath, OR) dissolved in 1 × Danio buffer containing 100 μM of a fluorescein labeled control morpholino was injected into either AB or transgenic fli-gfp embryos at the 1- to 4-cell stage.
The sequences of the injected morpholinos were as follows: VegfAa: GTATCAAATAAACAACCAAGTTCAT; VegfAb ATG: GGAGCACGCGAACAGCAAAGTTCAT; VegfAb-75: TCCGATGATAGGCGCACTTTAGCAA; Kdra: CCGAATGATACTCCGTATGTCACTT; Kdrb-m1: TATGCTCTATTAGATGCCTGTTTAA; and Kdrb-m2: CAGTTATGCTCTATTAGATGCCTGT.
Rescue of the VegfAb morpholino VegfAb-75 was accomplished by subcloning the complete open reading frames (ORFs) of both Vegfab isoforms in PCS2+, and production of capped mRNA of each isoform separately (Ambion, Austin, TX). RNA (50 pg of each) was then simultaneously injected with the morpholino. All injected embryos were photographed live on a Zeiss Axiovert (fluorescence) using a Zeiss Axiocam and Axiovision software.
Affymetrix microarrays
Embryos injected with either the VegfAa, VegfAb-75 morpholinos, or controls were grown in standard embryo medium until 24 hours after fertilization (hpf). Embryos were then disaggregated in Trizol. Isolation of cDNA, hybridization, and analysis of the microarrays were accomplished as previously described.40
Cell culture and Western blotting
Chinese hamster ovary (CHO) cells, African green monkey kidney cells (Cos7), and human embryonic kidney cells (HEK293) were purchased from American Type Culture Collection (ATCC, Manassas, VA). Cell culture and transfection were performed in principal as previously described.41 After 48 hours of incubation, conditioned medium of transfected cells was collected and the transfected cells were lysed with lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA) supplemented with protease inhibitor cocktail (Complete Mini; Roche, Indianapolis, IN). The conditioned medium and total cell lysates were resolved by 6% and 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) for Flk1 and VegfA, respectively. For detection of Flk1 or VegfA proteins, the blots were incubated with mouse monoclonal anti-V5 antibody (1:5000 dilution; Invitrogen, Frederick, MD)
Ligand-receptor binding
HEK 293 cells were transfected with Igκ-HA-vegfAa165-myc/pSecTag2, Igκ-HA-vegfAb171-myc/pSecTag2, soluble kdra-V5/pcDNA3.1, and soluble kdrb-V5/pcDNA3.1. After 4 days of incubation, the conditioned media were collected. The amount of VegfA and RTK was estimated by Western blotting with anti-Myc antibody (1:2000 dilution) and mouse monoclonal anti-V5 antibody, respectively. The conditioned media were mixed and incubated at 4°C overnight with agitation. VegfA proteins were pulled down by antirabbit polyclonal V5 antibody (Novus Biologicals, Littleton, CO) and the complexes resolved by 6% SDS-PAGE. For detection of VegfA bound to Flk1, the membranes were incubated with mouse monoclonal anti-Myc antibody.
Results
Cloning and analysis of VegfAb and Flk1
The zebrafish vegfA gene located on zebrafish chromosome 16 encodes multiple isoforms, but no apparent homologues of human VEGF145, VEGFA189, or VEGFA206. We postulated that a second, duplicate zebrafish VegfA gene might encode the missing isoforms. Using a standard tblastn algorithm, short sequence segments with homology to mammalian VEGFA and KDR were identified using the zebrafish genome assembly versions 2 and 3, available at the Sanger Center website (http://www.sanger.ac.uk/Projects/Drerio/). We found a duplicate vegfA gene, hereafter referred to as vegfAb to distinguish it from the previously cloned vegfA gene (now termed vegfAa). This DNA sequence then served as the starting point for isolating the complete cDNA from TU fish using a combination of RACE and RT-PCR. Radiation hybrid mapping of this gene placed vegfAb on zebrafish chromosome 4, whereas vegfAa is located on chromosome 16 (Yi Zhou, personal written communication), November 2003).
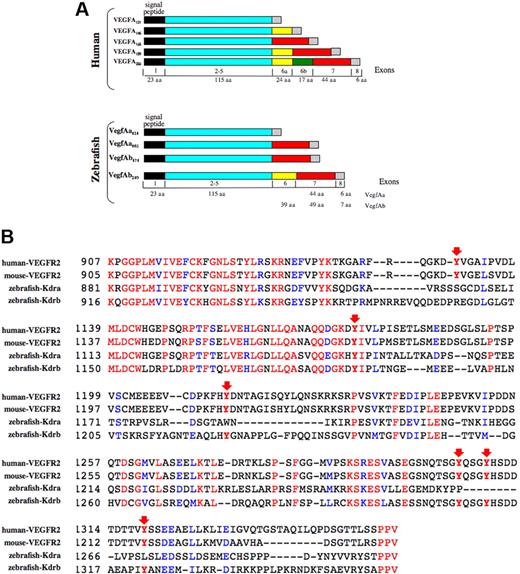
The full-length vegfAb cDNA contained 2814 nucleotides (nt's) and a predicted ORF of 233 aa's and encoded a putative protein with stronger homology to VEGFA than VEGFB, VEGFC, VEGFD, or PLGF (Figure 1). Therefore, the newly identified gene appeared to be a duplicated zebrafish vegfA gene. By RT-PCR, expression of 2 differentially spliced mRNAs was detected from the early embryo (8 hours after fertilization [hpf]) through 4 days after fertilization (dpf) (data not shown). The larger of 2 bands corresponded to an isoform of 210 aa's (VegfAb210), the smaller to a second isoform of 171 amino acids (VegfAb171). The 2 zebrafish VegfA proteins (VegfAa and VegfAb) were identical in size and exon structure to human VEGFA through the first 138 aa's (exons 1-5), suggesting common ancestry (Figure 2A); however, insertions and deletions occurred in the C-terminus, primarily in exons 6 and 7. Analysis of the genomic structure of the vegfA isoforms by blasting against the available zebrafish genome showed 6 exons for vegfAa121, 7 for vegfAa165 and vegfAb171, and 8 for vegfAb210; vegfAb171 and vegfAb210 share exons 1 to 5, 6, and 7 (Figure 2A). Figure 2 shows that exon use is highly conserved between humans and zebrafish, but also that exon 6a, which promotes mammalian VEGF binding to the basement membrane in VEGFA145, VEGFA189, and VEGFA206, is found only in vegfAb210. We suggest that zebrafish vegfAb210 may be the functional equivalent of these particular VEGFA isoforms and that vegfAb171 is orthologous to mammalian VegfA165.
Evolutionary relationship between human and zebrafish Vegfa isoforms and receptors. Protein sequences were aligned using ClustalW (European Bioinformatics Institute, Cambridge, United Kingdom) with the complete open reading frames. Species are indicated as follows: zf, zebrafish; m, mouse; and h, human. (A) Alignment of VegfA proteins shows a closer relationship of the newly isolated gene to VEGFA than VEGFB, VEGFC, VEGFD, and PLGF. (B) Alignment of RTK sequences. The zebrafish gene (Kdra) formerly referred to as flk1 shows a closer relationship to hFLT1 and mFlt1 than to hKDR (hFLK1) and mKDR (mFlk1). (C) Radiation hybrid mapping of zebrafish kdra, flt1, and kdrb and synteny comparison. Zebrafish kdrb is located on chromosome 20 in a region syntenic to human KDR, nearby the zebrafish orthologues of kit and pdgfra, which map near human KDR on human chromosome 4q12. Zebrafish kdra is on chromosome 14, and flt1 near cdk8 and ipf1 in a region on chromosome 24, which is syntenic to the human 13q12 region where FLT1, CDK8, and IPF1 are located.
Evolutionary relationship between human and zebrafish Vegfa isoforms and receptors. Protein sequences were aligned using ClustalW (European Bioinformatics Institute, Cambridge, United Kingdom) with the complete open reading frames. Species are indicated as follows: zf, zebrafish; m, mouse; and h, human. (A) Alignment of VegfA proteins shows a closer relationship of the newly isolated gene to VEGFA than VEGFB, VEGFC, VEGFD, and PLGF. (B) Alignment of RTK sequences. The zebrafish gene (Kdra) formerly referred to as flk1 shows a closer relationship to hFLT1 and mFlt1 than to hKDR (hFLK1) and mKDR (mFlk1). (C) Radiation hybrid mapping of zebrafish kdra, flt1, and kdrb and synteny comparison. Zebrafish kdrb is located on chromosome 20 in a region syntenic to human KDR, nearby the zebrafish orthologues of kit and pdgfra, which map near human KDR on human chromosome 4q12. Zebrafish kdra is on chromosome 14, and flt1 near cdk8 and ipf1 in a region on chromosome 24, which is syntenic to the human 13q12 region where FLT1, CDK8, and IPF1 are located.
Expression and structure of zebrafish VegfA genes and receptors. (A) Zebrafish vegfA exon/intron boundaries were predicted by blasting against the zebrafish genome. The structural similarities between vegfAb171, vegfAa165, and human VEGFA165 suggest that vegfAb171 is an orthologue of human VEGFA165 and similarly, that vegfAb210 is orthologous to human VEGFA189 and/or VEGFA206. (B) Alignment of the cytoplasmic domain of zebrafish Flk1 and human and mouse KDR. Conservation of critical tyrosine residues (arrows) shows that the flk1 RTK described here (Kdrb) is more closely related to KDR than the previously described putative flk1 gene (referred to now as kdra). See “Cloning and analysis of VegfAb and Flk1” for a full description of the critical residues and their function.
Expression and structure of zebrafish VegfA genes and receptors. (A) Zebrafish vegfA exon/intron boundaries were predicted by blasting against the zebrafish genome. The structural similarities between vegfAb171, vegfAa165, and human VEGFA165 suggest that vegfAb171 is an orthologue of human VEGFA165 and similarly, that vegfAb210 is orthologous to human VEGFA189 and/or VEGFA206. (B) Alignment of the cytoplasmic domain of zebrafish Flk1 and human and mouse KDR. Conservation of critical tyrosine residues (arrows) shows that the flk1 RTK described here (Kdrb) is more closely related to KDR than the previously described putative flk1 gene (referred to now as kdra). See “Cloning and analysis of VegfAb and Flk1” for a full description of the critical residues and their function.
By similar methods, we also obtained a 5360-nt kdrb cDNA that encoded a novel zebrafish KDR orthologue. Alignment of critical tyrosine residues between the zebrafish Kdrb cytoplasmic domain and human and mouse KDR (VEGFR2) showed that the flk1-related RTK described here is more closely related to KDR than the previously described flk1 gene.31 In humans, tyrosine residue Y1175 has been reported as a binding site for PLCγ1 and Src-homology 2 protein (Shb) in β-cells.42 The corresponding tyrosine residue in the mouse, Y1173, has recently been identified as an essential tyrosine residue for vasculogenesis and hematopoiesis, as knock-in mice substituting it with phenylalanine exhibited severe vascular and hematopoietic defects, similar to KDR-null mice.43 This critical tyrosine residue was conserved in both zebrafish Flk1 genes (Figure 2B). However, 4 important tyrosine residues in the C-terminal tail of KDR were conserved only in the zebrafish RTK gene reported here (Kdrb), but not in the previously identified RTK (Figure 2B). These residues, Y1214, Y1305, Y1309, and Y1319, mediate binding of focal adhesion kinase (FAK), which in turn interacts with PI3 kinase and paxillin.44-47 By radiation hybrid mapping (Yi Zhou, personal communication), the newly identified KDR gene was located on chromosome 20 near the zebrafish orthologues of KIT and PDGFRA, thereby establishing synteny between the zebrafish, human, and mouse KDR chromosomal regions (Figure 1C). Based on structural conservation, synteny, and biochemical data, we suggest that this RTK gene (Kdrb) is a duplicate of the previously cloned gene flka, which we suggest should be called Kdra. Another duplicated zebrafish type III RTK with homology to human FLT1 (Figure 1) is located on zebrafish chromosome 24, near the zebrafish orthologue of IPF and CDK8 (Yi Zhou, personal communication). Although this gene is not the subject of this paper, we suggest that it be called flt1 henceforth. The cloning and function of this FLT1 gene will be described elsewhere (Stefan Schulte-Merker, personal written communication, February 2005).
Expression of vegfAb and kdrb during embryogenesis
To analyze the temporal and spatial expression of the zebrafish vegfAb gene, we used whole-mount in situ hybridizations of wild-type embryos (Figure 3A). Both vegfAa isoforms were diffusely expressed at 12 somites, whereas at 24 hpf, although still broadly expressed, vegfAa165 was more evident in the somites compared with vegfAa121. At 2 dpf, expression of vegfAa165 was nearly absent, whereas vegfAa121 is still expressed in the developing heart and associated vasculature, head, and around the developing eye. There was no clear expression of vegfAa165 at 4 dpf, and vegfAa121 was restricted to developing pharyngeal structures and a subset of the developing CNS. A probe detecting both isoforms of vegfAb demonstrated significantly less expression at 12 somites compared with either vegfAa isoform. At 24 hpf, there was significant expression throughout the CNS and detectable, although significantly less than seen with vegfAa165, expression within the developing somites. At 2 dpf, expression was greatest surrounding the developing eye and retina, the pharyngeal arches, and glomeruli. At 4 dpf, expression persisted in vasculature surrounding the eye, and the glomeruli.
Expression of the zebrafish VegfA and Kdra genes in the developing embryo. (A) Whole-mount in situ hybridization with probes to vegfAa121, vegfAa165, or vegfAb. Both vegfAa121 and vegfAa165 are diffusely expressed at 12 somites, whereas vegfAb is not appreciably expressed at this time point. At 24 hours after fertilization (hpf), both isoforms of vegfAa were broadly expressed, although vegfAa165 was more clearly seen in the somites. vegfAa165 is more highly expressed in the developing CNS than vegfAb, whereas in the lens vegfAb is more highly expressed, and both are expressed in the developing somites ( ). At 2 dpf, vegfAa121 is expressed in the developing heart vasculature and pectoral fins. The aortic vasculature is also identified in vegfAb in situs, but in contrast, only vegfAb is expressed in the developing pronephros (◀). At 4 dpf, significant expression is restricted to vegfAb in the vasculature surrounding the eye. (B) Wild-type embryos were analyzed by whole-mount in situ hybridization with a probe to either kdrb (top panels) or kdra (bottom panels) at 16 somites (left panels), 2 dpf (middle panels), and 4 dpf (right panels). At 16 somites, expression is limited to the inner cell mass. By 2 dpf, the developing intersomitic vasculature expresses both kdrb and kdra. At 4 dpf, both genes are expressed in the developing subintestinal veins (SIVs) and in the remaining vasculature. See “In situ hybridization and photography” for image acquisition information.
). At 2 dpf, vegfAa121 is expressed in the developing heart vasculature and pectoral fins. The aortic vasculature is also identified in vegfAb in situs, but in contrast, only vegfAb is expressed in the developing pronephros (◀). At 4 dpf, significant expression is restricted to vegfAb in the vasculature surrounding the eye. (B) Wild-type embryos were analyzed by whole-mount in situ hybridization with a probe to either kdrb (top panels) or kdra (bottom panels) at 16 somites (left panels), 2 dpf (middle panels), and 4 dpf (right panels). At 16 somites, expression is limited to the inner cell mass. By 2 dpf, the developing intersomitic vasculature expresses both kdrb and kdra. At 4 dpf, both genes are expressed in the developing subintestinal veins (SIVs) and in the remaining vasculature. See “In situ hybridization and photography” for image acquisition information.
Expression of the zebrafish VegfA and Kdra genes in the developing embryo. (A) Whole-mount in situ hybridization with probes to vegfAa121, vegfAa165, or vegfAb. Both vegfAa121 and vegfAa165 are diffusely expressed at 12 somites, whereas vegfAb is not appreciably expressed at this time point. At 24 hours after fertilization (hpf), both isoforms of vegfAa were broadly expressed, although vegfAa165 was more clearly seen in the somites. vegfAa165 is more highly expressed in the developing CNS than vegfAb, whereas in the lens vegfAb is more highly expressed, and both are expressed in the developing somites ( ). At 2 dpf, vegfAa121 is expressed in the developing heart vasculature and pectoral fins. The aortic vasculature is also identified in vegfAb in situs, but in contrast, only vegfAb is expressed in the developing pronephros (◀). At 4 dpf, significant expression is restricted to vegfAb in the vasculature surrounding the eye. (B) Wild-type embryos were analyzed by whole-mount in situ hybridization with a probe to either kdrb (top panels) or kdra (bottom panels) at 16 somites (left panels), 2 dpf (middle panels), and 4 dpf (right panels). At 16 somites, expression is limited to the inner cell mass. By 2 dpf, the developing intersomitic vasculature expresses both kdrb and kdra. At 4 dpf, both genes are expressed in the developing subintestinal veins (SIVs) and in the remaining vasculature. See “In situ hybridization and photography” for image acquisition information.
). At 2 dpf, vegfAa121 is expressed in the developing heart vasculature and pectoral fins. The aortic vasculature is also identified in vegfAb in situs, but in contrast, only vegfAb is expressed in the developing pronephros (◀). At 4 dpf, significant expression is restricted to vegfAb in the vasculature surrounding the eye. (B) Wild-type embryos were analyzed by whole-mount in situ hybridization with a probe to either kdrb (top panels) or kdra (bottom panels) at 16 somites (left panels), 2 dpf (middle panels), and 4 dpf (right panels). At 16 somites, expression is limited to the inner cell mass. By 2 dpf, the developing intersomitic vasculature expresses both kdrb and kdra. At 4 dpf, both genes are expressed in the developing subintestinal veins (SIVs) and in the remaining vasculature. See “In situ hybridization and photography” for image acquisition information.
The expression of kdrb was indistinguishable from the expression of kdra (Figure 3B). Kdra was first expressed at about 7 to 8 somites in the bilateral stripes that contain the developing angioblasts (data not shown) and then localized to the intermediate cell mass (ICM) and the developing vasculature. By 30 hpf, kdra was expressed in the major trunk, head, and intersomitic vessels and persisted through 4 dpf. The expression patterns of kdra and kdrb in the major vessels are consistent with roles for both receptors in vascular development.
VegfAb morphants show defects in angiogenesis
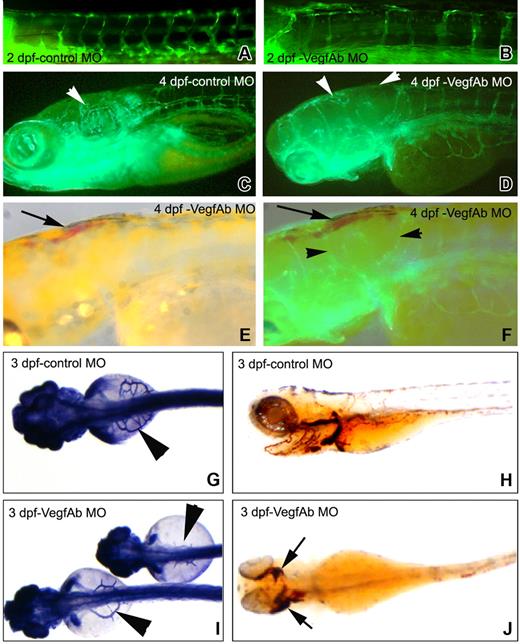
To determine whether VegfAb is required for vasculogenesis, angiogenesis, and hematopoiesis, either wild-type AB or transgenic fli1-gfp embryos were injected with morpholinos against vegfAb, and development of their vasculature and hematopoiesis was assayed. Injection of a vegfAb morpholino either against the start codon or the 5′ untranslated region (UTR) caused defects in angiogenesis evident at 2 dpf (Figure 4A), with extravasation of blood into the surrounding tissue beginning at about 3 to 4 dpf (Figure 4E,F,J), but had no effect on vasculogenesis, as judged by direct fluorescent visualization and alkaline phosphatase staining (data not shown). Indeed, red blood cells (RBCs) circulated unimpeded through the axial vasculature. The regular expression patterns of markers of early hematopoiesis in injected embryos, gata1 at 15 hpf and βE3 globin at 30 hpf (data not shown), suggest blood development is normal in VegfAb morphants.48
Vascular defects in VegfAb morphants. (A) Photomicrograph of a 5-bp mismatch vegfAb morpholino–injected (4.5 ng) fli1-gfp embryo at 2 dpf demonstrates no vascular defects (only one shown, although no phenotype was seen with either). (B) Defects in the formation of the ISVs anteriorly are clearly visible beginning at 2 dpf. (C) Photomicrograph of a 5-bp mismatch vegfAb control morpholino–injected (4.5 ng) fli1-gfp embryo at 4 dpf for comparison. (D-F) Beginning at 3 dpf, the number of circulating RBCs gradually decreased in 63% (162/255) of the embryos injected with 4.5 ng of the vegfAb ATG morpholino and in 47% (74/156) with 9 ng of the vegfAb-75 (5′UTR) morpholino (day-4 embryos are shown). Injection of an equivalent amount of 5-bp mismatch morpholinos did not affect vasculogenesis, and coinjection of the active vegfAb-75 (5′UTR) morpholino together with 50 pg each of vegfAb171 and vegfAb210 RNA reduced the number of embryos with defects in vasculogenesis by more than 50% (8/32 vs 22/40 abnormal embryo). VegfAb morphants showed decreased intersegmental vessel number and size ( , ◀ in panels D and F) and aberrant head vascular development. SIVs were severely reduced in number, size, and branching that was more pronounced anteriorly. (E) At 4 dpf, blood is apparent in the head and anterior embryos (
, ◀ in panels D and F) and aberrant head vascular development. SIVs were severely reduced in number, size, and branching that was more pronounced anteriorly. (E) At 4 dpf, blood is apparent in the head and anterior embryos ( ), which (F) corresponds to the areas with defects in angiogenesis. (G) Control morpholino–injected embryos demonstrate normal subintestinal vein (SIV) architecture by alkaline phosphatase staining, and (H) RBCs are shown by staining with o-dianisidine that stains hemoglobin reddish/brown. However, (I) injection of either morpholino targeting VegfAb leads to SIVs that are erratically placed and thin or nearly completely absent (only the start codon morpholino is shown) and (J) extravasation of RBCs in various structures, which is more pronounced anteriorly where the angiogenic defects are most visible (B-F). The results are combined from at least 3 separate experiments, and the photomicrographs are representative of the visible defects.
), which (F) corresponds to the areas with defects in angiogenesis. (G) Control morpholino–injected embryos demonstrate normal subintestinal vein (SIV) architecture by alkaline phosphatase staining, and (H) RBCs are shown by staining with o-dianisidine that stains hemoglobin reddish/brown. However, (I) injection of either morpholino targeting VegfAb leads to SIVs that are erratically placed and thin or nearly completely absent (only the start codon morpholino is shown) and (J) extravasation of RBCs in various structures, which is more pronounced anteriorly where the angiogenic defects are most visible (B-F). The results are combined from at least 3 separate experiments, and the photomicrographs are representative of the visible defects.
Vascular defects in VegfAb morphants. (A) Photomicrograph of a 5-bp mismatch vegfAb morpholino–injected (4.5 ng) fli1-gfp embryo at 2 dpf demonstrates no vascular defects (only one shown, although no phenotype was seen with either). (B) Defects in the formation of the ISVs anteriorly are clearly visible beginning at 2 dpf. (C) Photomicrograph of a 5-bp mismatch vegfAb control morpholino–injected (4.5 ng) fli1-gfp embryo at 4 dpf for comparison. (D-F) Beginning at 3 dpf, the number of circulating RBCs gradually decreased in 63% (162/255) of the embryos injected with 4.5 ng of the vegfAb ATG morpholino and in 47% (74/156) with 9 ng of the vegfAb-75 (5′UTR) morpholino (day-4 embryos are shown). Injection of an equivalent amount of 5-bp mismatch morpholinos did not affect vasculogenesis, and coinjection of the active vegfAb-75 (5′UTR) morpholino together with 50 pg each of vegfAb171 and vegfAb210 RNA reduced the number of embryos with defects in vasculogenesis by more than 50% (8/32 vs 22/40 abnormal embryo). VegfAb morphants showed decreased intersegmental vessel number and size ( , ◀ in panels D and F) and aberrant head vascular development. SIVs were severely reduced in number, size, and branching that was more pronounced anteriorly. (E) At 4 dpf, blood is apparent in the head and anterior embryos (
, ◀ in panels D and F) and aberrant head vascular development. SIVs were severely reduced in number, size, and branching that was more pronounced anteriorly. (E) At 4 dpf, blood is apparent in the head and anterior embryos ( ), which (F) corresponds to the areas with defects in angiogenesis. (G) Control morpholino–injected embryos demonstrate normal subintestinal vein (SIV) architecture by alkaline phosphatase staining, and (H) RBCs are shown by staining with o-dianisidine that stains hemoglobin reddish/brown. However, (I) injection of either morpholino targeting VegfAb leads to SIVs that are erratically placed and thin or nearly completely absent (only the start codon morpholino is shown) and (J) extravasation of RBCs in various structures, which is more pronounced anteriorly where the angiogenic defects are most visible (B-F). The results are combined from at least 3 separate experiments, and the photomicrographs are representative of the visible defects.
), which (F) corresponds to the areas with defects in angiogenesis. (G) Control morpholino–injected embryos demonstrate normal subintestinal vein (SIV) architecture by alkaline phosphatase staining, and (H) RBCs are shown by staining with o-dianisidine that stains hemoglobin reddish/brown. However, (I) injection of either morpholino targeting VegfAb leads to SIVs that are erratically placed and thin or nearly completely absent (only the start codon morpholino is shown) and (J) extravasation of RBCs in various structures, which is more pronounced anteriorly where the angiogenic defects are most visible (B-F). The results are combined from at least 3 separate experiments, and the photomicrographs are representative of the visible defects.
Beginning at 3 dpf, the number of circulating RBCs gradually decreased in 63% (162/255) of the embryos injected with 4.5 ng of the vegfAb start codon morpholino and in 47% (74/156) with 9 ng of the vegfAb 5′UTR morpholino. At the same time, RBCs were directly seen accumulating in the perivascular tissues, especially in the brain and in the eyes (Figure 4J), accounting for the drop in circulating RBCs. This loss was accompanied by increasing pericardial edema and death of the injected embryos between 5 to 7 dpf. Beginning at 2 dpf, injected embryos also showed defects in angiogenesis, particularly in the formation of intersegmental vessels and subintestinal veins (Figure 4B,G). The subintestinal vein defects ranged from near total absence to an erratically placed and thin vasculature (Figure 4I), the intersegmental vessels often missing or prematurely terminated (Figure 4B,D). These defects were more pronounced anteriorly, especially near the developing head and perhaps accounting for the tendency for blood accumulation in this area.
The VegfAa morphant phenotype differs significantly from that of VegfAb morphants. Whereas VegfAb morphants have normal vasculature and hematopoiesis until 2 dpf, VegfAa morphants have a nearly complete absence of the major intersegmental vessels at 2 dpf and reduced hematopoiesis (data not shown and Nasevicius et al22 ). Because injecting morpholinos against vegfAb should not alter the VegfAa121 and VegfAa165 isoforms, we wanted to test if coinjection of morpholinos against vegfAa and vegfAb might result in a more severe phenotype than that seen in VegfAa morphants. However, coinjection of morpholinos against vegfAa and vegfAb caused a vascular phenotype indistinguishable from that of VegfAa morphants (data not shown) and identical to the phenotype previously described for VegfAa morphants.20,22 These data suggest that that the role of VegfAa in angiogenesis is distinct from that of VegfAb.
Kdrb acts synergistically with Kdra
Targeted deletion of the Flk1 gene in the mouse leads to death around E9.5 with severe defects in both vasculogenesis and hematopoiesis.6 Because developmental pathways are often conserved between mice and zebrafish, we sought to determine whether the zebrafish Kdrb receptor, alone or in combination with Kdra, might also play a role in vasculogenesis or hematopoiesis. Embryos were first injected with 2 independent morpholinos directed against the 5′ UTR of kdrb. Injection of 4.5 ng of either morpholino did not lead to any demonstrable defect in angiogenesis. Variable defects in angiogenesis seen in 45 (40%) of 112 embryos injected at higher doses (> 9 ng/embryo) were also visualized in approximately 8% of an equal amount of 5-bp mismatch control morphants (14/184), suggesting this might due to nonspecific toxicity of the morpholinos. Hematopoiesis was unaffected in Kdrb morphants, as shown by whole-mount in situ hybridization with gata1 at 18 hpf and with βE3 globin at 30 hpf (data not shown).
Coinjecting kdrb and kdra morpholinos resulted in embryos in variable defects in the formation of the posterior vasculature, and most dramatically the intersegmental arteries (Figure 5C,F,I). These coinjection experiments suggested that Kdra and Kdrb cooperate in the growth, proliferation, and remodeling of the developing zebrafish embryonic vasculature, and particularly formation of the arteries. Compared with the murine Flk1 knockout phenotype, however, the phenotype of zebrafish coinjected with kdra and kdrb morpholinos was less severe and did not appear to affect hematopoiesis. Because of the large numbers of receptors mediating the VegfA signal and technical limitations on how many can be knocked down by morpholino injections, it is currently not possible to analyze vasculogenesis and angiogenesis in embryos lacking all VegfA activity. We therefore cannot determine whether the weaker phenotype resulted from residual Kdr-like activity or from species-dependent differences in receptor function.
Kdra and Kdrb cooperate to mediate vasculogenesis during zebrafish embryogenesis. Fli-gfp transgenic embryos were injected with combinations of morpholinos against kdra and kdrb or 5-bp mismatch control morpholinos. (A) Twenty-eight hpf embryos injected with either of 2 5-bp mismatch control morpholinos (4.5 ng each) corresponding to separate 5′UTR kdrb morpholinos (m1-kdrb [n = 32] and m2-kdrb [n = 44]) or the previously published kdra morpholino (m-kdra [n = 38]) had no vascular abnormalities. (B) Injection of 4.5 ng kdra or kdrb morpholino has no demonstrable effect on vasculogenesis at 28 hpf (107 embryos). Vascular defects were seen at higher concentrations of kdrb morpholinos; however, similar defects, but at a reduced penetrance, were also seen with equivalent amounts of the mismatched controls, suggesting these effects were not directly related to Kdrb function. (C) Coinjection of either 4.5 ng m1-kdrb or m2-kdrb with 4.5ng kdra morpholino caused variable loss of intersegmental arteries in 42 (24%) of 178 kdra/m1-kdrb or 53 (38%) of 138 kdra/m2-kdrb morpholino–injected embryos. (D) RBCs are seen in the anterior axial vasculature but cannot circulate. (E,F) Kdra/kdrb double morphant defects are still seen at 2 dpf. (G-I) At 4 dpf, injection of 4.5 ng kdra morpholino led to defects in the angiogenesis (◀,  ) in 34 (33%) of 101 injected embryos. Embryos coinjected with morpholinos targeting both kdra/b had severe axial vessel defects, although some subintestinal vasculature is apparent. The results are combined from at least 4 separate experiments and the photomicrographs are representative of the visible defects.
) in 34 (33%) of 101 injected embryos. Embryos coinjected with morpholinos targeting both kdra/b had severe axial vessel defects, although some subintestinal vasculature is apparent. The results are combined from at least 4 separate experiments and the photomicrographs are representative of the visible defects.
Kdra and Kdrb cooperate to mediate vasculogenesis during zebrafish embryogenesis. Fli-gfp transgenic embryos were injected with combinations of morpholinos against kdra and kdrb or 5-bp mismatch control morpholinos. (A) Twenty-eight hpf embryos injected with either of 2 5-bp mismatch control morpholinos (4.5 ng each) corresponding to separate 5′UTR kdrb morpholinos (m1-kdrb [n = 32] and m2-kdrb [n = 44]) or the previously published kdra morpholino (m-kdra [n = 38]) had no vascular abnormalities. (B) Injection of 4.5 ng kdra or kdrb morpholino has no demonstrable effect on vasculogenesis at 28 hpf (107 embryos). Vascular defects were seen at higher concentrations of kdrb morpholinos; however, similar defects, but at a reduced penetrance, were also seen with equivalent amounts of the mismatched controls, suggesting these effects were not directly related to Kdrb function. (C) Coinjection of either 4.5 ng m1-kdrb or m2-kdrb with 4.5ng kdra morpholino caused variable loss of intersegmental arteries in 42 (24%) of 178 kdra/m1-kdrb or 53 (38%) of 138 kdra/m2-kdrb morpholino–injected embryos. (D) RBCs are seen in the anterior axial vasculature but cannot circulate. (E,F) Kdra/kdrb double morphant defects are still seen at 2 dpf. (G-I) At 4 dpf, injection of 4.5 ng kdra morpholino led to defects in the angiogenesis (◀,  ) in 34 (33%) of 101 injected embryos. Embryos coinjected with morpholinos targeting both kdra/b had severe axial vessel defects, although some subintestinal vasculature is apparent. The results are combined from at least 4 separate experiments and the photomicrographs are representative of the visible defects.
) in 34 (33%) of 101 injected embryos. Embryos coinjected with morpholinos targeting both kdra/b had severe axial vessel defects, although some subintestinal vasculature is apparent. The results are combined from at least 4 separate experiments and the photomicrographs are representative of the visible defects.
Microarray analysis of VegfAa and VegfAb morphants
Although the expression patterns and morpholino analysis of vegfAa and vegfAb suggest that the 2 genes may function through different pathways, the magnitude of the differences is unclear. To help compare and contrast the pathways affected by the 2 genes, we determined the transcriptional profile of the 2 morphants by gene expression analysis. Affymetrix zebrafish GeneChips, containing approximately 14 900 zebrafish transcripts were used to examine the transcript expression of vegfAa and separately vegfAb morphants at 24 hpf, when the vegfAa morpholino phenotype is clearly evident and the embryos viable, and vegfAb was expressed. Two individual injections of vegfAb and vegfAa morphants compared with injected morpholino controls were made.
There was excellent correlation between the 2 injections of each morpholino (data not shown), and no obvious nonspecific toxicities were noted. We based further analyses on 302 genes in vegfAa morphants, 301 genes in vegfAb morphants, and 39 genes common to both morphants that were significantly regulated (Tables S1Table S2. Genes differentially regulated in VegfAb morphants (XLS, 1.56 MB)–S3, available on the Blood website; see the Supplemental Materials link at the top of the online article). When available through Zfin (http://zfin.org/cgi-bin/webdriver?MIval=aa-xpatselect.apg), the whole-mount in situ expression of the regulated gene is noted. A large number of genes affected in the vegfAa morphant that had a specific expression pattern were found to be expressed in the developing somites, myotomes, and CNS, while similarly, a number of genes expressed in the pharyngeal arches, CNS, eye, and lens were noted in vegfAb morphants. No clear expression pattern consensus emerged from the analyses of the commonly regulated vegfAa and vegfAb morphant genes.
We subsequently mapped our zebrafish expression values to human genes and then uploaded the mapped expression values for analysis with Ingenuity's Pathway Analysis program (IPA; Ingenuity Systems, Redwood City, CA), a human curated database of gene interactions. For the vegfAa morphant, the largest numbers of genes (37) have been implicated in “small molecule biochemistry,” followed by 30 in “skeletal and muscular system development and function.” In contrast, in vegfAb morphants the largest number of genes (71) are implicated in “cellular growth and proliferation.” A similar analysis of the 39 genes commonly altered in both vegfAa and vegfAb morphants did not clearly reveal particular pathway associations (data not shown).
Secretion and binding of VegfA and Flk1
VegfAa and vegfAb constructs encoding the entire ORF and fused to a 3′ terminal V5 tag were independently transfected into Chinese hamster ovary (CHO), COS7 (monkey kidney), and PAE (porcine aortic endothelial) cell lines (Figure 6). Although all 4 VegfA isoforms could be detected in cell lysates using a V5 antibody (Figure 6A), neither VegfAb171 nor VegfAb210 were secreted into the conditioned medium (Figure 6B), despite a well-defined signal peptide. Both Kdra and Kdrb can be detected in PAE cell lysates after transfection (Figure 6C).
VegfAb secretion and binding. Full-length vegfAa121, vegfAa165, vegfAb171, and vegfAb210 tagged with a V5 epitope at the N-termini were transfected into COS7 cells. (A) Western blot with a V5 antibody showed that although all 4 proteins are detectable in cell lysates (B) only VegfAa121 and VegfAa165 were secreted. (C) Transfected full-length kdra and kdrb fused to a V5 epitope were detectable in cell lysates of COS7 cells. (D) Replacing the endogenous signal peptide on all isoforms with the Igκ signal peptide resulted in secretion of both VegfAa and VegfAb. HEK 293 cells were transfected individually with Igκ-HA-vegfAa165-Myc, Igκ-HA-vegfAb171-Myc, soluble kdra-V5, and soluble kdrb-V5. Transfected cells were grown for 4 days, and conditioned media were collected, mixed, and incubated overnight. Immunoprecipitation (IP) with a V5 antibody (RTKs), blotting with Myc antibody (VegfAs), demonstrates that soluble Kdra bound to both VegfAa165 and VegfAb171; however, Kdrb did not appear to bind VegfAb171 as well as VegfAa165 in this assay. (E) Igκ-HA-vegfA-V5 constructs and Kdra-V5/pcDNA3.1 or Kdrb-V5/pcDNA3.1 were cotransfected into COS7 cells blotted with antiphosphotyrosine antibody, indicating activation of both RTKs by both VegfAa and VegfAb or (F) anti-V5 antibody demonstrating expression of the Kdra or Kdrb proteins.
VegfAb secretion and binding. Full-length vegfAa121, vegfAa165, vegfAb171, and vegfAb210 tagged with a V5 epitope at the N-termini were transfected into COS7 cells. (A) Western blot with a V5 antibody showed that although all 4 proteins are detectable in cell lysates (B) only VegfAa121 and VegfAa165 were secreted. (C) Transfected full-length kdra and kdrb fused to a V5 epitope were detectable in cell lysates of COS7 cells. (D) Replacing the endogenous signal peptide on all isoforms with the Igκ signal peptide resulted in secretion of both VegfAa and VegfAb. HEK 293 cells were transfected individually with Igκ-HA-vegfAa165-Myc, Igκ-HA-vegfAb171-Myc, soluble kdra-V5, and soluble kdrb-V5. Transfected cells were grown for 4 days, and conditioned media were collected, mixed, and incubated overnight. Immunoprecipitation (IP) with a V5 antibody (RTKs), blotting with Myc antibody (VegfAs), demonstrates that soluble Kdra bound to both VegfAa165 and VegfAb171; however, Kdrb did not appear to bind VegfAb171 as well as VegfAa165 in this assay. (E) Igκ-HA-vegfA-V5 constructs and Kdra-V5/pcDNA3.1 or Kdrb-V5/pcDNA3.1 were cotransfected into COS7 cells blotted with antiphosphotyrosine antibody, indicating activation of both RTKs by both VegfAa and VegfAb or (F) anti-V5 antibody demonstrating expression of the Kdra or Kdrb proteins.
To determine the ligand-receptor specificity, we analyzed the binding of soluble receptors tagged with V5 to VegfA isoforms tagged with Myc using the Igκ signal peptide in place of the VegfAb signal peptide, by which secretion of VegfAb in mammalian cells occurs (Figure 6D). HEK 293 cells were separately transfected with Igκ-HA-vegfAa165-Myc, Igκ-HA-vegfAb171-Myc, soluble kdra-V5, and soluble kdrb-V5. The conditioned VegfA and sRTK medias were pairwise mixed, incubated overnight, and then immunoprecipitated with anti-V5 antibody. Bound protein was detected by Western blot using an anti-Myc antibody (data not shown). Soluble Kdra bound to both VegfAa165 and VegfAb171; however, Kdrb did not appear to bind VegfAb171 as well as VegfAa165 in this assay. As human VegfA has been shown to bind multiple RTKs, we wanted to analyze if the different zebrafish VegfA genes and isoforms acted similarly.49 The zebrafish VegfA isoforms with an Igκ signal peptide and both HA and V5 tags were transfected into COS7 cells, and their expression was confirmed by blotting with an anti-V5 antibody (Figure 6F). Kdra and kdrb were separately transfected into these same cell lines with and without vegfAa165 or vegfAb171. Both vegfAa165 and vegfAb171 were able to activate both RTKs as demonstrated by antiphosphotyrosine antibody staining (Figure 6E). Therefore, the zebrafish VegfAa and VegfAb isoforms can both bind and activate Kdra and Kdrb.
Discussion
We report here the cloning and characterization of duplicated zebrafish genes that regulate vasculogenesis and angiogenesis. In addition to the reported VegfA locus, now renamed vegfAa, we find a second locus, vegfAb, which contains homologues of some of the mammalian VegfA isoforms not encoded by vegfAa. We also identified a second zebrafish RTK (Kdrb) with homology to mammalian KDR.
Expression of VegfAa and VegfAb during embryonic development
Whole-mount in situ hybridization of vegfAa and vegfAb revealed differing expression patterns of these 2 genes (Figure 3A). Whereas both vegfAa splice variants were broadly expressed at 12 somites, by 24 hpf, vegfAa165 was expressed in the CNS and the somites, whereas vegfAa121 RNA was more broadly distributed, although at 48 hpf it was expressed within/near the developing heart and vasculature. These data differ from the reported expression of VegfA during zebrafish embryogenesis. In that study, an unspecified probe that detected both splice variants was similarly seen in the prospective cardiac vasculature and somites but also in the pronephros (seen in the present studies with vegfAb165) and was not seen as diffusely within the CNS. The reasons for these differences are unclear but might be related to the differing time points analyzed and the probe chosen for use in these analyses. In the present studies, we separately used the ORF of each isoform, whereas the previous studies were done with an unspecified probe that recognized both alleles, and that perhaps included 3′UTR sequences as well. Our whole-mount in situ hybridization data also correlated with our microarray data. The expression of genes whose known embryonic whole-mount in situ pattern is within the developing CNS and myotomes was more likely to be altered in vegfAa morphants, whereas expression of genes in the CNS, eyes, lens, and pharyngeal arches was altered in vegfAb morphants. Interestingly, Hiratsuka et al (2005) examined the localization of VegfA by immunohistochemistry during murine development and found expression throughout the developing embryo, but especially in the anterior part of the embryo.50 This pattern more closely, but not completely, resembles the cumulative expression of both zebrafish VegfA genes than any one gene or splice variant. These expression patterns suggest that the various functions of mammalian VEGFA may be encompassed within more than just these 2 zebrafish VegfA genes.
The role of zebrafish Vegf and Kdr in vascular development
In VegfAb morphants, early vascular and hematopoietic development appeared normal at first, however as early as 2 dpf, morphants had defects in angiogenesis. Concomitant with the vascular phenotype, blood began to extravasate into the surrounding tissues beginning at 3 dpf (Figure 4). This extravasation of blood into the tissues surrounding the eye and CNS in vegfAb morphants is consistent with the later expression of vegfAb RNA in the vasculature surrounding the eye (Figure 3A), and CNS. Similar defects in angiogenesis are seen in Kdra morphants and the previously described zebrafish Kdra mutant.37 The intersomitic vascular defects seen in Kdra morphants appeared to encompass more of the embryo (Figure 5H) than those in either VegfAb morphant, whose defects were more pronounced anteriorly (Figure 4B,D). Whether this result is secondary to redundant signals activating Kdra or variable penetration of the VegfAb and Kdra morpholino phenotype is uncertain. Reducing all known VegfA signals by coinjection of morpholinos against vegfAa (previously described in Nasevicius et al22 ) and vegfAb resulted in the same vascular phenotype seen in VegfAa morphants. Loss of VegAa and VegfAb signaling in the zebrafish does not cause death in the developing embryo in the first few days of development, unlike in mice where hemizygous VegfA mutants show defective vasculogenesis and die between 11 and 12 days after conception (dpc).3-5 These differences are likely secondary to the ability of oxygen to diffuse directly into the embryo. Homozygous cloche embryos, which lack all endothelium and circulating erythrocytes do not die until 4 to 5 dpf.31,51,52 These results may also be due to activity of other molecules, such as Vegfc, that retain activity in these studies.53 However, these data are consistent with a model in which VegfAa and VegfAb have functionally diverged since arising from a common ancestor.
Beginning at 3 dpf, a delay in further maturation was seen in VegfAb morphants compared with controls (Figure 4C,D). However, 2 lines of evidence support our contention that the defects in vascular development were not caused by a general developmental delay in VegfAb morphants. The first is that the angiogenic defects were seen in both active VegfAb morpholinos, and not in their 5-bp mismatch injected controls. The second is that the defects in angiogenesis were visible at 2 dpf, when no overall differences in appearance were seen between the morphants and their controls. We further believe these defects are specific to the loss of VegfAb functioning in the injected embryos. Injection of equivalent amounts of 5-bp mismatch morpholinos did not affect vascular development, and coinjection of the active VegfAb-75 morpholino that targets the 5′UTR upstream of the start codon, with 50 pg each vegfAb171 and vegfAb210 RNA, reduced the number of embryos with severe defects in angiogenesis by more than 50% (8/32 vs 22/40 abnormal embryo).
Multiple RTK receptors are required for normal zebrafish vascular development
Morpholinos targeting both Kdra and Kdrb resulted in defects in vasculogenesis, especially of the intersegmental arteries, supporting a common pathway resulting from RTK activation leading to embryonic zebrafish vascular development. Yet compared with the knockout mouse, the phenotype of the combined morphant is less severe, possibly because of residual activity of the zebrafish flt1 gene, or possibly because other zebrafish genes may have acquired roles in vasculogenesis and angiogenesis. Lending support to these possibilities is that zebrafish treated with the type III RTK VEGF receptor inhibitor PTK787/ZK222584, which blocks all VEGF receptor activity, lack all major blood vessels.54
An alternative explanation for the differences seen between the murine KDR knockout and the zebrafish KDR morphant phenotypes is because of evolutionary changes to the molecular function of zebrafish RTKs and other genes. For example, the zebrafish phospholipase C γ-1 (plcγ1) mutant displays only a subset of the defects seen in Plcγ1 knockout mice. Significantly, the zebrafish plcγ1 mutant fails to replicate the complete defect of erythropoiesis.55,56
The overlapping expression patterns of Kdra and Kdrb, combined with the differential knockdown phenotypes seen in these studies, suggest that it is not the expression of these RTKs that generates differential development of the vascular system. We instead speculate that differences in ligand expression and/or other not yet described RTKs in the zebrafish account for these differences. These data are essentially similar to those seen by Covassin et al in their studies of these 2 RTK receptors.57 In their report, they described the morphant phenotypes of Kdra and Kdrb and extended their observations of the interaction between these 2 RTKs to their differing interactions in the formation of veins and arteries. With these experiments, we have extended their findings to demonstrate that multiple Vegfs can bind and activate these 2 RTKs. The successful in vitro expression and activation of both zebrafish Kdrs by both VegfA genes support a model in which both VegfA loci may signal through multiple RTKs. We can further hypothesize that these interactions may account for some of the complexities of VegfA signaling seen in developmental and pathological studies.
Transfection of native zebrafish vegfA constructs into mammalian cell lines showed that VegfAa121 and VegfAa165 are secreted, but VegfAb171 and VegfAb210 are not, even though a well-defined signal peptide is present. Although it is possible to speculate that the VegfAb isoforms are secreted by a novel zebrafish-specific mechanism in vivo, this effect may be due to the nature of zebrafish signal peptide recognition in a mammalian cell culture system.
Because of the ease with which zebrafish genes can be targeted and studied, we believe that the zebrafish will make important contributions to understanding these pathways. In addition, newly available mutant alleles and chemical inhibitors will enable a more complete block of VegfA signaling and identify the individual contributions of the different, redundant signal molecules and their receptors to vasculogenesis and hematopoiesis. As it becomes increasingly clear that vasculogenesis and angiogenesis act not only in embryonic development, but also in the adult to modulate wound healing, tissue regeneration, and progression of human diseases, the VegfA signaling pathway will become an important target for new molecular medicines.58-60 Because of its unique characteristics, the zebrafish system is an excellent system to model human development and disease and a detailed understanding of vasculogenesis and angiogenesis in the zebrafish will help make new medicines in this area a reality.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by grants from the Crohn's and Colitis Foundation of America, March of Dimes, National Institutes of Health (NIH) K08 HL03777 (N.B.), and NIH/National Cancer Institute 45548 (M.K.).
National Institutes of Health
Authorship
Contribution: N.B. designed and performed research, analyzed data, and wrote the paper; K.G. designed and performed research and analyzed data; C.S. performed research, analyzed data, and wrote the paper; G.W. performed research and analyzed data; J.L., C.A.S., and S.S.B. performed research; and M.K. and L.I.Z. designed research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nathan Bahary, University of Pittsburgh, 3501 5th Ave, 5058 BST3, Pittsburgh, PA 15260; e-mail:bahary@pitt.edu.



![Figure 5. Kdra and Kdrb cooperate to mediate vasculogenesis during zebrafish embryogenesis. Fli-gfp transgenic embryos were injected with combinations of morpholinos against kdra and kdrb or 5-bp mismatch control morpholinos. (A) Twenty-eight hpf embryos injected with either of 2 5-bp mismatch control morpholinos (4.5 ng each) corresponding to separate 5′UTR kdrb morpholinos (m1-kdrb [n = 32] and m2-kdrb [n = 44]) or the previously published kdra morpholino (m-kdra [n = 38]) had no vascular abnormalities. (B) Injection of 4.5 ng kdra or kdrb morpholino has no demonstrable effect on vasculogenesis at 28 hpf (107 embryos). Vascular defects were seen at higher concentrations of kdrb morpholinos; however, similar defects, but at a reduced penetrance, were also seen with equivalent amounts of the mismatched controls, suggesting these effects were not directly related to Kdrb function. (C) Coinjection of either 4.5 ng m1-kdrb or m2-kdrb with 4.5ng kdra morpholino caused variable loss of intersegmental arteries in 42 (24%) of 178 kdra/m1-kdrb or 53 (38%) of 138 kdra/m2-kdrb morpholino–injected embryos. (D) RBCs are seen in the anterior axial vasculature but cannot circulate. (E,F) Kdra/kdrb double morphant defects are still seen at 2 dpf. (G-I) At 4 dpf, injection of 4.5 ng kdra morpholino led to defects in the angiogenesis (◀, ) in 34 (33%) of 101 injected embryos. Embryos coinjected with morpholinos targeting both kdra/b had severe axial vessel defects, although some subintestinal vasculature is apparent. The results are combined from at least 4 separate experiments and the photomicrographs are representative of the visible defects.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/10/10.1182_blood-2006-04-016378/6/m_zh80230709790005.jpeg?Expires=1765986463&Signature=ABmPt2kZsoI0oBSeN0qsUgCPNrnxkhbGtEmQQLaFGFtZeb2i6~-4NqONwUK2f7vDN88A3oQl4jSeNuURX1SfTax14hKWA6oUr~7dkiUhj4ohMe5hmiET9tPxk~AQmMXZAy5Y4G-rPOKYakD3UlQSqs41H4jepdzwZcHYnvnngbjZf2jNVLfJNo8Bl3T2dRZP0OnKCs1QXU3bqFSTPQs6lg9D7EapUupDMZsY1wrLvFwT2GYzj95yPgmW2O7AXvE7XdWbEZFjoTRMEuIcow37-QWV5VcfzRpswS8NPOp9RHJCEV~f8Lbs8XEmhBjXN5Bl0KxHoA8lvsg6-a5AJRkBrA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

