Abstract
Analysis of the recombinatorial diversity of rearranged T cell receptor genes in mature T cells is an essential tool in the evaluation of immune status and immune reconstitution in hematopoietic cell transplantation studies. While the spectratyping technique is available in human clinical research and mouse models, the canine genome has not been sufficiently annotated to simply implement the assay on the dog TCRB locus. To this end, we have annotated by bioinformatics and experimentally all canine TCRBV segments, as well as TCRBD, TCRBC and most of TCRBJ segments. Of all 31 canine TCRBV found, 23 were functional and 8 were pseudogenes. A multiplex PCR-based assay was further designed to analyze the entire TCRBV spectratype in a set of 4 reactions each containing 4 to 5 V-segment specific forward primers and a common C-segment specific reverse primer. Direct sequencing of RT-PCR products confirmed that all amplified genes originated from predicted V-segments and that the designed V-specific PCR primers did not cross-react with other TCRBV family members. The usefulness of the spectratyping technique for canine model of transplantation was further demonstrated in analysis of T-cell repertoire reconstitution of irradiated dogs at different time points of recovery. Moreover, our simple and rapid V (J) annotation strategy relies on internet resources open to the general public and does not require specialized training in bioinformatics. It can be readily applied for de novo identification and mapping of TCRB gene families in other animal species where genome sequence drafts become available.
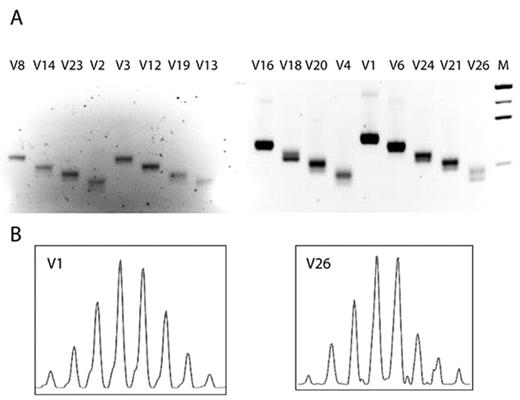
TCRB spectratype of fetal canine thymus. (A) RT-PCR using TCRBV family-specific forward primers (V1 through V26) and a common C region-specific reverse primer. (B) Amplification products specific for family V1 (high abundance) and V26 (low abundance) were copied in run-off reactions with the fluorescent C-specific primer and resolved on capillary gel.
TCRB spectratype of fetal canine thymus. (A) RT-PCR using TCRBV family-specific forward primers (V1 through V26) and a common C region-specific reverse primer. (B) Amplification products specific for family V1 (high abundance) and V26 (low abundance) were copied in run-off reactions with the fluorescent C-specific primer and resolved on capillary gel.
Author notes
Disclosure: No relevant conflicts of interest to declare.