Abstract
Proteinase 3 (PR3), a serine proteinase contained in neutrophil azurophilic granules, is considered a risk factor for vasculitides and rheumatoid arthritis when expressed on the outer leaflet of neutrophil plasma membrane and is the preferred target of antineutrophil cytoplasm autoantibodies (ANCA) in Wegener granulomatosis. ANCA binding to PR3 expressed at the surface of neutrophils activates them. Evidence is provided that neutrophil apoptosis induced significantly more membrane PR3 expression without degranulation (but no enhanced membrane CD35, CD66b, CD63, myeloperoxidase, or elastase expression). This observation was confirmed on cytoplasts, a model of granule-free neutrophils. We hypothesized that PR3 could interact with proteins involved in membrane flip-flop (eg, phospholipid scramblase 1 [PLSCR1]). PR3-PLSCR1 interaction in neutrophils was demonstrated by confocal microscopy and coimmunoprecipitation. In the RBL-2H3 rat mast-cell line stably transfected with PR3 or its inactive mutant (PR3S203A), PR3 externalization depended on PLSCR1, as shown by less PR3 externalization in the presence of rPLSCR1 siRNA, but independently of its serine-proteinase activity. Finally, apoptosis-externalized PR3 decreased the human macrophage-phagocytosis rate of apoptotic PR3 transfectants. Therefore, in addition to ANCA binding in vasculitis, the proinflammatory role of membrane PR3 expression may involve interference with macrophage clearance of apoptotic neutrophils.
Introduction
Proteinase 3 (PR3), also called myeloblastin,1 belongs to the family of neutrophil microbicidal serine proteinases that are stored within azurophilic granules.2 They are considered proinflammatory proteinases because they mediate deleterious effects on host tissues during inflammation.3,4 Intriguingly, among the numerous proteins contained within azurophilic granules, only PR3 and myeloperoxidase (MPO) are the main targets of antineutrophil cytoplasm antibodies (ANCA) associated with systemic vasculitides.5,6 More than 80% of Wegener granulomatosis patients have anti-PR3 ANCA, whereas only 10% have anti-MPO ANCA. By contrast, microscopic polyangiitis, Churg-Strauss syndrome, and pauci-immune crescentic glomerulosclerosis are generally associated with anti-MPO ANCA.7 Unlike MPO, whose subcellular localization is restricted to azurophilic granules, PR3 has been detected on secretory vesicle membranes and the outer leaflet of plasma membranes.8,9 ANCA have a pathophysiologic role because they activate neutrophils when ANCA antigens are expressed at the neutrophil membrane.7 We and others demonstrated that high percentages of membrane PR3-positive neutrophils could favor the development or progression of chronic inflammatory diseases, namely vasculitides and rheumatoid arthritis.10-12 Moreover, enhanced membrane PR3 expression was observed on neutrophils from patients with sepsis.13 Taken together, these data strongly suggest that membrane PR3 expression constitutes a pathogenic factor in ANCA-associated vasculitis and other inflammatory diseases involving neutrophils.
Because of the unexpected complex subcellular localization of PR3 in neutrophils, we previously investigated whether it could have specific molecular substrates and, consequently, unanticipated functions. To explore this possibility, we used the rat mast cell lines (RBL-2H3) stably transfected with human neutrophil elastase (HNE), PR3, or its inactive mutant PR3S203A and demonstrated that PR3, unlike its homolog HNE, was able to access cytosolic (eg, the cyclin-dependent kinase inhibitor p21/waf1)14 or membrane substrates (eg, procaspase-3).15 We also observed that PR3 could be expressed at the cell surface during apoptosis, independently of degranulation, and was strongly associated with phosphatidylserine (PS) externalization,16 which is an “eat-me” signal recognized by macrophages.17,18 Thus, it became evident, at least in our model of stable transfectants, that PR3 was located outside the granules.
The aim of the present study was (1) to identify the molecular mechanisms leading to membrane PR3 expression and (2) to compare PR3 to MPO, another ANCA target, and to HNE, its close serine-proteinase homolog.
Herein, we provide evidence that PR3 was externalized during apoptosis and was associated with human phospholipid scramblase-1 (hPLSCR1), a protein that has been implicated in the bidirectional movement of plasma-membrane phospholipids in response to high cytosolic calcium levels, injury, or apoptotic insult.19 We also show that apoptosis-induced PR3 externalization decreased the macrophage-phagocytosis rate of apoptotic cells, independently of its enzymatic activity.
Materials and methods
Approval was obtained from the INSERM Institutional Review Board for these studies. Informed consent from control blood donors to provide blood for research purposes was obtained in accordance with the Declaration of Helsinki.
Neutrophil and RBL-2H3 culture conditions and apoptosis induction
Human neutrophils were isolated from ethylenediaminetetraacetic acid (EDTA)–anticoagulated healthy donor blood from the Etablissement Français du Sang (Paris, France) using density-gradient centrifugation through polymorphoprep (Nycomed, Munich, Germany) as previously described.9 To trigger in vitro degranulation, neutrophils were preincubated with cytochalasin B (CB; 10 μg/mL; Sigma, St Louis, MO) in Hanks balanced salt solution (HBSS) containing Ca2+/Mg2+ (Gibco, Carlsbad, CA) for 5 minutes and were stimulated with 1 μM N-formyl-methionyl-leucyl-phenylalanine (f-MLP; Sigma) for 15 minutes. To induce physiologic apoptosis, neutrophils were resuspended at 2 × 106/mL in RPMI medium (Gibco) supplemented with 10% fetal calf serum (FCS) and kept for 16 hours at 4°C before being incubated for 6 hours at 37°C in a humidified 5% CO2 atmosphere. When indicated, neutrophil apoptosis was also potentiated with gliotoxin (0.1 μg/mL; Sigma), which inhibits NF-κB signaling.20
RBL-2H3 cells were transfected with pcDNA3 plasmid alone, pcDNA3/PR3, or pcDNA3/PR3S203A and were cultured as previously described.14 Apoptosis was induced by 16 hours of incubation at 37°C with 25 μM etoposide (Dakota Pharma, Bismarck, ND), which triggers apoptosis via the mitochondrial pathway.15
Neutrophils or RBL-2H3 cells labeled with phycoerythrin (PE)–conjugated annexin-V and 7-amino-actinomycin D (7-AAD) were used to assess apoptosis21 and necrosis,22 respectively, as previously described.16 Cells were analyzed using a FACSCalibur flow cytometer and CELLQuest version 3.3 software (Becton Dickinson Immunocytometry Systems, San Jose, CA).
Flow-cytometry analysis of PR3, HNE, MPO, CD35, CD66b, CD63, or PLSCR1 membrane expression
After incubating neutrophils (5 × 105) with heat-aggregated goat IgG–5% FCS to block membrane Fc receptors, cells were incubated for 30 minutes with either mouse anti-PR3 CLB (Central Laboratory of the Netherlands Red Cross Blood Transfusion Service; 2 μg/mL; CLB-12.8 clone),23 mouse anti-MPO (2 μg/mL; CLB), or mouse anti-HNE (37 μg/mL; Biogenesis clone 39A) monoclonal antibodies, followed by FITC-conjugated antimouse IgG (2 μg/mL; Immunotech, Marseille, France) for 30 minutes.9 Membrane expressions of CD35, CD66b, and CD63 were assessed using anti-CD35–FITC (25 μg/mL), anti-CD66b–FITC (50 μg/mL), or anti-CD63–PE (8 μg/mL) (Immunotech), respectively. Transfected RBL-2H3 cells were analyzed with mouse anti-PR3 and mouse anti–rat (r)PLSCR1 (clone 129.2; 5 μg/mL)24 monoclonal antibodies. Cytoplasts were also labeled with the mouse anti-PR3 monoclonal antibody (2 μg/mL) followed by biotinylated antimouse IgG and Alexa 488–conjugated streptavidin.
Cytoplast preparation
Cytoplasts or granule-free neutrophils were prepared from 3 × 108 neutrophils as described.25 Briefly, neutrophils were centrifuged through a discontinuous Ficoll-70 (Sigma) gradient (12.5%, 16%, 25%) containing CB (5 μg/mL) for 30 minutes at 34°C at 81 000g. The upper band of cellular material was composed of 99% cytoplasts. After several washes in phosphate-buffered saline (PBS), cytoplasts were resuspended at a final concentration of 8 × 106/mL in RPMI–10% FCS and incubated for 16 hours at 4°C (basal conditions) or at 37°C in a humidified 5% CO2 atmosphere to induce physiologic apoptosis with or without 0.1 μg/mL of gliotoxin.
Confocal-microscopy analysis of immunofluorescence labeling of PR3, PLSCR1, or αII-spectrin
Neutrophils, cytoplasts, or RBL-2H3 cells were induced to adhere on poly-L-lysine–precoated coverslips; fixed in PBS-3% para-formaldehyde (PFA); and permeabilized with either methanol or streptolysin-O (SLO)15 as indicated. After saturation with PBS-1% BSA, cells were incubated for 45 minutes with the primary antibody and incubated for 30 minutes with the secondary antibody as follows: rabbit polyclonal anti-MPO (1.4 μg/mL; Calbiochem, San Diego, CA) followed by FITC–antirabbit IgG (6 μg/mL; Jackson ImmunoResearch Laboratories, Bar Harbor, ME); mouse monoclonal anti-PR3 (2 μg/mL) followed by Alexa 488–antimouse IgG (6.7 μg/mL; Molecular Probes, Eugene, OR); rabbit polyclonal anti–αII-spectrin (2 μg/mL)26 followed by Alexa 555–conjugated antirabbit IgG (13 μg/mL; Molecular Probes); rabbit anti-hPLSCR1 (2 μg/mL) produced against a peptide mapping the 306–318 amino acids (CESTGSQEQSSGVW)27,28 followed by Alexa 555–conjugated antirabbit IgG (13 μg/mL). The specificity of the antibody has been confirmed in neutrophils by fluorescence microscopy after competition experiments in the presence of the peptide (data not shown). To label cytoplast PR3, mouse anti-PR3 monoclonal antibody (8 μg/mL) was followed by biotinylated antimouse IgG (20 μg/mL; Dako, Carpinteria, CA) and Alexa 488–conjugated streptavidin (10 μg/mL; Molecular Probes). To label rPLSCR1 on RBL-2H3 cells, rhodamine-conjugated mouse anti-rPLSCR1 monoclonal antibody (3 μg/mL; clone 129.2)24 was used. The specificity of the 129.2 antibody was confirmed by confocal microscopy (data not shown) and Western-blot analysis (Figure 6B) in rPLSCR1 siRNA experiments. To perform double PR3 and rPLSCRI immunolabeling in RBL cells, immunopurified rabbit polyclonal anti-PR3 raised against the whole protein (3 μg/mL, from the laboratory of Dr Jürg Wieslander, Wieslab, Lund, Sweden) specific for neutrophil PR3 was used. Slides were mounted using Vectashield mounting medium (Vector Laboratories, Burlingame, CA) and analyzed either by fluorescence with a Leica DMRD and an Olympus camera or confocal microscopy with a Zeiss LSM-5 confocal scanning laser microscope. Zeiss LSM Image Browser version 3.5.0.376 software was used for image acquisition and image processing. Percentages of colocalization were determined using LSM 510 (Laser Scanning Microscope) version 3.2 SP2 (Zeiss).
Coimmunoprecipitation experiments and Western-blot analysis
Coimmunoprecipitation experiments were performed with neutrophils under basal or apoptotic conditions.24 Briefly, 2.5 × 106 cells were lysed in 500 μL of lysis buffer (50 mM NaCl, 50 mM NaF, 1 mM sodium ortho-vanadate, 1% Chaps, 50 mM N-2-hydroxyethylpiperazine-N-2-ethanesulfonic acid (HEPES), and antiproteinases [4 mM phenylmethylsulfonyl fluoride, 400 μM leupeptin, 400 μM pepstatin, 2 mM EDTA]) for 10 minutes at 4°C and centrifuged to obtain the soluble fraction. PLSCR1 was immunoprecipitated with rabbit anti-hPLSCR1 polyclonal antibody directly coupled to Sepharose-4B beads (Pharmacia, Uppsala, Sweden). Mouse anti-PR3 monoclonal antibody (4 μg) was mixed with 70 μL of Sepharose–protein G beads (Amersham, Arlington Heights, IL) and 400 μL of lysis buffer on a rotating wheel at 4°C for 4 hours. Antibody-coupled beads were incubated for 2 hours at 4°C with 500 μL of cell lysate to immunoprecipitate PR3. Controls consisted of incubating equivalent amounts of each lysate with isotypic control antibody coupled to Sepharose beads. Bound material was eluted in 50 μL of Laemmli sample buffer 3× and analyzed by Western blotting as previously described.29 PR3 and hPLSCR1 were detected using a rabbit polyclonal anti-PR3 peptide1,14 and a rabbit polyclonal anti-hPLSCR1,27 respectively, followed by horseradish peroxidase–conjugated antirabbit IgG (1:5000; Nordic Immunology, Tilburg, The Netherlands), using the SuperSignal West Pico detection kit (Pierce, Rockford, IL).
rPLSCR1 siRNA experiments
RBL/PR3 cells were transfected by electroporation with small-interfering RNA (siRNA) twice at a 24-hour interval. Cells were resuspended at 10 × 106/mL in electroporation buffer (120 mM KCl; 10 mM NaCl; 1 mM KH2PO4; 10 mM glucose; 20 mM HEPES, pH 7) and 900 μL of cell suspension was electroporated (Eurogentec) at 224 V, 2100 μF. Immediately, 750 nM of control siRNA (UGC-UAG-UGA-CCU-GCC-AUA-A; Eurogentec, Seraing, Belgium) or rPLSCR1 siRNA (GGC-AGG-ACG-UUC-UAA-AGG-U) was added to the electroporated cells and incubated for 15 minutes at room temperature. Cells (0.5 × 106/mL) were then deposited into a 6-well plate and incubated for 16 hours in DMEM-10% FCS at 37°C without antibiotics. Rhodamine-conjugated control siRNA was used to detect intracellular siRNA by flow cytometry and monitor transfection efficiency. Apoptosis was induced by 16 hours of incubation with 25 μM etoposide. RBL/PR3 cells transfected with specific rPLSCR1 siRNA or control siRNA were lysed and rPLSCR1 expression was analyzed by Western blotting using the mouse anti-rPLSCR1 monoclonal antibody. The membrane was stripped by incubation in Restore Buffer Solution (Pierce) and reprobed with anti-FcϵRIβ (1 μg/mL; clone 30.9)30 as the loading control. Blots were quantified after scanning of the films.
Quantification of phagocytosis of RBL-2H3 by macrophages
Human monocytes were isolated from blood, incubated for 24 hours in DMEM-10% FCS containing 1 ng/mL of phorbol-miristate acetate (PMA; Sigma), then washed twice in PBS and cultured for 7 days in DMEM-10% FCS. The phagocytosis assay was performed on day 8 after PMA treatment. Control RBL-2H3, RBL-2H3/PR3, or RBL-2H3/PR3S203A cells (0.5 × 106/mL) were incubated for 16 hours with 25 μM etoposide to induce apoptosis. Control and apoptotic RBL-2H3 transfectants were loaded with 50 μg/mL tetramethyl-6-carboxyrhodamine (TAMRA; Molecular Probes) for 2 hours at 37°C. After removing the culture medium, 1.5 × 106 RBL-2H3 transfectants were incubated with 0.5 × 106 macrophages for 2 hours at 37°C in DMEM-10% FCS to assess phagocytosis. After 2 washes in PBS, cells were detached from the plates, labeled with FITC-conjugated anti-CD14 monoclonal antibody (Immunotech), fixed, and cytocentrifuged. Phagocytosis was analyzed by confocal fluorescence microscopy by counting the number of CD14+ macrophages (green) containing a TAMRA-loaded (red) RBL-2H3 transfectant. Phagocytosis of apoptotic neutrophils is described in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Statistical analyses
Statistical analyses were performed using Statview for Windows version 5.0 software (SAS Institute, Cary, NC). Comparisons were made by analysis of variance (ANOVA) or Student t test when appropriate. Differences were considered significant when the P value was less than .05.
Results
Apoptotic human neutrophils externalize PR3 without degranulation
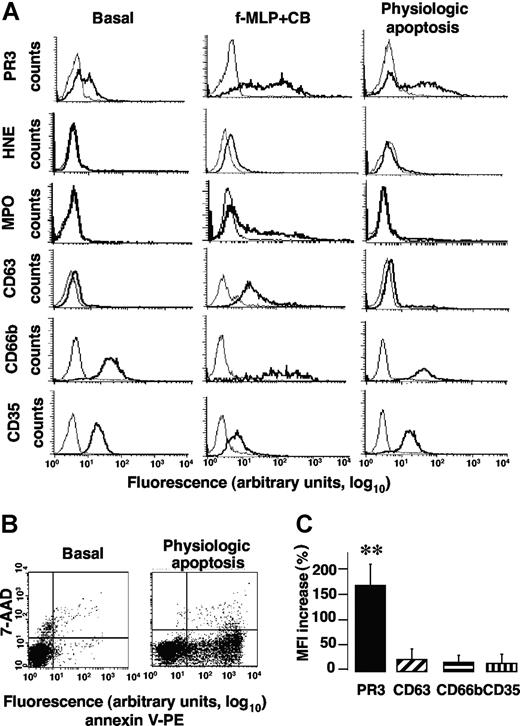
We first sought to determine the mechanisms by which PR3 was translocated to the neutrophil surface compared with proteins localized in azurophilic granules, namely, HNE, MPO, and CD63. Their membrane expressions were assessed by flow cytometry after neutrophil immunolabeling following either degranulation or apoptosis and compared with basal conditions. Exocytosis of specific granules and secretory vesicles was evaluated as membrane CD66b or CD35 expression, respectively.31
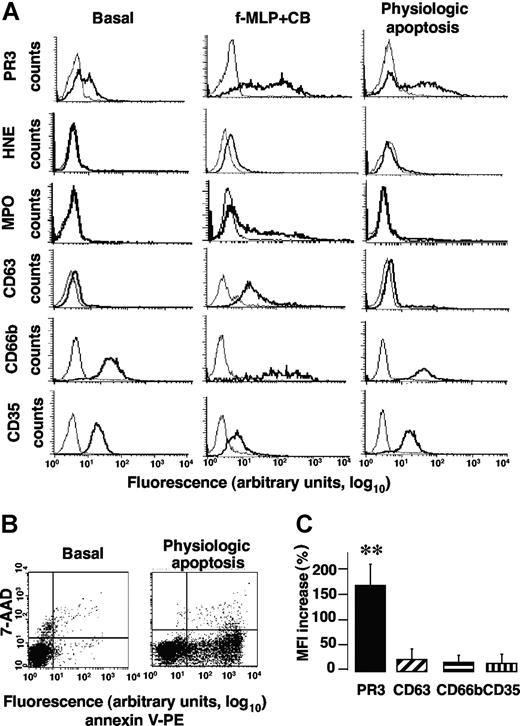
As previously described,10 a subset of freshly isolated neutrophils expressed PR3 (so-called mPR3+ subset) at their surface. In the representative experiment reported in Figure 1A, 46% of neutrophils expressed PR3 at their membrane under basal conditions. By contrast, no HNE, MPO, or CD63 could be detected but CD66b and CD35 were present. After f-MLP triggered azurophilic degranulation in the presence of CB, the percentage of PR3-expressing neutrophils remained constant but the mean fluorescence intensity (MFI) of membrane PR3 expression was significantly increased (note the shift to the right) versus basal conditions (38.8 ± 12 vs 7.2 ± 1.2, n = 6, **P < .01, respectively). As expected, compared with basal conditions, f-MLP + CB-induced neutrophil degranulation (Figure 1A) led to the significantly higher membrane expression of CD63 (MFI 31.3 ± 8 vs 4.5 ± 0.4, n = 5, **P < .01) and CD66b (MFI 114.3 ± 8 vs 42.2 ± 2.8, n = 5, **P < .01) and MPO (MFI 60.3 ± 8 vs 3.5 ± 0.4, n = 5, **P < .01) detection. However, membrane HNE expression rose only slightly under these conditions, thereby confirming our previous findings obtained with neutrophils9 and stably HNE-transfected RBL-2H3 cells, demonstrating that HNE was released as a soluble serine proteinase but was not expressed at the plasma membrane.16 Note the significant decline of membrane CD35 expression after azurophilic degranulation versus basal conditions (MFI 9.7 ± 0.3 vs 23.2 ± 2.2, n = 5, **P < .01, respectively). This observation is not surprising, as surface CD35 down-regulation after azurophilic degranulation attributed to intracellularly stored proteinases released during mobilization has been reported.9,32
Apoptosis increased membrane PR3 expression without degranulation of isolated human neutrophils. (A) Flow-cytometry analysis of membrane expression of PR3, HNE, MPO, CD63, CD66b, and CD35 (bold lines) or control IgG (thin lines) on isolated resting neutrophils (basal) or after f-MLP plus CB-triggered degranulation or physiologic apoptosis induction. This representative experiment used neutrophils from 1 donor; comparable results were obtained with neutrophils from 4 others. (B) Evaluation of PS externalization on apoptotic neutrophils. This typical experiment shows PS externalization detected with PE-conjugated annexin-V binding, plotted versus cell necrosis evaluated by 7-AAD labeling. (C) Variations of the membrane PR3, CD63, CD66b, or CD35 expression before and after apoptosis. After inducing apoptosis, mean fluorescence intensity (MFI) of membrane PR3 expression increased significantly compared with basal conditions (**P < .01 using ANOVA), whereas MFI of CD63, CD66b, or CD35 remained unchanged. Results are means (± SEM) from 5 independent experiments with neutrophils from 5 different healthy donors.
Apoptosis increased membrane PR3 expression without degranulation of isolated human neutrophils. (A) Flow-cytometry analysis of membrane expression of PR3, HNE, MPO, CD63, CD66b, and CD35 (bold lines) or control IgG (thin lines) on isolated resting neutrophils (basal) or after f-MLP plus CB-triggered degranulation or physiologic apoptosis induction. This representative experiment used neutrophils from 1 donor; comparable results were obtained with neutrophils from 4 others. (B) Evaluation of PS externalization on apoptotic neutrophils. This typical experiment shows PS externalization detected with PE-conjugated annexin-V binding, plotted versus cell necrosis evaluated by 7-AAD labeling. (C) Variations of the membrane PR3, CD63, CD66b, or CD35 expression before and after apoptosis. After inducing apoptosis, mean fluorescence intensity (MFI) of membrane PR3 expression increased significantly compared with basal conditions (**P < .01 using ANOVA), whereas MFI of CD63, CD66b, or CD35 remained unchanged. Results are means (± SEM) from 5 independent experiments with neutrophils from 5 different healthy donors.
Physiologic apoptosis induction by 6 hours of incubation at 37°C was confirmed by the increase of PS externalization measured by flow cytometry after PE-conjugated annexin-V labeling (Figure 1B). Negative freshly isolated neutrophils became positive after apoptosis induction with respective mean membrane PS expression: 0% versus 41.4% ± 3.4% (n = 5). After physiologic apoptosis induction, membrane expression of proteins from azurophilic granules (eg, CD63), specific granules (eg, CD66b), or secretory vesicles (eg, CD35) was not enhanced compared with basal conditions, demonstrating the absence of apoptosis-associated neutrophil granule exocytosis. Moreover, neither HNE nor MPO was detected on neutrophil surface. By contrast, membrane PR3 expression increased significantly in the mPR3+ subset versus basal conditions (Figure 1C). Thus, higher membrane PR3 expression was detected on neutrophils after apoptosis without degranulation and this feature was specific to PR3, as it was not seen for other granule proteins.
Apoptosis-enhanced PR3 expression on cytoplasts
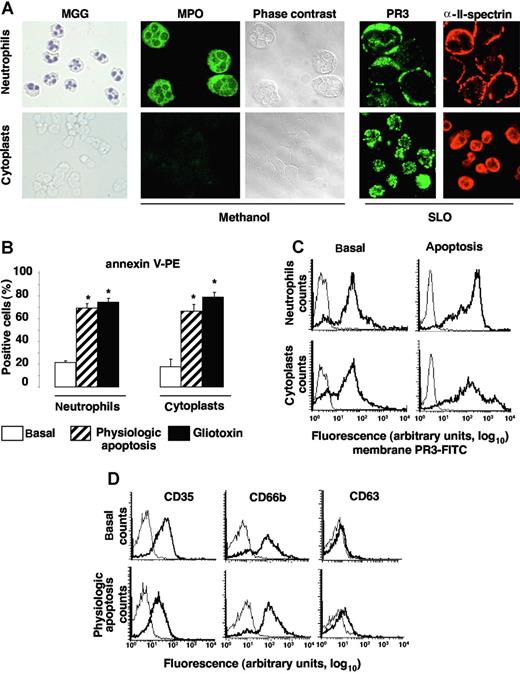
To demonstrate that apoptosis and degranulation are 2 distinct mechanisms leading to PR3 mobilization to the neutrophil surface, we used cytoplasts as cellular models of granule-free neutrophils.25 After May-Grünwald-Giemsa (MGG) staining, cytoplasts were recognized by (1) their absence of nuclei, unlike neutrophils, and (2) their MPO negativity, confirmed by immunolabeling after methanol permeabilization (Figure 2A). PR3 expression under resting conditions was observed by confocal microscopy after SLO permeabilization, which precludes granule protein labeling. For cytoplasts, similar results were obtained after methanol permeabilization because they do not contain granules (data not shown). αII-spectrin is a ubiquitous protein associated with cytoskeletal molecules that form a complex scaffolding under the lipid bilayers.33 Immunolabeling with anti–αII-spectrin was used to verify that cytoplast plasma membranes were intact (Figure 2A). Another element indicating their integrity was the exclusion of trypan-blue dye by 99% of cytoplasts (data not shown). Apoptosis was induced by an overnight incubation at 37°C with or without gliotoxin and yielded comparable percentages of PS externalization on cytoplasts and neutrophil surfaces (Figure 2B). The percentages of annexin-V–positive neutrophils and their corresponding cytoplasts were comparable for all donors tested (Figure 2B). PR3 expression rose comparably in neutrophils and their corresponding cytoplasts (Figure 2C). Moreover, physiologic apoptosis induction did not significantly change CD35, CD66b, or CD63 expression, indicating, as expected, the absence of degranulation by granule-free cytoplasts (Figure 2D). Thus, granular PR3 pool is not required for PR3 externalization during neutrophil apoptosis. From these experiments using cytoplasts, it could be concluded that (1) PR3 was mobilized to the neutrophil surface upon apoptosis in the total absence of degranulation and (2) apoptosis and degranulation appeared to be 2 distinct mechanisms leading to membrane PR3 expression on neutrophils.
Apoptosis-induced membrane PR3 expression on cytoplasts: a model of granule-free neutrophils. (A) Morphology of cytospin, freshly isolated neutrophils and neutrophil-derived cytoplasts. Nuclei are visible in MGG-stained neutrophils but not cytoplasts (magnification ×40). After methanol permeabilization of all membranes, including those of granules, MPO was detected only in neutrophils by immunolabeling and phase-contrast microscopy. PR3 and αII-spectrin immunolabeling of neutrophils and cytoplasts after SLO permeabilization. Slides were analyzed by confocal microscopy with a Zeiss LSM-5 confocal scanning laser microscope version 3.2 SP2 (Zeiss), equipped with an argon laser and helium-neon lasers, using a plan-aprochromat ×63/1.40 NA oil immersion objective lens and a crop ×2, at room temperature. Green fluorescence was observed with a 505-530–nm band-pass emission filter under 488-nm laser illumination and red fluorescence with a 560 long-pass emission filter under 543 laser illumination. Pinhole diameters were set to get 0.8-μm thick optical slices, and images were collected every 0.4 μm along the z-axis. Imaging corresponding to alexa 488, alexa 555, and rhodamin fluorescence were obtained using the multitrack mode. (B) Quantification of PS externalization assessed by annexin-V positivity (and 7-AAD negativity) of neutrophils and cytoplasts by flow cytometry, under basal conditions or after physiologic or gliotoxin-induced apoptosis. Under basal conditions almost no PS externalization was detected on either neutrophils or cytoplasts, whereas after apoptosis induction, significantly more (*P < .05 ANOVA) annexin-V–labeled neutrophils and cytoplasts were observed. Data are mean (± SEM) of the percentage of positive cells in neutrophils from 6 distinct donors. (C) Flow-cytometry analysis of membrane PR3 expression (bold lines) on neutrophils and cytoplasts versus control IgG1 (thin lines). In this representative experiment, under resting conditions (basal), 82% of neutrophils or their corresponding cytoplasts expressed PR3 at their cell surface. Under basal and apoptotic conditions, respectively, the MFIs of membrane PR3 expression were 46 and 190 on neutrophils and 51 and 116 on their corresponding cytoplasts. (D) Flow-cytometry analysis of degranulation markers CD35, CD66b, and CD63 (bold lines) versus isotypic control (thin lines) on cytoplasts under basal or apoptotic conditions. Panels A, C, and D report representative results for neutrophils from a given donor and their corresponding cytoplasts, which were confirmed in independent experiments performed with 3 different cytoplast preparations from 3 different donors.
Apoptosis-induced membrane PR3 expression on cytoplasts: a model of granule-free neutrophils. (A) Morphology of cytospin, freshly isolated neutrophils and neutrophil-derived cytoplasts. Nuclei are visible in MGG-stained neutrophils but not cytoplasts (magnification ×40). After methanol permeabilization of all membranes, including those of granules, MPO was detected only in neutrophils by immunolabeling and phase-contrast microscopy. PR3 and αII-spectrin immunolabeling of neutrophils and cytoplasts after SLO permeabilization. Slides were analyzed by confocal microscopy with a Zeiss LSM-5 confocal scanning laser microscope version 3.2 SP2 (Zeiss), equipped with an argon laser and helium-neon lasers, using a plan-aprochromat ×63/1.40 NA oil immersion objective lens and a crop ×2, at room temperature. Green fluorescence was observed with a 505-530–nm band-pass emission filter under 488-nm laser illumination and red fluorescence with a 560 long-pass emission filter under 543 laser illumination. Pinhole diameters were set to get 0.8-μm thick optical slices, and images were collected every 0.4 μm along the z-axis. Imaging corresponding to alexa 488, alexa 555, and rhodamin fluorescence were obtained using the multitrack mode. (B) Quantification of PS externalization assessed by annexin-V positivity (and 7-AAD negativity) of neutrophils and cytoplasts by flow cytometry, under basal conditions or after physiologic or gliotoxin-induced apoptosis. Under basal conditions almost no PS externalization was detected on either neutrophils or cytoplasts, whereas after apoptosis induction, significantly more (*P < .05 ANOVA) annexin-V–labeled neutrophils and cytoplasts were observed. Data are mean (± SEM) of the percentage of positive cells in neutrophils from 6 distinct donors. (C) Flow-cytometry analysis of membrane PR3 expression (bold lines) on neutrophils and cytoplasts versus control IgG1 (thin lines). In this representative experiment, under resting conditions (basal), 82% of neutrophils or their corresponding cytoplasts expressed PR3 at their cell surface. Under basal and apoptotic conditions, respectively, the MFIs of membrane PR3 expression were 46 and 190 on neutrophils and 51 and 116 on their corresponding cytoplasts. (D) Flow-cytometry analysis of degranulation markers CD35, CD66b, and CD63 (bold lines) versus isotypic control (thin lines) on cytoplasts under basal or apoptotic conditions. Panels A, C, and D report representative results for neutrophils from a given donor and their corresponding cytoplasts, which were confirmed in independent experiments performed with 3 different cytoplast preparations from 3 different donors.
hPLSCR1 as a molecular partner of membrane PR3
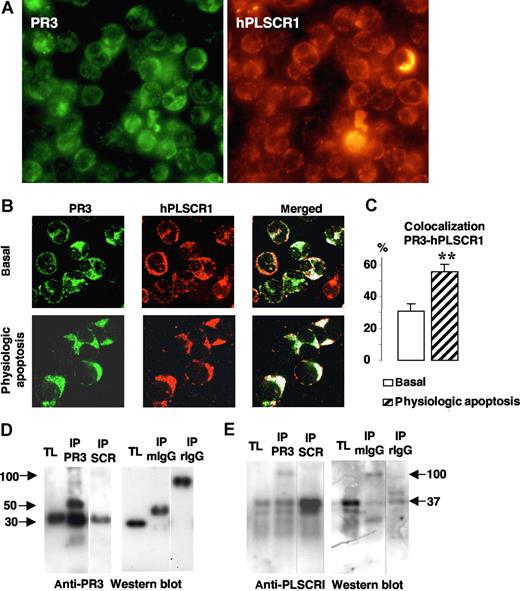
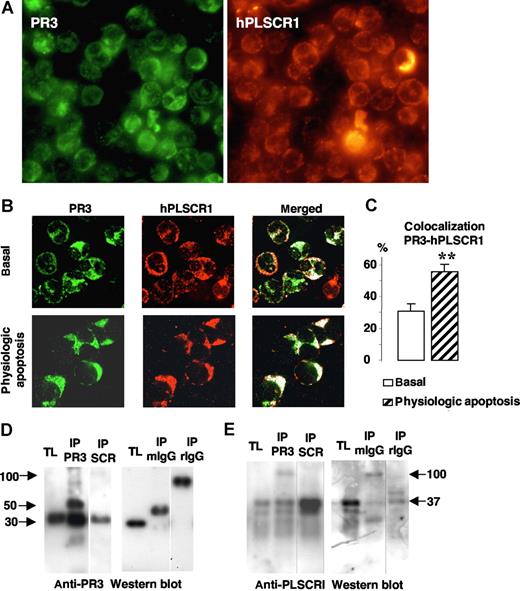
To understand how membrane PR3 could be involved in PS externalization during apoptosis, we hypothesized that PR3, which has been detected as being present in lipid rafts,34 could interact with other proteins involved in PS flip-flop movements. hPLSCR1, one of these putative proteins, was localized to neutrophil membrane lipid rafts.28 Therefore, we examined the colocalization of PR3 with hPLSCR1 in neutrophils by immunofluorescence labeling after SLO permeabilization to preclude detection of the granular PR3 pool, as previously described.15 Immunofluorescence double-labeling experiments were performed (mouse monoclonal anti-PR3 and rabbit polyclonal anti-hPLSCR1 antibodies) under basal conditions or after physiologic apoptosis induction. As a first step, we evaluated the expression of both proteins using fluorescence microscopy. As shown in Figure 3A, all neutrophils expressed PR3 and PLSCR1. To precisely determine colocalization, we next used confocal fluorescence microscopy. This analysis provided evidence that in resting neutrophils, PR3 (green) and hPLSCR1 (red) were partially localized in the cytosol but also on membranes (Figure 3B). PR3 and hPLSCR1 colocalization, calculated with LSM software, rose significantly after physiologic apoptosis induction (Figure 3C), suggesting a modification of subcellular protein localizations facilitating PR3 and hPLSCR1 interaction.
Colocalization and molecular association of PR3 and hPLSCR1 in neutrophils. (A) Immunofluorescence microscopy analysis of neutrophils after PR3 (left) and hPLSCR1 (right) immunolabeling (magnification × 63). (B) Confocal microscopy analysis of indirect fluorescence immunolabeling of PR3 (green) and hPLSCR1 (red) under basal conditions and after physiologic apoptosis induction. Images were obtained as described in Figure 2. Under resting conditions, punctuated membrane PR3 labeling was easily detected in SLO-permeabilized neutrophils. hPLSCR1-immunofluorescence patterns were similar to those observed for PR3. The merged fluorescence (white) showed cytoplasmic and membrane colocalizations of PR3 and hPLSCR1 in resting and apoptotic neutrophils. Control experiments with mouse or rabbit IgG antibodies alone were not labeled (data not shown). (C) Percentages of PR3-hPLSCR1 colocalizations calculated with LSM software. PR3 labeling colocalized with the hPLSCR1 (**P < .01) based on counts of 401 basal and 112 apoptotic neutrophils from 5 different donors. Data are mean (± SEM). (D,E) Western blots of reciprocal coimmunoprecipitation from total neutrophil lysates (TL). Immunoprecipitates (IP) were analyzed by 10% sodium dodecylsulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride membranes. PR3 was detected as a single band at 30 kDa in TLs and in both PR3 (IP PR3) and hPLSCR1 (IP SCR) immunoprecipitates by a rabbit polyclonal anti-PR3 (D); hPLSCR1 was detected as a single band at 37 kDa in TLs and in both hPLSCR1 (IP SCR) and PR3 (IP PR3) immunoprecipitates by a rabbit polyclonal anti-hPLSCR1 (E). TLs were also immunoprecipitated with mouse (IP mIgG) or rabbit (IP rIgG) control IgG and no PR3 or hPLSCR1 was detected. Vertical lines have been inserted to indicate a repositioned gel lane.
Colocalization and molecular association of PR3 and hPLSCR1 in neutrophils. (A) Immunofluorescence microscopy analysis of neutrophils after PR3 (left) and hPLSCR1 (right) immunolabeling (magnification × 63). (B) Confocal microscopy analysis of indirect fluorescence immunolabeling of PR3 (green) and hPLSCR1 (red) under basal conditions and after physiologic apoptosis induction. Images were obtained as described in Figure 2. Under resting conditions, punctuated membrane PR3 labeling was easily detected in SLO-permeabilized neutrophils. hPLSCR1-immunofluorescence patterns were similar to those observed for PR3. The merged fluorescence (white) showed cytoplasmic and membrane colocalizations of PR3 and hPLSCR1 in resting and apoptotic neutrophils. Control experiments with mouse or rabbit IgG antibodies alone were not labeled (data not shown). (C) Percentages of PR3-hPLSCR1 colocalizations calculated with LSM software. PR3 labeling colocalized with the hPLSCR1 (**P < .01) based on counts of 401 basal and 112 apoptotic neutrophils from 5 different donors. Data are mean (± SEM). (D,E) Western blots of reciprocal coimmunoprecipitation from total neutrophil lysates (TL). Immunoprecipitates (IP) were analyzed by 10% sodium dodecylsulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride membranes. PR3 was detected as a single band at 30 kDa in TLs and in both PR3 (IP PR3) and hPLSCR1 (IP SCR) immunoprecipitates by a rabbit polyclonal anti-PR3 (D); hPLSCR1 was detected as a single band at 37 kDa in TLs and in both hPLSCR1 (IP SCR) and PR3 (IP PR3) immunoprecipitates by a rabbit polyclonal anti-hPLSCR1 (E). TLs were also immunoprecipitated with mouse (IP mIgG) or rabbit (IP rIgG) control IgG and no PR3 or hPLSCR1 was detected. Vertical lines have been inserted to indicate a repositioned gel lane.
Coimmunoprecipitation experiments from neutrophil lysates were carried out next to determine whether a physical interaction exists between PR3 and hPLSCR1. PR3 and hPLSCR1 were immunoprecipitated. Each immunoprecipitation was validated by Western-blot analysis using anti-PR3 (Figure 3D) and anti-hPLSCR1 (Figure 3E). The reciprocal detection of the 2 proteins validated the protocol and supported the notion of PR3-hPLSCR1 association. Of note, neither PR3 nor hPLSCR1 was immunoprecipitated with control IgG. This PR3-hPLSCR1 interaction was also confirmed by reciprocal coimmunoprecipitation from apoptotic neutrophil lysates (data not shown).
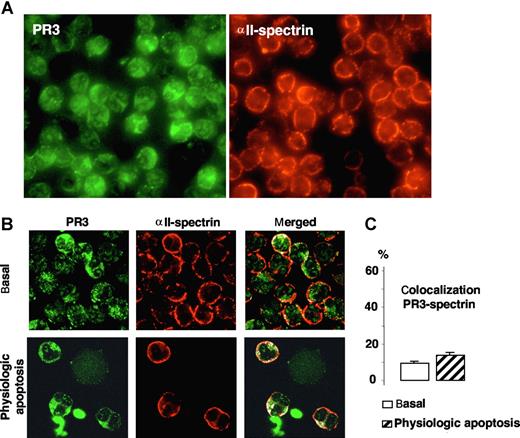
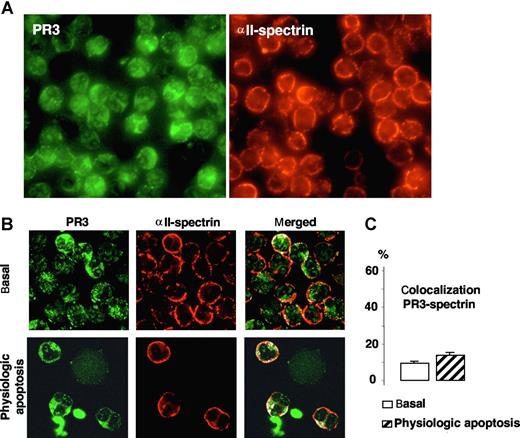
To assess the specificity of the colocalization of PR3 with PLSCR1, we examined whether PR3 could colocalize with αII-spectrin, a protein found at the inner face of the plasma membrane that interacts with actin microfilaments and is necessary for cell integrity. As shown in Figure 4A, immunofluorescence microscopy analysis of αII-spectrin labeling demonstrated a homogeneous expression in neutrophils mostly located, as expected, at the plasma membrane. Confocal microscopy analysis confirmed this membrane localization and showed that PR3 colocalized to only a very limited extent with αII-spectrin in the neutrophil membrane under basal conditions and after physiologic apoptosis induction (Figure 4B), as evidenced by low colocalization percentages (Figure 4C). Hence, PR3-hPLSCR1 colocalization was probably not the fortuitous result of their being in the same cellular compartment but rather reflected a molecular interaction.
Immunofluorescence and confocal microscopy analysis of PR3 and αII-spectrin in neutrophils. (A) Immunofluorescence microscopy analysis of neutrophils after PR3 (left) and αII-spectrin (right) immunolabeling (magnification ×63). (B) Confocal microscopy of indirect immunolabeling of αII-spectrin (red) in resting or apoptotic neutrophils. Images were obtained as described in Figure 2. Under both conditions, αII-spectrin was localized at the inner face of the neutrophil plasma membrane and gave a typical membrane pattern different from that of PR3. No evidence of marked colocalization (white) under basal or apoptotic conditions was found. (C) Percentages of PR3–αII-spectrin colocalization calculated with LSM software, based on 407 resting and 85 apoptotic neutrophils from 4 different donors. Data are mean (± SEM).
Immunofluorescence and confocal microscopy analysis of PR3 and αII-spectrin in neutrophils. (A) Immunofluorescence microscopy analysis of neutrophils after PR3 (left) and αII-spectrin (right) immunolabeling (magnification ×63). (B) Confocal microscopy of indirect immunolabeling of αII-spectrin (red) in resting or apoptotic neutrophils. Images were obtained as described in Figure 2. Under both conditions, αII-spectrin was localized at the inner face of the neutrophil plasma membrane and gave a typical membrane pattern different from that of PR3. No evidence of marked colocalization (white) under basal or apoptotic conditions was found. (C) Percentages of PR3–αII-spectrin colocalization calculated with LSM software, based on 407 resting and 85 apoptotic neutrophils from 4 different donors. Data are mean (± SEM).
Functional interaction between PR3 and rPLSCR1 in RBL/PR3
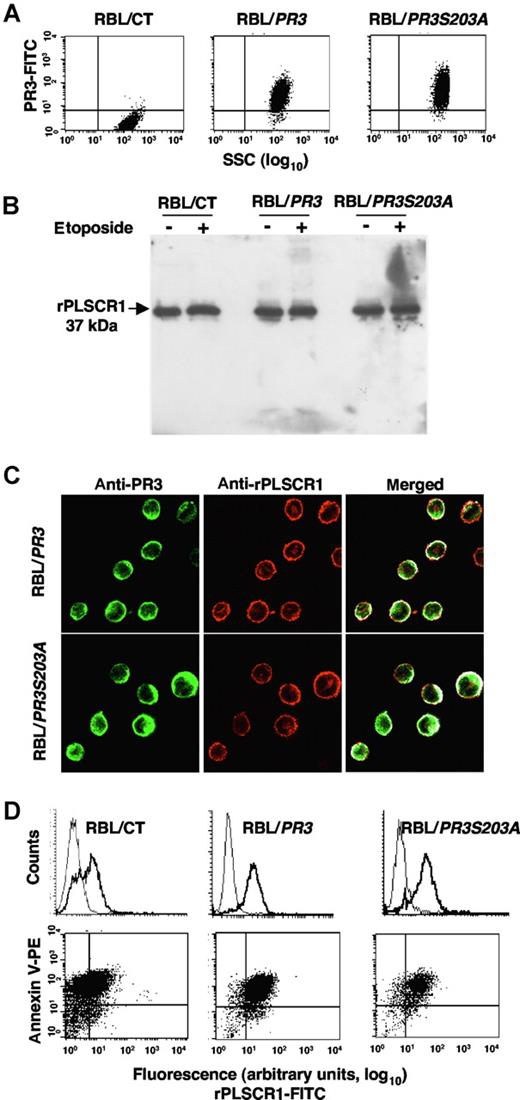
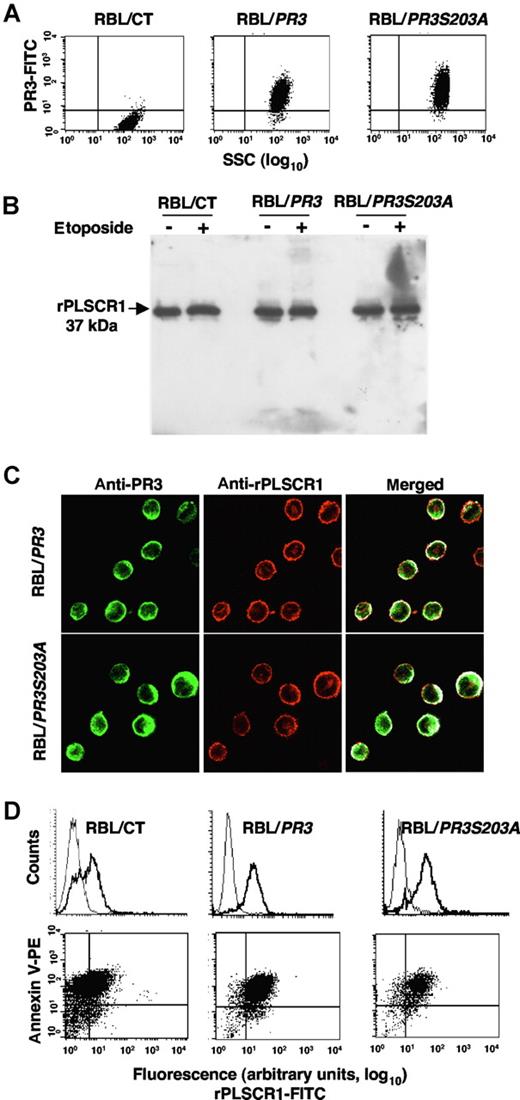
We next investigated whether the PR3-PLSCR1 association might influence apoptosis-induced PR3 externalization. However, because it was impossible to modulate specifically PR3 or PLSCR1 in mature neutrophils, we used our cellular model of RBL-2H3 cells stably transfected with human PR3 cDNA or its enzymatically inactive mutant PR3S203A.14-16 We verified by flow-cytometry analysis of intracellular PR3 that RBL/PR3 and RBL/PR3S203A but not RBL/CT expressed recombinant human PR3 (Figure 5A). Western-blot analysis of rPLSCR1 showed that it was detected at an apparent molecular mass of 37 kDa in RBL/CT, RBL/PR3, and RBL/S203A, thus demonstrating an absence of rPLSCR1 proteolysis by PR3 in RBL/PR3 under either basal or apoptotic conditions (Figure 5B).
rPLSCR1 expression in RBL-2H3 lines expressing recombinant PR3 or its inactive PR3S203A mutant. (A) Intracellular expression of PR3 and PLSCR1 in RBL/CT, RBL/PR3, and RBL/PR3S203A. Flow-cytometry analysis of intracellular expression of PR3 in RBL transfectants. (B) Western-blot analysis of PLSCR1 (using the mouse monoclonal 129.2) in lysates under resting conditions or after etoposide-induced apoptosis. RBL cells were lysed in lysis buffer and 10 μg of protein was loaded/well. (C) Double immunofluorescence labeling of PR3 and rPLSCR1 in RBL/PR3 and RBL/PR3S203A after SLO permeabilization. The merged fluorescence showed a membrane PR3-rPLSCR1 colocalization under resting conditions. Images were obtained as described in Figure 2. (D) Flow-cytometry analysis of rPLSCR1 membrane expression (bold line) compared with the isotypic control (thin line) in RBL/CT, RBL/PR3, and RBL/PR3S203A after etoposide-induced apoptosis (top panels). Double labeling of annexin-V and PLSCR1 is shown in the bottom panels. A representative experiment of 4 different experiments giving the same result is shown.
rPLSCR1 expression in RBL-2H3 lines expressing recombinant PR3 or its inactive PR3S203A mutant. (A) Intracellular expression of PR3 and PLSCR1 in RBL/CT, RBL/PR3, and RBL/PR3S203A. Flow-cytometry analysis of intracellular expression of PR3 in RBL transfectants. (B) Western-blot analysis of PLSCR1 (using the mouse monoclonal 129.2) in lysates under resting conditions or after etoposide-induced apoptosis. RBL cells were lysed in lysis buffer and 10 μg of protein was loaded/well. (C) Double immunofluorescence labeling of PR3 and rPLSCR1 in RBL/PR3 and RBL/PR3S203A after SLO permeabilization. The merged fluorescence showed a membrane PR3-rPLSCR1 colocalization under resting conditions. Images were obtained as described in Figure 2. (D) Flow-cytometry analysis of rPLSCR1 membrane expression (bold line) compared with the isotypic control (thin line) in RBL/CT, RBL/PR3, and RBL/PR3S203A after etoposide-induced apoptosis (top panels). Double labeling of annexin-V and PLSCR1 is shown in the bottom panels. A representative experiment of 4 different experiments giving the same result is shown.
Immunofluorescence double-labeling experiments were performed on RBL/PR3 and RBL/PR3S203A, using rabbit anti-PR3 (directed against the whole PR3 molecule) and rhodamine-conjugated mouse anti-rPLSCR1 monoclonal antibodies. Confocal fluorescence microscopy showed a partial colocalization of PR3 and rPLSCR1 under basal conditions in both transfected cell lines, with respective colocalization percentages of 46% plus or minus 4.7% and 54% plus or minus 3.6% (Figure 5C). Interestingly, after etoposide-induced apoptosis, flow cytometry performed on 7-AAD–negative cells detected immunolabeled rPLSCR1 at the cell surface without permeabilization, strongly suggesting that, like PR3, rPLSCR1 was externalized upon apoptosis (Figure 5D). The RBL cells used for the analysis of the membrane expression of rPLSCR1 were not necrotic and had an intact plasma membrane, thus precluding an artefactual access of the antibody to intracellular PLSCR1. Moreover, rPLSCR1 and annexin-V double-immunolabeling demonstrated a strong relationship between rPLSCR1 and PS externalization on RBL-2H3 lines, which paralleled that of PR3 and PS externalization on RBL/PR3 and in RBL/PR3S203A that we described previously.16
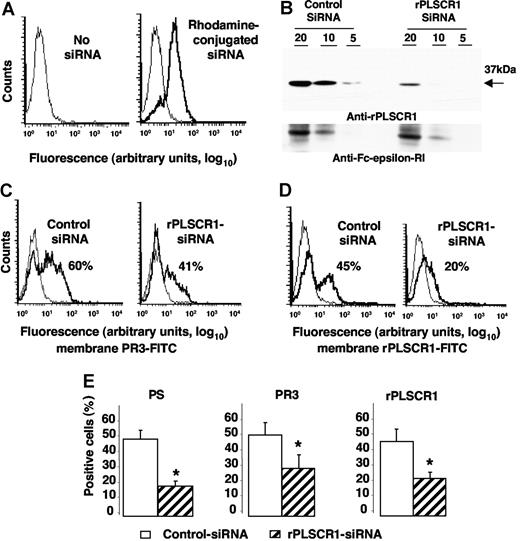
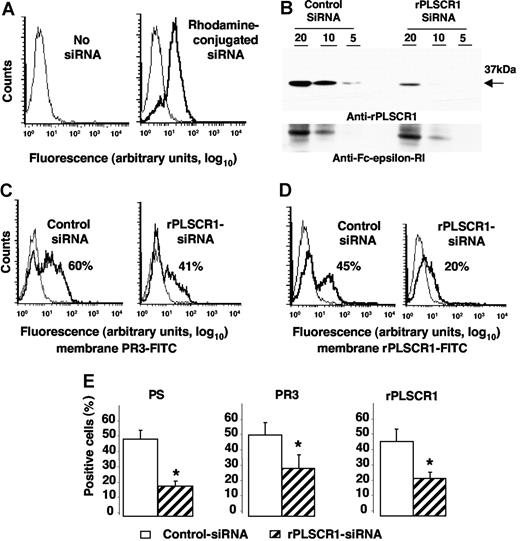
To test whether PLSCR1-PR3 interaction might facilitate its membrane expression, we used siRNA technology to inhibit specifically rPLSCR1 expression in RBL/PR3 cells. RBL/PR3 cells were electroporated with a duplex siRNA targeting the specific degradation of rPLSCR1 mRNA. First, we checked that control PE-conjugated siRNA was successfully internalized by cells, with 85% plus or minus 5% (n = 4) positive cells (Figure 6A). As expected, rPLSCR1 was detected by Western-blot analysis after electroporation of control siRNA but was markedly lower (> 50% extinction; using Image J version 1.34q software [Wayne Rasband, National Institutes of Health, Bethesda, MD]) in cells transfected with rPLSCR1 siRNA (Figure 6B). No modulation of the control FcϵRIβ was detected. No difference in cell viability assessed by 7-AAD labeling was observed between cells transfected with control siRNA or rPLSCR1 siRNA, before or after etoposide-induced apoptosis, thus strongly suggesting that rPLSCR1 did not modify RBL survival (data not shown). After etoposide-induced apoptosis, membrane PR3 expression measured on 7-AAD–negative cells declined on cells transfected with rPLSCR1 siRNA (40%) compared with those transfected with control siRNA (60%; Figure 6C). Concomitantly, a significant decrease in rPLSCR1 membrane expression was also observed on cells transfected with rPLSCR1 siRNA (20%) compared with those transfected with control siRNA (45%) in the typical experiment shown in Figure 6D. Moreover, in parallel with the decreased membrane expression of PR3 and rPLSCR1, annexin-V–positive RBL/PR3 cells after rPLSCR1 siRNA transfection were significantly lower than after control siRNA transfection, thus demonstrating decreased PS externalization after rPLSCR1 protein extinction (Figure 6E). These data strongly suggest that rPLSCR1 is involved in membrane PR3 expression during apoptosis-induced phospholipid redistribution between the 2 plasma membrane leaflets.
Impaired membrane PR3 expression in RBL/PR3 transfected with rPLSCR1/siRNA after etoposide-induced apoptosis. (A) Flow-cytometry analysis after rhodamine-conjugated siRNA electroporation. Typical experiment showing that 85% of the cells emitted a positive signal after siRNA electroporation (bold line), whereas no fluorescence was detected in the absence of siRNA (thin line). (B) Western-blot analysis of rPLSCR1 and FcϵRIβ expressions in RBL/PR3 transfected with control siRNA or rPLSCR1 siRNA. Different amounts of cell lysates (20, 10, or 5 μg) were loaded per well. Most rPLSCR1 protein disappeared 48 hours after electroporation with siRNA but not control siRNA. No FcϵRIβ modulation was observed. (C) Flow-cytometry analysis of membrane PR3 expression (bold line) versus control IgG1 (thin line) of RBL/PR3 transfected with either control siRNA or with rPLSCR1 siRNA after etoposide-induced apoptosis. (D) Flow-cytometry analysis of membrane rPLSCR1 expression (bold line) versus control IgG1 (thin line) of RBL/PR3 transfected with either control siRNA or with rPLSCR1 siRNA after etoposide-induced apoptosis. (E) Decreased PS externalization and membrane PR3 and rPLSCR1 expression after rPLSCR1 siRNA transfection. Percentages of annexin-V labeling and membrane PR3 and rPLSCR1 expression were significantly lower (data are means ± SEM; *P < .05 Student t test, n = 4) for apoptotic RBL/PR3 transfected with rPLSCR1 siRNA than control siRNA.
Impaired membrane PR3 expression in RBL/PR3 transfected with rPLSCR1/siRNA after etoposide-induced apoptosis. (A) Flow-cytometry analysis after rhodamine-conjugated siRNA electroporation. Typical experiment showing that 85% of the cells emitted a positive signal after siRNA electroporation (bold line), whereas no fluorescence was detected in the absence of siRNA (thin line). (B) Western-blot analysis of rPLSCR1 and FcϵRIβ expressions in RBL/PR3 transfected with control siRNA or rPLSCR1 siRNA. Different amounts of cell lysates (20, 10, or 5 μg) were loaded per well. Most rPLSCR1 protein disappeared 48 hours after electroporation with siRNA but not control siRNA. No FcϵRIβ modulation was observed. (C) Flow-cytometry analysis of membrane PR3 expression (bold line) versus control IgG1 (thin line) of RBL/PR3 transfected with either control siRNA or with rPLSCR1 siRNA after etoposide-induced apoptosis. (D) Flow-cytometry analysis of membrane rPLSCR1 expression (bold line) versus control IgG1 (thin line) of RBL/PR3 transfected with either control siRNA or with rPLSCR1 siRNA after etoposide-induced apoptosis. (E) Decreased PS externalization and membrane PR3 and rPLSCR1 expression after rPLSCR1 siRNA transfection. Percentages of annexin-V labeling and membrane PR3 and rPLSCR1 expression were significantly lower (data are means ± SEM; *P < .05 Student t test, n = 4) for apoptotic RBL/PR3 transfected with rPLSCR1 siRNA than control siRNA.
Apoptosis-induced membrane PR3 expression impaired macrophage phagocytosis of RBL/PR3 and RBL/PR3S203A
To explore (1) whether apoptosis-induced membrane PR3 expression could interfere with the mechanism of clearance by macrophages and (2) if so, whether PR3 enzymatic activity was required, we determined the phagocytosis rates of apoptotic RBL/PR3 or RBL/PR3S203A. As we previously described, etoposide-induced apoptosis resulted in (1) PS externalization on control RBL, RBL/PR3, or RBL/PR3S203A and (2) membrane PR3 expression on RBL/PR3 or RBL/PR3S203A (Figure 7A). Apoptosis-induced membrane PR3 expression was closely associated with PS externalization on RBL/PR3 and RBL/PR3S203A. This relationship was independent of PR3 enzymatic activity, since the same results were obtained for RBL/PR3 and inactive RBL/PR3S203A.
Membrane PR3 expression decreased human macrophage phagocytosis of RBL/PR3 and RBL/PR3S203A independently of its serine-proteinase activity. (A) Analysis of membrane PS and PR3 externalizations by flow cytometry after etoposide-induced apoptosis (bold line) compared with untreated cells (thin line). This representative experiment shows PS externalization on RBL/CT, RBL/PR3, and RBL/PR3S203A after etoposide-induced apoptosis; PR3 was expressed on apoptotic RBL/PR3 and RBL/PR3S203A but not apoptotic RBL/CT. Double labeling of membrane PR3 and annexin-V showed their close relationship. No relationship was observed under basal conditions because no annexin-V labeling was observed (data not shown). (B) Confocal fluorescence microscopy quantification of human macrophage phagocytosis of apoptotic RBL-2H3. TAMRA-labeled apoptotic RBL (red) were incubated with FITC-conjugated anti-CD14–labeled macrophages derived from human monocytes (green). Phagocytosis yielded a red-labeled apoptotic RBL engulfed by a green-labeled macrophage. Images were obtained as described in Figure 2. (C) Quantification of annexin-V–positive RBL cells after etoposide-induced apoptosis. All apoptotic RBL transfectants showed significantly enhanced PS externalization (**P < .01, ANOVA). However, there was no significant difference in PS externalization between all apoptotic transfectants. (D) Quantification of macrophage phagocytosis. The phagocytosing macrophages were identified by confocal microscopy to ensure that the RBL cells had been engulfed and were not just sitting on the macrophage surface. Phagocytosis rates were calculated as the ratio between the number of phagocytosing macrophages on total macrophages. As expected, phagocytosis rates of apoptotic RBL/CT, RBL/PR3, or RBL/PR3S203A were higher than viable RBL lines (P < .001, for all transfectants using ANOVA; not indicated in the figure). The rates of apoptotic RBL/PR3 and RBL/PR3S203A phagocytosis were significantly lower than that of apoptotic RBL/CT (**P < .01, ANOVA) but RBL/PR3 and RBL/PR3S203A rates were comparable (not significant [NS], ANOVA). Data are means (± SEM) from 8 independent experiments.
Membrane PR3 expression decreased human macrophage phagocytosis of RBL/PR3 and RBL/PR3S203A independently of its serine-proteinase activity. (A) Analysis of membrane PS and PR3 externalizations by flow cytometry after etoposide-induced apoptosis (bold line) compared with untreated cells (thin line). This representative experiment shows PS externalization on RBL/CT, RBL/PR3, and RBL/PR3S203A after etoposide-induced apoptosis; PR3 was expressed on apoptotic RBL/PR3 and RBL/PR3S203A but not apoptotic RBL/CT. Double labeling of membrane PR3 and annexin-V showed their close relationship. No relationship was observed under basal conditions because no annexin-V labeling was observed (data not shown). (B) Confocal fluorescence microscopy quantification of human macrophage phagocytosis of apoptotic RBL-2H3. TAMRA-labeled apoptotic RBL (red) were incubated with FITC-conjugated anti-CD14–labeled macrophages derived from human monocytes (green). Phagocytosis yielded a red-labeled apoptotic RBL engulfed by a green-labeled macrophage. Images were obtained as described in Figure 2. (C) Quantification of annexin-V–positive RBL cells after etoposide-induced apoptosis. All apoptotic RBL transfectants showed significantly enhanced PS externalization (**P < .01, ANOVA). However, there was no significant difference in PS externalization between all apoptotic transfectants. (D) Quantification of macrophage phagocytosis. The phagocytosing macrophages were identified by confocal microscopy to ensure that the RBL cells had been engulfed and were not just sitting on the macrophage surface. Phagocytosis rates were calculated as the ratio between the number of phagocytosing macrophages on total macrophages. As expected, phagocytosis rates of apoptotic RBL/CT, RBL/PR3, or RBL/PR3S203A were higher than viable RBL lines (P < .001, for all transfectants using ANOVA; not indicated in the figure). The rates of apoptotic RBL/PR3 and RBL/PR3S203A phagocytosis were significantly lower than that of apoptotic RBL/CT (**P < .01, ANOVA) but RBL/PR3 and RBL/PR3S203A rates were comparable (not significant [NS], ANOVA). Data are means (± SEM) from 8 independent experiments.
We then hypothesized that PR3 could modulate macrophage phagocytosis of apoptotic cells because PS externalization is an eat-me signal recognized by phagocytes. Macrophages were immunolabeled with an FITC-conjugated anti-CD14 monoclonal antibody and RBL-2H3 cells were labeled with TAMRA. Red phagocytosed RBL-2H3 cells in green membrane-labeled macrophages were quantified by confocal fluorescence microscopy (Figure 7B). As expected, etoposide-induced apoptosis triggered PS externalization and significantly increased the percentages of phagocytosis of all transfectants (Figure 7C). It should be noted that PS externalization was comparable for the RBL-2H3 transfectants. Phagocytosis of RBL/PR3, which expressed membrane PR3 and PS, decreased significantly compared with RBL/CT (**P < .01), thereby demonstrating that PR3 impaired phagocytosis mechanisms. Interestingly, this effect was independent of PR3 enzymatic activity because it was also observed for RBL/PR3S203A expressing the inactive PR3 mutant. It could be postulated that PR3 might bind to some partner protein(s) independently of its enzymatic activity and interfere with either macrophage recognition or effector mechanisms. Interestingly, the rates of phagocytosis of apoptotic neutrophils and RBL/PR3 were not significantly different (Figure S1).
Discussion
Although the principal PR3 pool is stored within the azurophilic granules of neutrophils, we previously reported that a fraction of this serine proteinase was expressed at the plasma membrane of resting neutrophils, which could be explained by PR3 presence in the membranes of the secretory vesicles that are easily mobilized to the surface.9 The data presented herein identified PLSCR1 as a molecular partner for PR3, which is then able to translocate during apoptosis-induced membrane flip-flop independently of degranulation. In contrast, MPO and HNE were not detected at the cell surface during apoptosis.
To date, very little is known about neutrophil-membrane expression of the 2 ANCA antigens PR3 and MPO during apoptosis. Our results differ from those of Gilligan et al,35 who reported that both molecules were externalized during apoptosis because of the translocation of azurophilic granules to the plasma membrane. This phenomenon seems to occur when neutrophils undergo late apoptosis and secondary necrosis,36 leading to expression of all granule proteins on the cell surface. Indeed, to avoid degranulation, we used a 6-hour incubation, which was sufficient to induce significant PS exposure. Notably, the authors of all studies that achieved modulation of membrane PR3 or MPO expression during apoptosis had always used tumor necrosis factor–alpha, which is a priming agent known to up-regulate both ANCA antigens on neutrophils.37,38
Thus, to the best of our knowledge, the results of this study are the first to differentiate the respective effects, namely physiologic apoptosis, secondary necrosis, and degranulation, on the membrane expression of these antigens. Notably, we were first able to demonstrate that the mechanisms leading to membrane PR3 expression are clearly distinct from those involved in membrane MPO expression. Absolutely no membrane MPO expression was seen on neutrophils undergoing physiologic apoptosis. Thus, this new finding, that PR3 can be mobilized to the neutrophil plasma membrane after apoptosis induction in the total absence of degranulation, strongly suggests that a PR3 pool was externalized by apoptosis-induced membrane flip-flop and that it differs from the intragranular pool. Our present data obtained in mature neutrophils are in agreement with our previous study in RBL/PR3, showing apoptosis-induced PR3 externalization in the absence of degranulation. Therefore, apoptosis-induced PR3 membrane expression does not result from release of intracellular PR3 followed by rebinding to the plasma membrane.16
To further elucidate the existence of this PR3 pool, we used an artificial model of granule-free neutrophils, called cytoplasts,25 which are ideal to explore degranulation-independent phenomena, as they contain one third of the plasma membrane and about one half of the cytoplasm present in intact neutrophils.39 However, their cytosol and plasma membrane retain the apparatus for phagocytosis, generation of bactericidal oxygen compounds, and killing bacteria.25 First, we confirmed that cytoplasts externalized PS after apoptosis induction at the same rate as neutrophils. In fact, although cytoplasts lack nuclei, mitochondria, and granules, they can display some features of apoptosis such as caspase activation or PS externalization.40 Subsequently, we also verified that cytoplasts too had apoptosis-induced membrane PR3 expression. Taken together, these results confirmed the existence of a PR3 pool independent of the granular one that can be mobilized during apoptosis.
We were also able to demonstrate that PLSCR1 is a membrane partner of PR3. The association of these 2 proteins may contribute to their externalization during the membrane rearrangement that accompanies apoptosis. In apoptotic RBL cells, we showed that both PR3 and PLSCR1 transit to the cell surface; we have performed this analysis on annexin-V–positive but 7-AAD–negative cells to make sure of apoptotic cells' membrane integrity. Moreover, we have previously demonstrated, in elastase-transfected RBL cells, that there is no artefactual membrane expression of an intracellular protein, for instance elastase, during etoposide-induced apoptosis.16 The observation that PLSCR1 was externalized during apoptosis is of interest, as it may be related to the ability of PLSCR1 to promote redistribution of phospholipids between the 2 leaflets and may reflect a general property of apoptotic membranes.
It can be hypothesized that PR3 and PLSCR1 are part of a network of membrane flip-flop–associated proteins. Indeed, in neutrophils, both PR334 and PLSCR1 have been localized in lipid rafts.28 Interestingly, PLSCR1 was also found in the membranes of secretory vesicles where we also detected PR3. PLSCR1 function is not restricted to apoptosis-induced flip-flop because it was expressed at the membrane of f-MLP–activated neutrophils and was detected on neutrophil uropods during migration.28 It could be relevant to determine whether PLSCR1 and PR3 are associated in apoptosis-independent PLSCR1 functions, like migration or activation, in which we foresee that they might cooperate. We have not yet elucidated the mechanisms by which PR3 in cooperation with PLSCR1 could translocate and cross the plasma membrane in order to be expressed at the cell surface. Indeed, PR3 does not have a transmembrane domain, but it does harbor various hydrophobic regions. Although the results of 1 study showed that PR3 association with the neutrophil membrane was only through ionic interactions,41 we found that membrane PR3 expression could not be suppressed by high salt concentration.9 Moreover, in liposomes, PR3 partially inserts into the hydrophobic region of the lipid membranes, thus strongly suggesting that PR3 behaves as a peripheral membrane protein.42 PR3 association with the plasma membrane might involve strong hydrophobic interactions with lipids and most likely requires some protein partners such as CD177 (NB1), which has been recently described.43,44
Finally, the impaired macrophage phagocytosis of apoptotic RBL-2H3 expressing surface PR3 that we observed might explain the proinflammatory role of neutrophil PR3 membrane in vasculitis and rheumatoid arthritis patients. Our results show that PR3 enzymatic activity was not required for this effect, thereby demonstrating that no proteolysis was necessary for this inhibition of apoptotic cell recognition and/or engulfment. Hence, this mechanism might be distinct from that described for HNE, which inhibited macrophage removal of neutrophils by cleaving the PS receptor.45 For several years, PS externalization has been accorded a critical role in apoptotic cell clearance, as it is recognized by its receptor on macrophages,46 but this concept has now become controversial.47 Hence, the mechanisms of apoptotic cell recognition and clearance are complex,48 and the PR3-mediated effect might also involve several pathway(s) and/or protein interaction(s). Taking into account PR3 affinity for lipids, one could foresee that PR3 itself might be a direct PS-binding protein and thus interfere with PS-mediated phagocytosis. This hypothesis is in keeping with the fact that the enzymatic activity of PR3 is not required for the inhibitory effect of membrane PR3 on macrophage phagocytosis. Further investigations will be required to fully address this critical issue.
It must be emphasized that anti-PR3 ANCA present in Wegener granulomatosis might also interfere with PS externalization and/or macrophage clearance of neutrophils.49 Indeed, it was previously described that ANCA accelerated tumor necrosis factor–alpha–induced apoptosis with uncoupling of nuclear and surface membrane changes, yielding to less phagocyte recognition and engulfment.37 Apoptosis-induced membrane PR3 expression goes against the current dogma of the anti-inflammatory role of apoptosis and appears to be a paradox. A better understanding of the mechanisms involved in membrane PR3 expression and molecular analysis of PR3-PLSCR1 interaction might lead to novel and innovative therapeutic strategies, such as targeting PR3 by interfering with its membrane expression in ANCA-associated vasculitides and other systemic inflammatory diseases.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Researchers were individually funded by fellowships from the Association Vaincre La Mucoviscidose (C.K.), the Association de Recherche sur la Polyarthrite and Fondation pour la Recherche Médicale (M.P.-R.), and Ministère de l'Enseignement (O.A.-M.). This study was also supported by unrestricted grants from BAXTER (Extramural Grant Programm), Leg Poix, ABCF Mucoviscidose, and AMGEN.
The authors thank Meriem Garfa-Traoré (IFR94, Faculté de Medécine Necker-Enfants Malades, Paris) and Cécile Pouzet (IFR2, Faculté de Médecine Bichat, Paris) for excellent technical assistance in obtaining the confocal microscopy data.
Authorship
Contribution: C.K., M.P.-R., V.G.-D., and O.A.-M. performed experiments and analyzed data; M.-C.L. and I.C.M. contributed vital new reagents; M.B. and V.W.-S. designed the research project and wrote the paper. C.K. and M.P.-R. contributed equally to this work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Véronique Witko-Sarsat, INSERM, U845, Hôpital Necker, 149 rue de Sèvres, 75743 Paris Cedex 15, France; e-mail: witko-sarsat@necker.fr.


![Figure 7. Membrane PR3 expression decreased human macrophage phagocytosis of RBL/PR3 and RBL/PR3S203A independently of its serine-proteinase activity. (A) Analysis of membrane PS and PR3 externalizations by flow cytometry after etoposide-induced apoptosis (bold line) compared with untreated cells (thin line). This representative experiment shows PS externalization on RBL/CT, RBL/PR3, and RBL/PR3S203A after etoposide-induced apoptosis; PR3 was expressed on apoptotic RBL/PR3 and RBL/PR3S203A but not apoptotic RBL/CT. Double labeling of membrane PR3 and annexin-V showed their close relationship. No relationship was observed under basal conditions because no annexin-V labeling was observed (data not shown). (B) Confocal fluorescence microscopy quantification of human macrophage phagocytosis of apoptotic RBL-2H3. TAMRA-labeled apoptotic RBL (red) were incubated with FITC-conjugated anti-CD14–labeled macrophages derived from human monocytes (green). Phagocytosis yielded a red-labeled apoptotic RBL engulfed by a green-labeled macrophage. Images were obtained as described in Figure 2. (C) Quantification of annexin-V–positive RBL cells after etoposide-induced apoptosis. All apoptotic RBL transfectants showed significantly enhanced PS externalization (**P < .01, ANOVA). However, there was no significant difference in PS externalization between all apoptotic transfectants. (D) Quantification of macrophage phagocytosis. The phagocytosing macrophages were identified by confocal microscopy to ensure that the RBL cells had been engulfed and were not just sitting on the macrophage surface. Phagocytosis rates were calculated as the ratio between the number of phagocytosing macrophages on total macrophages. As expected, phagocytosis rates of apoptotic RBL/CT, RBL/PR3, or RBL/PR3S203A were higher than viable RBL lines (P < .001, for all transfectants using ANOVA; not indicated in the figure). The rates of apoptotic RBL/PR3 and RBL/PR3S203A phagocytosis were significantly lower than that of apoptotic RBL/CT (**P < .01, ANOVA) but RBL/PR3 and RBL/PR3S203A rates were comparable (not significant [NS], ANOVA). Data are means (± SEM) from 8 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/12/10.1182_blood-2007-03-080457/4/m_zh80240710040007.jpeg?Expires=1765908302&Signature=CbZe45BxJDhmv0cijFkwk47ZjMJmKsxNgP0qS~qT~bM6bG~bv9gXbItksmO2ppdPbMchnfqu2q9o~k4J-3YAzKPpDtBS819MarPKXiYxpgrZ4bKBigDjIZNUbs6acM21it~7V4EeVY0Tf9v-YHNZQcTHioTNv-bj7FF8znJAmHyB7T~6-5yXmHfJPDwvJTIje34zwrxdCVeAhN95aIMs3XiSU3LyPIde8hZUG6GRhvhOSLmgMZIxEBnJTiqW8g5ZY8hEWNcx8wMd~W2wDFNkV1ailSut46cuN3jC51~30j7x~~DnwDG4eLMSKYgk9~~8Bu1oZW8cDAdR3I72vzsIpQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 7. Membrane PR3 expression decreased human macrophage phagocytosis of RBL/PR3 and RBL/PR3S203A independently of its serine-proteinase activity. (A) Analysis of membrane PS and PR3 externalizations by flow cytometry after etoposide-induced apoptosis (bold line) compared with untreated cells (thin line). This representative experiment shows PS externalization on RBL/CT, RBL/PR3, and RBL/PR3S203A after etoposide-induced apoptosis; PR3 was expressed on apoptotic RBL/PR3 and RBL/PR3S203A but not apoptotic RBL/CT. Double labeling of membrane PR3 and annexin-V showed their close relationship. No relationship was observed under basal conditions because no annexin-V labeling was observed (data not shown). (B) Confocal fluorescence microscopy quantification of human macrophage phagocytosis of apoptotic RBL-2H3. TAMRA-labeled apoptotic RBL (red) were incubated with FITC-conjugated anti-CD14–labeled macrophages derived from human monocytes (green). Phagocytosis yielded a red-labeled apoptotic RBL engulfed by a green-labeled macrophage. Images were obtained as described in Figure 2. (C) Quantification of annexin-V–positive RBL cells after etoposide-induced apoptosis. All apoptotic RBL transfectants showed significantly enhanced PS externalization (**P < .01, ANOVA). However, there was no significant difference in PS externalization between all apoptotic transfectants. (D) Quantification of macrophage phagocytosis. The phagocytosing macrophages were identified by confocal microscopy to ensure that the RBL cells had been engulfed and were not just sitting on the macrophage surface. Phagocytosis rates were calculated as the ratio between the number of phagocytosing macrophages on total macrophages. As expected, phagocytosis rates of apoptotic RBL/CT, RBL/PR3, or RBL/PR3S203A were higher than viable RBL lines (P < .001, for all transfectants using ANOVA; not indicated in the figure). The rates of apoptotic RBL/PR3 and RBL/PR3S203A phagocytosis were significantly lower than that of apoptotic RBL/CT (**P < .01, ANOVA) but RBL/PR3 and RBL/PR3S203A rates were comparable (not significant [NS], ANOVA). Data are means (± SEM) from 8 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/12/10.1182_blood-2007-03-080457/4/m_zh80240710040007.jpeg?Expires=1766015634&Signature=00qtIV5EOSBj5CJtvM2umDsEztikMBRWDX3Djs8T6JEN7gtHrEK4cabEwqp4Lvv4575Zmj4I7GwcZPh1S1zBlSN0iuDcYKHM-gHCzsFpEaY7pHEopTer7bKYFGaQWBO1g8o2BqFGyqInkd5vK5j2nL6kn3Fd~OndQafRsi5ZPOqJOj39bloTZ-3T3wRPrO4Qbfs0ajY~rc55TRYL2PVbkrDIJ85mXnawn~lJFfAAWO76QZvU~TSCeuKFQyG8h62lXQVblQ7iTv8UyUP1UJd-n490bsSGNKMRAP5M~nLulPbsmdrNkqvXM6TQFfyUNvGKlrcMLHIIMlpyBCpKy4nPyA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)