Abstract
Clinical and histopathological characteristics have limited prognostic value for children with anaplastic large-cell lymphoma (ALCL). We evaluated the presence, extent, and prognostic impact of circulating tumor cells in bone marrow (BM) and peripheral blood (PB) of children and adolescents with NPM-ALK–positive ALCL at diagnosis using qualitative and quantitative polymerase chain reaction (PCR) for NPM-ALK. Numbers of NPM-ALK transcripts were normalized to 104 copies ABL (NCNs). BM was analyzed from 80 patients and PB from 52. BM was positive for NPM-ALK in 47.5% of patients, and positivity was significantly correlated with clinical stage, mediastinal or visceral involvement, microscopic BM involvement, and histologic subtype. Qualitative and quantitative PCR results in BM and PB strongly correlated. BM PCR was associated with the cumulative incidence of relapses (CI-Rs): CI-R was 50% ± 10% for 38 PCR-positive and 15% ± 7% for 42 PCR-negative patients (P < .001). Sixteen patients with more than 10 NCNs NPM-ALK in BM had a CI-R of 71% ± 14% compared with a CI-R of 18% ± 6% for 59 patients with 10 or fewer NCNs (P < .001). PB PCR results led to a similar grouping. Thus, quantitative PCR in BM or PB allows identification of 20% of patients experiencing 60% of all relapses with an event-free survival of 20%.
Introduction
Anaplastic large-cell lymphoma (ALCL) accounts for 10% to 15% of pediatric and adolescent non-Hodgkin lymphomas.1 The relapse rate of children and adolescents with ALCL still reaches 30% even after use of current, intensive short-pulse chemotherapy strategies.2-6 Analyses of cumulative data from European national studies identified the clinically based poor prognostic factors of mediastinal involvement, visceral involvement, and skin lesions, each with a relative risk of failure of approximately 2.7 Atypical histologic subtypes also may indicate a higher risk of relapse.2,8 Despite the emergence of promising new treatment options for high-risk ALCL, such as allogeneic stem cell transplantation or monoclonal antibodies,9-11 the paucity of identified risk factors prohibits defining a group of poor-risk patients for such interventions in first remission
More than 80% of children and adolescents with ALCL carry the translocation t(2;5)(p23;q35) resulting in the specific fusion gene NPM-ALK.12-17 The fusion gene transcript can be readily and sensitively detected using reverse-transcriptase polymerase chain reaction (RT-PCR) technology,18 allowing not only a more accurate diagnosis of this lymphoma entity but also enabling detection of disease extension at a submicroscopic level.
Mussolin et al recently described the prognostic meaning of minimal bone marrow involvement measured by qualitative RT-PCR for NPM-ALK in 35 children with ALCL.19 Absence of NPM-ALK positivity in bone marrow was associated with a very low risk of failure. However, patients with bone marrow positive for NPM-ALK still had a progression-free survival of more than 40%.
Quantifying the number of cells expressing NPM-ALK in bone marrow of ALCL patients at diagnosis may enhance the prognostic strength in defining a patient group with a very poor prognosis. The quantification of fusion gene transcripts has been made possible by TaqMan-based quantitative real-time PCR (RQ-PCR).20,21
We report on the measurement of circulating NPM-ALK–positive cells by real-time PCR in the bone marrow of 80 children and adolescents with NPM-ALK–positive ALCL treated according to a uniform Berlin-Frankfurt-Münster (BFM) strategy.4,22 Bone marrow was screened for NPM-ALK positivity by qualitative RT-PCR. Positive results were quantified by a TaqMan-based RQ-PCR for NPM-ALK, which was developed according to the standardized criteria of the Europe Against Cancer (EAC) program.20,23 Peripheral blood was compared with bone marrow as the medium for the measurement of minimal systemic disease in 52 patients. The prognostic impact of the extent of circulating NPM-ALK–positive cells in bone marrow and peripheral blood was evaluated.
Patients, materials, and methods
The studies NHL-BFM95 and ALCL99 were approved by the Justus-Liebig-University IRB. Both IRB approvals explicitly included the use of patient material for the study of new prognostic markers. All patients/their parents signed an informed consent in accordance with the Declaration of Helsinki to use the material for the study of prognostic factors (in ALCL99 even more specified: minimal systemic disease).
Eligibility
Patients from study NHL-BFM95 with a diagnosis of ALCL and German patients enrolled in the European intergroup trial ALCL99 were potentially eligible for this study. Both studies were approved by the institutional ethics committee of the primary investigator of the BFM group (A.R.). Eligibility was confirmed with demonstration of NPM-ALK positivity of the ALCL by either positive NPM-ALK PCR and/or positive 2-color fluorescence in situ hybridization for t(2;5) and/or nuclear and cytoplasmic staining for ALK in the tumor biopsy; in addition, bone marrow had to be available at diagnosis for PCR analyses. Immunohistochemical staining of ALK in both nucleus and cytoplasm is restricted to ALCL harboring the NPM-ALK fusion protein; all other known ALK fusion proteins are located exclusively in the cytoplasm.17 Histopathological diagnosis and ALK staining were confirmed using national reference pathology. Patients who had undergone significant steroid therapy for more than one week before diagnosis were excluded.
Patients
Between April 1996 and November 2005, 96 patients were registered into NHL-BFM95 and 127 German patients into ALCL99. Of these, bone marrow was available for 80, who were eligible for this study after informed consent. Patients were stratified according to stage (St Jude's system) and the involvement of risk organs.4,7 Staging procedures included bone marrow aspiration cytology and spinal tap. Bone marrow biopsies were done in only less than 20% of patients. Bone marrow involvement was defined by morphologically detectable ALCL cells in the aspiration cytology, irrespective of numbers. All bone marrow smears were reviewed by 2 experienced hematologists at the NHL-BFM study center. The treatment strategy was based on protocol NHL-BFM90, as described previously.4 All patients except those with stage I/II–resected disease received 6 5-day chemotherapy courses over a period of 4 to 6 months; those with stage I/II–resected disease received 3. Ten patients from study ALCL99 were randomized to receive weekly vinblastine maintenance; the last patient finished this therapy in April 2005.
The median age of the 80 study patients at diagnosis was 12.8 years (range, 1.1-17.9 years). The male-to-female ratio was 2.1 to 1. The clinical characteristics of the patients are shown in Table 1. There was no significant difference between the clinical characteristics of the 80 patients analyzed and the 143 German patients with an ALCL registered in the studies NHL-BFM95 and ALCL99 but not included in this study (Table S1, available on the Blood website; see the Supplemental Table link at the top of the online article). Central review by at least 2 experienced lymphoma pathologists according to the WHO classification led to the following distribution of histologic subtypes: 39 common, 13 mixed, 14 lymphohistiocytic, 6 small cell, 1 giant cell, and 2 not further classified; 2 cases were diagnosed cytologically, and there was no more material available for the remaining 3 patients for subclassification. All cases were CD30 and ALK positive and were negative for the B-cell markers CD20 and CD79a. Thirty-nine percent expressed CD3.
Bone marrow (BM) was obtained from all patients before the start of therapy and sent to the NHL-BFM study center within 24 hours. From 52 of the 80 study patients, peripheral blood (PB) could be evaluated for the presence of NPM-ALK–expressing cells as well. Mononuclear cells from BM and PB were obtained by density gradient centrifugation and immediately frozen at −80°C.
RT-RCR
RNA was extracted by Trizol reagent (Invitrogen, Carlsbad, CA). cDNA synthesis was performed with 1 μg total RNA, 5 μM random hexamer primer, 0.5 mM of each dNTP, and 200 U Superscript reverse transcriptase (Invitrogen). NPM-ALK cDNA was amplified in a 2-step PCR reaction. The following primers were used for the first round of amplification of the NPM-ALK transcript: 5′-TCCCTTGGGGGCTTTGAAATAACACC-3′ and 5′-GCCAGCAAAGCAGTAGTTGGGGTTG-3′. The primers 5′-CCAGTGGTCTTAAGGTTGAAG-3′ and 5′ TTGTACTCAGGGCTCTGCAGC-3′ were used for the second round of amplification.24 A single-step RT-PCR of ABL with the primers 5′-CGGCCAGTAGCATCTGACTTT G-3′ and 5′-CCTTGGCCATTTTTGGTTTGG-3′ was used as control for cDNA amplification. The first round of PCR amplification was performed with 0.08 μM of each primer and 1 U Taq DNA polymerase (Invitrogen). Primers were annealed at 64°C. Nested PCR was carried out with 1 μL of the first-round amplification product and 0.4 μM of each primer. The annealing temperature for the primers was 62°C.
The PCR products were analyzed on an agarose gel and visualized by UV illumination after ethidium bromide staining.
lp;&7q;1Primers and probes
Real-time quantitative PCR (RQ-PCR) was performed using TaqMan technology.
The oligonucleotide probe was labeled with a 5′ fluorescent reporter dye FAM and a 3′ quencher dye TAMRA. The primers for the ABL control gene were forward 5′-CAACACTGCTTCTGATGGCAA-3′ and reverse 5′-CGGCCACCGTTGAATGAT-3′ (MWG Biotech, Ebersberg, Germany).21 The sequence for the probe was 5′-FAM-CAACACCCTGGCCGAGTTGGTTCAT-TAMRA-3′ (Applied Biosystems, Weiterstadt, Germany). Primers and probes for NPM-ALK were forward 5′-CAGTGCATATTAGTGGACAGCACTTAG-3′, reverse 5′-TGATGGTCGAGGTGCGGA-3′, and probe 5′-FAM-CACCAGGAGCTGCAAGCCATGCA-TAMRA-3′.
Standard plasmids and cell lines
For the generation of the standard curves, plasmids containing NPM-ALK and ABL were used.21,25 The NPM-ALK plasmid was linearized with the XhoI restriction enzyme, and the ABL plasmid was treated with NotI (New England Biolabs, Beverly, MA). Both plasmids were dephosphorylated with alkaline phosphatase (New England Biolabs) and purified with a Qiagen PCR Purification Kit (Qiagen, Hilden, Germany). After quantification at 260 nm, the plasmids were diluted in H2O to a concentration of 107 copies/μL. Ten-fold serial dilutions in 1 mM Tris (pH 8), 0.1 mM EDTA, and 50 μg/mL E coli tRNA (Roche, Mannheim, Germany) were carried out to obtain standard plasmid concentrations from 106 to 101 copies/μL.
Karpas 299 NPM-ALK–positive ALCL and DG 75 Burkitt lymphoma cell lines were obtained from the German Collection of Microorganisms and Cell Cultures (http://www.dsmz.de; DSMZ, Braunschweig, Germany). Cells were routinely maintained in RPMI1640 medium supplemented with 10% fetal bovine serum.
RQ-PCR
A quantitative RQ-PCR for NPM-ALK was developed according to the standardized protocol of the EAC program for TaqMan-based RQ-PCR.20 ABL was selected as the control gene to compensate for variations in RNA integrity and differences in the RT-PCR step.23,26 Therefore, NPM-ALK and the ABL control gene were amplified separately from the same cDNA. All samples were analyzed before quantitative PCR by RT-PCR. NPM-ALK–positive RT-PCR samples were subsequently analyzed by RQ-PCR. The RQ-PCR analysis was carried out in duplicates of both undiluted and 1:2 diluted cDNA.
The RQ-PCR was set up with 1 × TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA), 300 nM of each primer, 200 nM NPM-ALK or ABL probe, and 1 μL cDNA in a total volume of 25 μL. For the standard curve, 1 μL of each standard plasmid dilution was used.
cDNA from the leukemic cell line HL60 and H2O were used as the “no amplification control” and the “no template control,” respectively. The thermal cycler conditions were 2 minutes at 50°C for uracil N-glycosylase treatment and 10 minutes at 95°C for inactivation of uracil N-glycosylase and activation of AmpliTaq Gold Polymerase, followed by 50 cycles of 15 seconds at 95°C and 1 minute at 60°C. All reactions were performed on the ABI PRISM 7700 Sequence Detection System. To compare RQ-PCR assays from different runs, the threshold was set at 0.1 and the baseline at 3 to 12 for NPM-ALK and ABL. Samples with fewer than 2000 copies of ABL were regarded as poor-quality RNA and not further analyzed.
The NPM-ALK fusion transcript copy numbers were normalized to the copy numbers of the ABL gene. The absolute copy numbers of NPM-ALK and ABL were estimated with their corresponding standard curves. The normalized copy numbers (NCNs) were expressed as copy numbers of NPM-ALK per 104 copies ABL.20
Statistical analysis
Analysis of event-free survival (EFS) and survival was performed using the Kaplan-Meier method with differences compared by the log-rank test; standard error was calculated according to Greenwood.27-29 EFS was calculated from the date of diagnosis to the first event (nonresponse, death from any cause, tumor progress, or second malignancy) or to the date of last follow-up, survival from the date of diagnosis to death from any cause, or last follow-up. Cumulative incidence functions for relapse were constructed following the method of Kalbfleisch and Prentice.30 Functions were compared with Gray test.31
The prognostic effect of circulating tumor cells on treatment outcome was compared with other known prognostic factors in patients with ALCL using Cox regression analysis.32
Standard methods were used for descriptive statistics. All analyses were performed using SAS (SAS-PC, version 9.1; SAS Institute, Cary, NC). Data were updated as of March 2006.
Results
Sensitivity and specificity of the RT-PCR and RQ-PCR for NPM-ALK
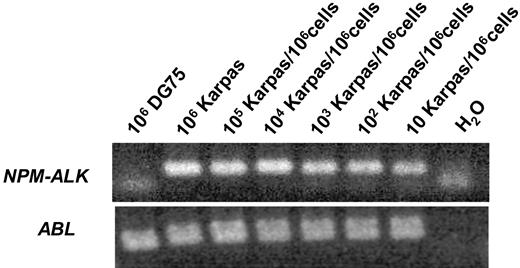
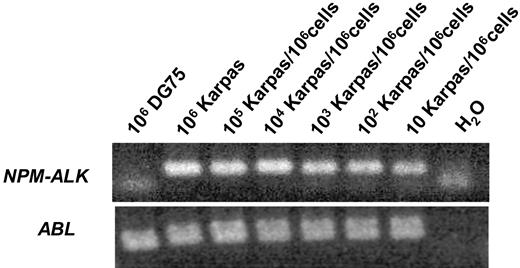
The sensitivity and specificity of the RT-PCR was tested with Karpas299 NPM-ALK–positive ALCL cells diluted in DG 75 Burkitt lymphoma cells and various NPM-ALK–negative cell lines as well as peripheral blood from 25 healthy blood donors. The sensitivity was 10−5NPM-ALK–positive cells/control cells (Figure 1). Nonspecific amplification was not observed within other NHL or leukemia cell lines, healthy blood donors, NPM-ALK–negative patients, or human genomic DNA (data not shown).
Sensitivity of the qualitative RT-PCR for NPM-ALK.NPM-ALK–positive Karpas 299 cells were diluted in DG 75 Burkitt lymphoma cells.
Sensitivity of the qualitative RT-PCR for NPM-ALK.NPM-ALK–positive Karpas 299 cells were diluted in DG 75 Burkitt lymphoma cells.
The absolute copy numbers of NPM-ALK and the ABL control gene were determined by 10-fold dilutions of the appropriate standard curve. In serial cell dilution experiments with Karpas 299 (Su-DHL-1, SR786) in NPM-ALK–negative DG 75 (Table 2) peripheral blood or bone marrow, the method detected fewer than 10 NPM-ALK–positive cells/106 control cells, indicating a sensitivity of at least 10−5 on the cellular level. The RQ-PCR method reliably measured 0.2 to 1 copies of NPM-ALK/104 copies of ABL in patient material provided that sufficient copies of ABL were detected.
Characteristics of patients with positive bone marrow PCR for NPM-ALK
The bone marrow (BM) was positive for the NPM-ALK transcript in 38 (47.5%) of the 80 patients. The PCR result in BM significantly correlated with the clinical stage. Only 2 (9%) of the 22 patients with stage I or II disease showed a positive PCR compared with 36 (62%) of the 58 patients with stage III or IV disease (P < .001, Table 1). There also was a significant association of a positive BM NPM-ALK PCR result with the known clinical risk features of involvement of mediastinum and visceral organs7 ; the involvement of skin and CNS did not correlate with a positive RT-PCR (Table 1). Eight patients showed microscopically detectable ALCL cells in the BM (percentage of ALCL cells: 2 × 1%, 2%, 5.5%, 7%, 9%, 20%, and 41%). They all had a positive PCR result for NPM-ALK in BM. A positive BM PCR correlated with an atypical histopathological subtype as defined by noncommon histology (Table 1). The BM PCR result was not associated with the ALCL immunophenotype as defined by expression of the T-cell marker CD3, which was evaluated throughout the whole study period as part of the national reference pathology review.
From 32 of the 38 patients with a positive BM RT-PCR material of sufficient quality with more than 2000 copies ABL was available for quantitative TaqMan PCR. Between 0.2 and more than 5000 copies of NPM-ALK /104 copies ABL (NCNs) were detected.
There was a significant association between microscopic BM infiltration and a BM NPM-ALK NCN of more than 10. Five of the 16 patients with a BM NPM-ALK NCN higher than 10 had detectable ALCL cells in BM compared with only 2 of 58 patients with a lower NCN (P < .01). The NPM-ALK NCN in PCR-positive and cytologically positive patients varied between 2 and 5600, thus overlapping with PCR-positive but cytologically negative patients (0.2 to 534), suggesting that BM involvement might easily be missed by microscopy in ALCL.
Comparison of simultaneous NPM-ALK PCR results from bone marrow and peripheral blood
PB mononuclear cells could be analyzed in addition to BM in 52 of the 80 patients. NPM-ALK mRNA was detected in 52% (27/52) of the PB samples. In 45 (86.5%) of the 52 patients, the NPM-ALK RT-PCR yielded concordant results between BM and PB (24 negative/negative; 21 positive/positive; Kappa coefficient = 0.73, P < .001).
Regarding the 7 patients with PCR results diverging between BM and PB, 1 patient with positive BM PCR was PB PCR negative, and 6 patients were PB positive and BM negative. Quantitative NPM-ALK PCR data were available from 5 of the 6 PCR-positive PB samples with a corresponding PCR-negative BM sample. NCN was 1 or less in 3 patients and 2 and 4 in the remaining 2 patients. One NCN NPM-ALK was measured in the single positive BM sample corresponding to a negative PB PCR.
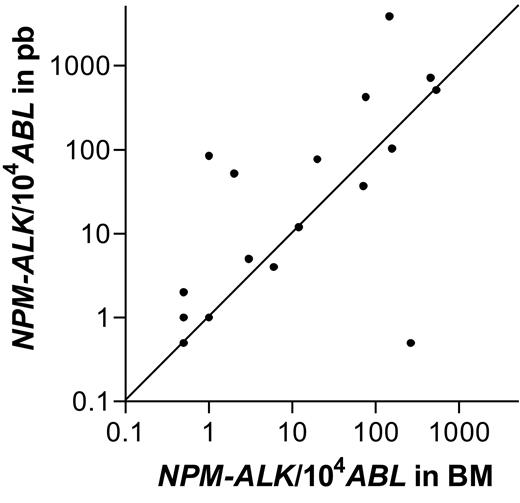
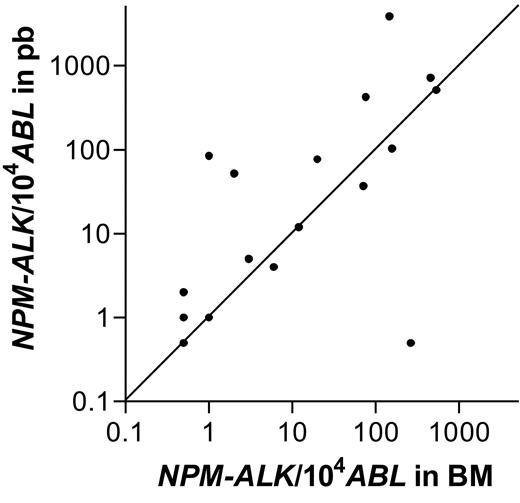
The NPM-ALK NCN in BM and PB could be compared in 19 of the 21 patients with a positive qualitative RT-PCR in both BM and PB. The results were either identical or showed slight variation within one log in 15 of the 19 patients, indicating a high concordance of NPM-ALK positivity at the quantitative level, as well (Spearman correlation coefficient = 0.84, P < .001). The NPM-ALK NCN between PB and BM varied slightly more than 1 log in 3 patients and more than 2 logs in 1 patient; NCN was higher in PB compared with BM in 3 of these 4 patients (Figure 2).
Comparison of quantitative NPM-ALK PCR results between bone marrow and peripheral blood. Number of NPM-ALK copies/104 copies ABL in bone marrow and peripheral blood of 19 patients with a positive RT-PCR in bone marrow and blood.
Comparison of quantitative NPM-ALK PCR results between bone marrow and peripheral blood. Number of NPM-ALK copies/104 copies ABL in bone marrow and peripheral blood of 19 patients with a positive RT-PCR in bone marrow and blood.
All together, the comparison of both qualitative and quantitative PCR for NPM-ALK between BM and PB showed high concordance. However, more patients were positive in PB, and positive patients tended to have higher copy numbers in PB compared with BM.
Prognostic impact of a positive qualitative PCR for NPM-ALK in BM and PB
The median follow-up was 3.2 years. Only 2 patients had a follow-up of less than a year from diagnosis. Because relapse/progression occur early in pediatric ALCL, follow-up was sufficient to allow detailed analyses. Overall survival (OS) and event-free survival (EFS) for the whole group of 80 patients analyzed by PCR were 74% ± 6% and 61% ± 6%, respectively. The cumulative incidence of relapse (CI-R) was 32% ± 6%. The comparable data for EFS, CI-R, and OS for the 143 German ALCL patients from trials NHL-BFM95 and ALCL99 but not included in this study were 72% ± 4%, 25% ± 4%, and 90% ± 3%, respectively. EFS and the CI-R of these 143 patients were statistically not significantly different from the 80 analyzed patients (P = .12 for comparison of EFS and P = .32 for comparison of CI-R), while OS was (P = .02).
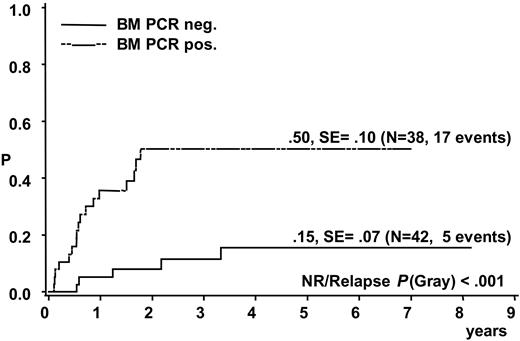
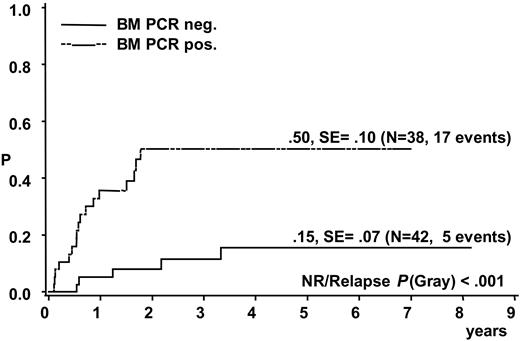
A total of 17 of the 38 patients with a positive BM PCR for NPM-ALK relapsed compared with 5 of the 42 patients with negative BM PCR (CI-R 50% ± 10% versus 15% ± 7%, P < .001) (Figure 3). Patients with a positive NPM-ALK PCR result in BM had an EFS and OS at 5 years of 38% ± 9% and 60% ± 9%, respectively, compared with 82% ± 7% and 86% ± 7%, respectively, for patients without detection of NPM-ALK in BM (P < .001 for EFS and P < .006 for OS).
Cumulative incidence of relapse of 80 analyzed ALCL patients according to qualitative PCR results for NPM-ALK in bone marrow.
Cumulative incidence of relapse of 80 analyzed ALCL patients according to qualitative PCR results for NPM-ALK in bone marrow.
The prognostic impact of the BM PCR depended on the stage of disease: Only 2 of 22 patients with stage I and II disease were BM PCR positive, and both remained free of disease. However, among the 58 patients with stage III or IV disease, 17 of 36 patients with positive PCR relapsed compared with 2 of 22 with negative BM PCR (cumulative incidence 54% ± 11% versus 11.6% ± 8%, P = .002).
Because of the high concordance of BM and PB PCR results in the 52 parallel patient samples, a positive PCR in PB also was associated with a high risk of failure. Three of the 25 patients with negative PCR in PB relapsed compared with 10 of 27 patients with detection of NPM-ALK by PCR in PB (cumulative incidence 20% ± 11% versus 41% ± 11%, P = .03) resulting in a 5-year EFS of 46% ± 10% and 80% ± 11% for patients with and without positive PCR in PB, respectively (P = .004).
Only 1 of the 7 patients with PCR results diverging between PB and BM relapsed; the patient was positive in PB. The 3 patients with relapse of a PB PCR-negative ALCL also were negative for NPM-ALK mRNA in BM, suggesting that application of PB in addition to BM does not enhance the negative predictive value of the qualitative PCR. In the 3 cases, NPM-ALK positivity was confirmed by nuclear and cytoplasmic ALK staining in the tumor cells. Moreover, in relapse PB samples of these 3 patients tested positive for NPM-ALK, demonstrating that the systemic involvement in relapse could be detected by PCR.
Prognostic impact of quantification of NPM-ALK transcripts in BM and PB
Outcome analyses with inclusion of quantitative PCR data for PCR-positive samples could be performed on 74 patients with BM PCR, 51 patients with PB PCR.
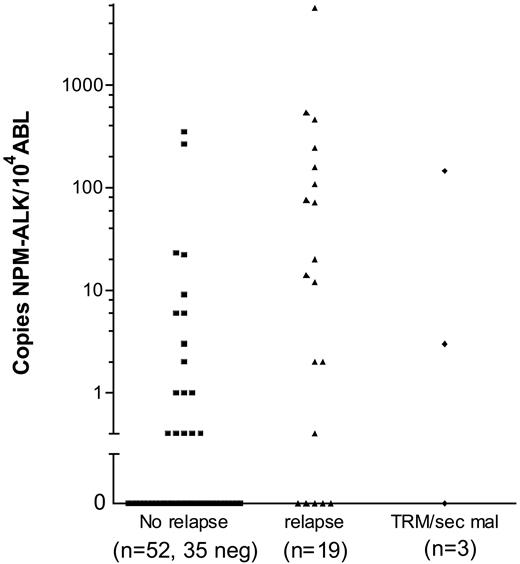
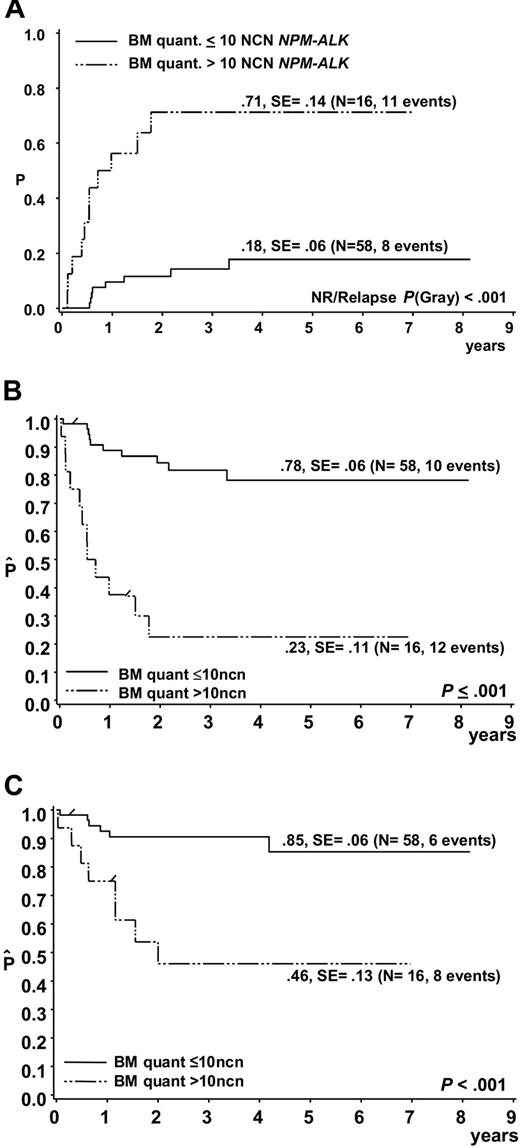
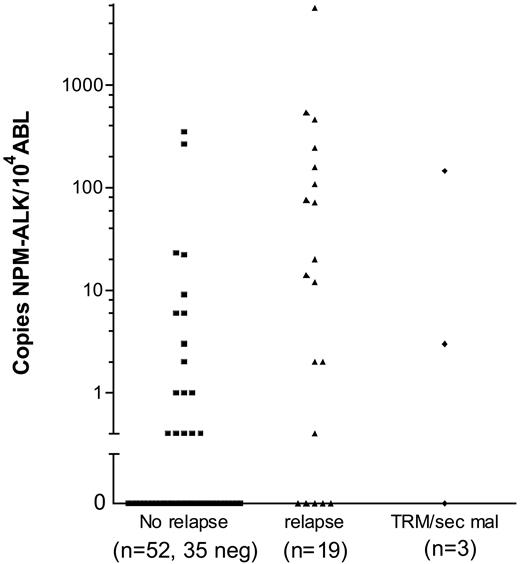
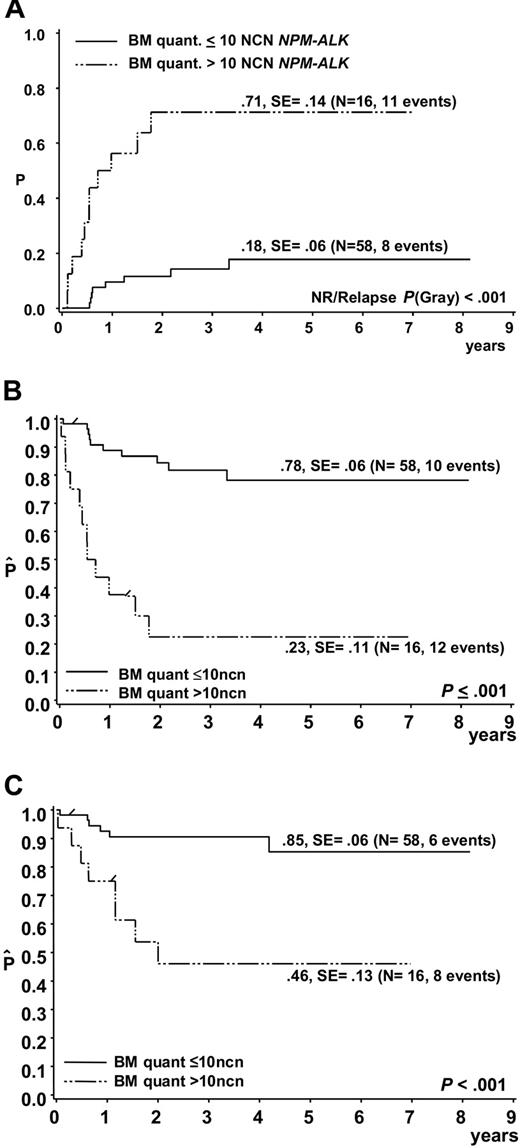
Of the 74 patients, 19 suffered a relapse, 2 patients died due to therapy and 1 due to a second malignancy. In 11 of 19 patients who suffered a relapse, more than 10 NCNs NPM-ALK were detected in initial BM samples, while only 4 of 52 patients who stayed in first complete remission had more than 10 NCNs NPM-ALK in BM at diagnosis (Figure 4). The cumulative incidence of relapse among the 16 patients with more than 10 NCNs NPM-ALK in BM was 71% ± 14% (Figure 5A). Only 4 patients stayed in long-term remission, resulting in an EFS of only 23% ± 11% compared with an EFS of 78% ± 6% for 58 patients with NCN less than 10 (P < .001) (Figure 5B). OSs were 46% ± 13% and 85 ± 6% for patients with more than 10 and 10 or fewer NCNs in BM, respectively (Figure 5C). Five of these 16 patients had cytological BM involvement, and only 1 of them stayed in complete remission (CR). Likewise, only 3 of the 11 patients with NCNs NPM-ALK more than 10 but without cytological BM involvement stayed in CR, suggesting that at the submicroscopic level, a high extent of BM involvement is as associated with a poor prognosis as microscopic detection of tumor cells. Microscopic BM infiltration, however, detects only one third of the very high risk patients compared with quantitative PCR. A positive qualitative BM PCR, on the other hand, cannot substitute for quantification of NPM-ALK transcripts because patients with positive PCR but low NCN (< 10) had a similar risk of relapse (N = 16, CI-R 20% ± 10%) as those with no detectable disease (N = 42, CI-R 15% ± 7%).
Quantitative NPM-ALK PCR results in bone marrow according to the outcome of the ALCL patients. Normalized NPM-ALK copy numbers (copies NPM-ALK/104 copies ABL) in bone marrow of the 74 patients with either a negative qualitative BM PCR (= 0 copies NPM-ALK) or a positive qualitative BM PCR using quantitative PCR results. TRM indicates treatment related mortality; sec mal, second malignancy.
Quantitative NPM-ALK PCR results in bone marrow according to the outcome of the ALCL patients. Normalized NPM-ALK copy numbers (copies NPM-ALK/104 copies ABL) in bone marrow of the 74 patients with either a negative qualitative BM PCR (= 0 copies NPM-ALK) or a positive qualitative BM PCR using quantitative PCR results. TRM indicates treatment related mortality; sec mal, second malignancy.
Outcome of ALCL patients according to quantitative PCR results for NPM-ALK in bone marrow. (A) Cumulative incidence of relapse and Kaplan-Meier estimates of (B) event-free survival and (C) overall survival of the 74 patients with either a negative qualitative BM PCR or a positive qualitative BM PCR using quantitative PCR results and a NPM-ALK cutoff copy number of 10/104 copies ABL (NCN).
Outcome of ALCL patients according to quantitative PCR results for NPM-ALK in bone marrow. (A) Cumulative incidence of relapse and Kaplan-Meier estimates of (B) event-free survival and (C) overall survival of the 74 patients with either a negative qualitative BM PCR or a positive qualitative BM PCR using quantitative PCR results and a NPM-ALK cutoff copy number of 10/104 copies ABL (NCN).
A high number of circulating NPM-ALK–positive cells in PB was associated with a similar high risk of relapse. The 12 patients with more than 10 NCNs NPM-ALK in PB had an EFS of 19% ± 12% and only 3 patients stayed in remission, compared with an EFS of 77% ± 9% of the 39 patients with copy numbers below that threshold or without detectable NPM-ALK (P < .001).
The pattern of relapse was compared between the 11 patients with more than 10 NCNs NPM-ALK in initial BM samples and the 8 with 10 or fewer NCNs. The sites of relapse did not vary between the 2 patient groups (relapse site: 11 patients with > 10 NCNs in BM: 3 local, 4 local + new sites, 2 local + new + BM, 1 local + new + CNS, 1 CNS; 8 patients with ≤ 10 copies: 2 local, 4 local + new sites, 2 local + new + BM). However, the time of relapse differed between patients when grouped according to quantitative PCR results: 5 of the 11 patients with NCN NPM-ALK more than 10 who experienced a relapse progressed during initial therapy. In contrast, none of the 8 patients with a relapse but NCN of 10 or less in initial BM progressed during initial therapy. The outcome did not differ significantly among these 2 groups of patients (11 relapse patients with > 10 NCN: 4 CR, 4 death of disease [DOD], 2 treatment-related death [TRM], 1 death due to a second malignancy; 8 patients with ≤ 10 NCNs: 3 CR, 4 DOD, 1 TRM). However, relapse therapy for patients with progression during first-line therapy was more aggressive including allogeneic blood stem cell transplantation in most instances compared with patients with relapse after initial therapy.
Correlation of positive NPM-ALK PCR results in BM or PB with other prognostic factors
Both known clinical risk features (mediastinal, visceral, or skin involvement)7 and a noncommon histology influenced the risk of relapse in addition to the NPM-ALK-PCR in BM in univariate analysis (Table 3). In a Cox regression analysis with the covariables “histologic subtype ‘not common’” and clinical risk features, PCR positivity in BM and atypical histology were revealed to be prognostic factors with similar risk ratios (Table 3). Clinical risk features had no additional significant influence in the multivariate analyses. In a multivariate analysis including quantification of minimal bone marrow involvement, the highest risk of relapse was conferred by NCN NPM-ALK more than 10 measured by quantitative PCR in BM and PB (Tables 4–5); the histologic subtype had no significant prognostic value.
Discussion
Our RT- and RQ-PCR approach enabled us to detect specifically circulating NPM-ALK–positive ALCL cells in BM and PB with a sensitivity of 10−5 and to quantify the extent of circulating tumor cells relative to the control gene ABL.
The prevalence of a positive RT-PCR in BM of 48% in our series of 80 children and adolescents with NPM-ALK–positive ALCL is along the lines of the 61% observed by Mussolin et al, confirming the high percentage of circulating tumor cells in BM in ALCL.19 The slight methodological differences in the RT-PCR assay between the 2 studies are unlikely to have caused the higher percentage of BM PCR positivity in the Italian cohort of patients. Because clinical stage and BM PCR results strongly correlated in our group of patients (Table 1), the slightly higher percentage of submicroscopic BM involvement in the Italian cohort may be explained by the noticeably higher percentage of advanced-stage patients included in their series.
We detected a significant correlation of a positive BM RT-PCR with the clinical risk features of mediastinal and visceral organ involvement and with advanced-stage disease in our cohort of 80 patients, suggesting that the detection of circulating tumor cells reflects the extent of the disease. The correlation between qualitative BM PCR and disease stage might have been missed in the Italian cohort of 41 patients because of the low number of patients with stage I and II disease.19 However, a positive NPM-ALK PCR in BM did not correlate with the clinical risk factor of skin lesions.3,7 Only 4 patients were diagnosed with CNS involvement, precluding statistical analysis despite BM PCR positivity in 3 of the 4 patients. There was a strong interrelationship among qualitative BM PCR, clinical risk features, and atypical histologic subtypes suggesting that an uncommon histology may reflect different biologic behavior of the tumor cells that manifests in widespread clinical and subclinical dissemination.
The high concordance of both qualitative and quantitative NPM-ALK PCR results between BM and PB suggests that BM PCR positivity in ALCL may, in analogy to T-cell acute lymphoblastic leukemia,33,34 be regarded as a reflection of circulating tumor cells rather than of true BM micrometastasis, which can often be detected in solid tumors.
The relapse rate for patients with negative BM PCR still reached 15%, a risk comparable with that of patients without clinical risk features and with a common histologic subtype.7 In addition, those patients with positive BM PCR but NCN NPM-ALK of 10 or less in the quantitative analysis have a similar low relapse rate than BM PCR–negative patients and could be added to this group. However, in contrast to the data of Mussolin et al,19 a very low risk group of patients amenable to therapy reduction could not be defined by measuring circulating tumor cells. In our cohort of patients, a group of approximately one third of the patients with an EFS of 95% and an OS of 100% could be identified only by combining the positive factors of negative BM PCR and common histologic subtype. But this finding has to be confirmed by a larger study.
Quantitative PCR in BM and PB, on the other hand, allowed identification of a poorer risk group of patients. While a positive qualitative PCR in BM and PB already conferred a 50% risk of relapse, the risk rose to 70% for patients with more than 10 NCNs NPM-ALK in BM or PB. Methodological differences between this study and that with an Italian population might explain why the prognostic impact of a high tumor cell load measured by quantitative PCR in the current work could not be detected by the Italian group in bone marrow samples.19 Tumor cells were enriched in the mononuclear cell fraction by density gradient centrifugation in our study, allowing a higher sensitivity than the measurement in nuclear cells (C.D.-W. et al, unpublished data, June 2006). We used the control gene ABL, which we and others have shown to have the lowest variation between lymphoid and normal cells as well as between patients and normal bone marrow compared with GAPDH.21,23,26,35 The exclusion of material with fewer than 2000 copies of the control gene in our study, as suggested by the EAC guidelines, might also have contributed to enhancing the predictive value of the quantitative PCR assay.23
Accumulating evidence suggests that significant BM involvement in NPM-ALK–positive ALCL is a surrogate for highly aggressive biologic disease. This is underscored by our observation that relapses occurred even during initial treatment in patients with more than 10 NCNs NPN-ALK. Qualitative positive BM PCR, quantitative PCR, and microscopic BM infiltration detected by aspiration cytology were associated with a poor prognosis in our cohort of patients from 2 consecutive clinical trials. The same was true for qualitative PCR in the Italian cohort19 and BM histology with CD30 and EMA staining.36 Earlier analyses within national groups, however, did not identify the poor prognostic significance of BM cytology partly because of the low number of BM-positive patients in each trial.2,4,5,7 In addition, usually only low numbers of ALCL cells infiltrate the BM, and they are difficult to detect by morphology only.36,37 It is still not established whether BM immunohistology using the ALK antibody could be an option to accurately and sensitively detect circulating tumor cells. The available data from Mussolin et al with 35 patients and using both qualitative BM PCR and BM immunohistology do not hint at this possibility.19 In our patient group, more than 3 times as many patients had more than 10 NCNs NPM-ALK in BM than had a positive BM cytology with an identical high risk of failure, suggesting that an accurate quantitative method at the submicroscopic level is needed to identify patients with high-risk disease.
The establishment of a quantitative PCR assay according to the quality standards of the EAC in several laboratories with the application of interlaboratory quality control measures might allow use of this technology for stratification criteria in future international trials on ALCL. This technology could enable the identification of 20% of NPM-ALK–positive patients early after diagnosis with a relapse rate of 70%, representing 60% of all relapses.
In conclusion, our results confirm the prognostic importance of submicroscopic BM infiltration in patients with NPM-ALK–positive ALCL above all other known clinical risk features. Patients with a high number of circulating tumor cells defined by more than 10 NCNs NPM-ALK in BM and/or PB have an extremely poor prognosis. These patients might constitute a group amenable to altered therapy in future studies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the BMBF competence network pediatric Oncology and Hematology–MRD in leukemia and lymphoma (01GI9963/2).
We thank S. Morris for providing the plasmid pcDNA3 NPM-ALK. We thank the members of the national reference pathology panel (R. Parwaresch, A. C. Feller, M. L. Hansmann, P. Möller, H. Müller-Hermelink, and H. Stein). We especially acknowledge L. Brugieres for critically reading the paper. A special thanks to all children and their parents for taking part in this study and to the physicians and nurses caring for these children.
Authorship
Contribution: C.D.-W., K.B., S.V., and J.S. designed the PCR assays and performed the research; I.O. and W.K. were responsible for the central histopathological review; K.B., J.H., W.W., and A.R. designed and supervised the research-project; C.D.-W., B.B., A.R., and W.W. collected, interpreted, and analyzed the data; M.Z. performed statistical analyses; C.D.-W. and W.W. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: A. Reiter, Department of Pediatric Hematology and Oncology, University Children's Hospital; Feulgenstr. 12, D-35392 Giessen; Germany; e-mail: alfred.reiter@paediat.med.uni-giessen.de.