Abstract
Rare cases of histiocytic and dendritic cell (H/DC) neoplasms have been reported in patients with follicular lymphoma (FL), but the biologic relationship between the 2 neoplasms is unknown. We studied 8 patients with both FL and H/DC neoplasms using immunohistochemistry, fluorescence in situ hybridization (FISH) for t(14;18), and polymerase chain reaction (PCR)/sequencing of BCL2 and IGH rearrangements. There were 5 men and 3 women (median age, 59 years). All cases of FL were positive for t(14;18). The H/DC tumors included 7 histiocytic sarcomas, 5 of which showed evidence of dendritic differentiation, and 1 interdigitating cell sarcoma. Five H/DC tumors were metachronous, following FL by 2 months to 12 years; tumors were synchronous in 3. All 8 H/DC tumors showed presence of the t(14;18) either by FISH, or in 2 cases by PCR with the major breakpoint region (MBR) probe. PCR and sequencing identified identical IGH gene rearrangements or BCL2 gene breakpoints in all patients tested. All H/DC tumors lacked PAX5, and up-regulation of CEBPβ and PU.1 was seen in all cases tested. These results provide evidence for a common clonal origin of FL and H/DC neoplasms when occurring in the same patient, and suggest that lineage plasticity may occur in mature lymphoid neoplasms.
Introduction
Cells of the hematopoietic system are derived from common precursors that differentiate into lineages with distinct morphologic, immunophenotypic, and functional characteristics.1,2 The lineage of the cell of origin is the major criterion used to classify hematopoietic neoplasms.3 Most theories of hematopoietic cell differentiation have proposed that as cells differentiate fully, they become “lineage committed.” However, some clinical data have shown that 2 hematopoietic populations in the same patient may share identical genetic changes or abnormalities, raising the possibility that tumors expressing the phenotype of one hematopoietic lineage might “transdifferentiate” into a genetically similar but phenotypically distinct tumor of a different lineage. For example, we recently reported examples of patients with lymphoblastic leukemias or lymphomas who also had histiocytic or dendritic cell (H/DC) tumors and demonstrated that the lymphoblastic neoplasm and the H/DC tumor from each patient carried identical genetic changes, indicating a clonal relationship between the 2 tumors.4,5 Because lymphoblastic neoplasms are tumors of precursor cells, these cells might display more lineage plasticity than mature lymphoid tumors. For example, it has been shown that human pro-B cells can give rise to macrophages in vitro.6
Rare cases of H/DC neoplasms have been reported in patients with mature B-cell neoplasms. Vasef et al reported 3 patients with dendritic cell tumors associated with low-grade B-cell malignancies,7 including a 59-year-old woman who developed a dendritic cell sarcoma 9 years following a diagnosis of follicular lymphoma (FL), grade 1. The other 2 patients had dendritic tumors associated with chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL). Similarly, there have been other isolated case reports of H/DC cell tumors occurring in patients with FL, CLL/SLL, and extranodal marginal zone B-cell lymphoma of mucosa-associated lymphoid tissue (MALT lymphoma).8-12 The biologic relationships between the B-cell lymphomas and dendritic cell tumors in these cases were not studied.
Langerhans cell histiocytosis has been reported to occur in association with FL, and is usually an incidental finding of no clinical consequence.13-18 Magni et al described a patient with FL in an inguinal lymph node and synchronous Langerhans cell histiocytosis on the scalp,19 and reported identical clonal immunoglobulin gene rearrangements in both lesions. Anjuère et al suggested that Langerhans cells may develop from a “lymphoid-committed” precursor, based on evidence in mice that epidermal Langerhans cells can be generated from the same CD4low precursors that generate B cells, T cells, and natural killer (NK) cells.20
From 1994 to 2006 in the course of practice, we encountered 8 patients with documented FL who also developed a H/DC tumor, either synchronously or, in most cases, at a later point in time. This series of novel cases provided us with the opportunity to study the biologic relationships between the FLs and the H/DC tumors, and to examine possible mechanisms of this association.
Methods
This study was approved by the Institutional Review Boards of the National Cancer Institute, Stanford University, University of Barcelona, and Mayo Clinic. Informed consent was obtained in accordance with the Declaration of Helsinki from all patients enrolled in treatment protocols.
Histology and immunohistochemistry
Eight patients with both FL and H/DC neoplasms were identified from Stanford University Hospital/Alta Bates Summit Medical Center (1), Mayo Clinic (1), the University of Barcelona (1), and the National Cancer Institute (5). Frozen and paraffin-embedded tissue blocks were analyzed using morphologic assessment and immunohistochemistry. Diagnostic criteria for both FL and H/DC neoplasms were those of the WHO.3 Dendritic cell differentiation was used to designate those neoplasms that expressed S100 protein and in 2 cases, CD1a, in addition to manifesting evidence of histiocytic differentiation based on staining for CD68 and CD163. For immunohistochemistry, paraffin-embedded tissue sections were stained using an automated immunostainer (Ventana Medical Systems, Tucson, AZ) according to the company's protocols, with minor modifications. Antigen retrieval was performed using a Tender Cooker (Nordicware, Minneapolis, MN) with citrate buffer. Antibodies included CD20 (L26, 1:200; Dako, Carpinteria, CA), CD3 (F7.2.38, 1:50; Dako), CD10 (56C6, 1:40; Novocastra, Newcastle upon Tyne, United Kingdom), Bcl6 (PG-B6p, 1:20; Dako), Bcl2 (123, 1:20; Dako), CD68 (KP-1, 1:50; Dako), CD163 (10D6, 1:10; Novocastra), lysozyme (EC3.2.1.17, 1:2000; Dako), S100 (15E2E2, 1:4000; BioGenex, San Ramon, CA), CD1a (O10, undiluted; Immunotech, Marseilles, France), PAX5 (1:100 dilution; BD Transduction Laboratories, Lexington, KY), C/EBP-beta (clone H, 1:400 dilution; Santa Cruz, Santa Cruz, CA), and PU.1 (clone G148-74, 1:150 dilution; Pharmingen, San Diego, CA).
Fluorescence in situ hybridization
Fluorescent in situ hybridization (FISH) for t(14;18) was successfully performed using breakapart probe (BAP) and/or dual-fusion probe (D-FISH) methods on both FL and H/DC tumors in 6 of 8 cases. Hybridization reactions were unsuccessful in cases 6 and 8. Whole-tissue FISH studies were performed on 25-μm paraffin section. For samples with the 2 tumors in an individual section, the distinct lymphoma and histiocytic tumors were marked, and 2 cores were then extracted from these preidentified areas. All the samples were deparaffinized in 3 solutions of 100 μL xylene, washed one time each in 100 μL of 100% ethanol, 75% ethanol, and 50% ethanol. Disaggregated tissue from the core samples was digested in 100 μL of 0.005% (30 μ/mg) freshly prepared proteinase K solution. The samples were then incubated at 37°C for 30 minutes, followed by washing in PBS. Nuclei were fixed in a freshly prepared 3:1 methanol–acetic acid solution. The nuclei suspensions were placed on a slide within a 18-mm circle and air dried. The slide was then treated with 0.2 N HCL, rinsed with water, and pretreated with Vysis (Downers Grove, IL) pretreatment reagent for 30 minutes, at 80°C, followed by another rinse with water. The slide was then digested with 10% pepsin/0.2N HCL for 10 minutes at 37°C and then rinsed with water, followed by incubation with 10% buffered formalin for 10 minutes at 37°C, rinsing with water, dehydrating, and drying. The BCL2/IGH dual-fusion probes or BCL2 breakapart probes (Vysis) were then applied. Slides were incubated at 80°C for 8 minutes to denature the probes and hybridized at 37°C overnight. Slides were washed once with 0.4 × SSC/0.3% NP40 at 73°C for 2 minutes and once with 2 × SSC/0.1% NP40 at room temperature for 1 minute, and 15 μL DAPI/antifade was applied.
Polymerase chain reaction and sequencing
Polymerase chain reaction (PCR) and sequencing of IGH gene rearrangements (4 cases) or the BCL2/JH breakpoint region (1 case) were successfully analyzed in 5 of 8 cases. In 6 patients, the FL and H/DC were present in different specimens, and there was no morphologic or immunophenotypic evidence of FL cells infiltrating the H/DC tumors. In 1 patient (case 7), both tumors were present in the same specimen and comparison of the sizes and sequences of the PCR products could not be performed reliably. In a second case (case 4), segregation of the 2 components in a single mass and subsequent isolated recurrence of the H/DC tumor permitted analysis. PCR for the MBR of the BCL2 gene was successfully performed in both FL and H/DC tumors in 6 of 8 cases.
Initial IGH PCR analysis was performed using commercially available primers directed against FR1, FR2, and FR3 VDJ regions of IGH(InVivo Scribe, Carlsbad, CA). PCR products were analyzed by capillary electrophoresis. To further compare the sequences of the 2 tumors, genomic DNA was isolated according to Qiagen's DNA mini kit handbook (Valencia, CA).21 PCR was performed using primers IGHV3-FR1 (5′-CTGGGGGGTCCCTGAGACTCTCCTG-3′) and J region consensus primer (5′-CTTACCTGAGGAGACGGTGACC-3′). The PCR mixture contained 5 μL 10 × PCR buffer, 1.5 μL of 50 mM MgCl2, 1 μL of 10 mM dNTP, 0.5 μL Platinum Taq DNA Polymerase, 1 μL of 10 μm of each primer, and 2 μL of 1 μg/μL template DNA with water added to form a total reaction volume of 50 μL. The PCR products were amplified by the touchdown method with cycling conditions of 1 cycle at 95°C for 2 minutes; 25 cycles starting at 95°C for 30 seconds, followed by 65°C for 30 seconds with decreasing temperatures (− 0.5°C/cycle), and at 72°C for 1 minute; and 15 cycles starting at 95°C for 30 seconds, followed by 55°C for 1 minute and 72°C for 1 minute. The last extension cycle was at 72°C for 5 minutes. The PCR products were then purified before performing the sequencing reaction.
The sequencing reaction included the following: 4 μL BigDye Terminator v3.1, 2μL 5 × sequencing buffer (Applied Biosystems, Foster City, CA), 1 μL IGHV3-FR1 primer or J region consensus primer, 9 μL H2O, and 4 μL of the purified PCR product prepared as described in the preceding paragraph. Thermal cycling conditions were as follows: denaturing at 96°C for 1 minute, followed by 25 cycles at 96°C for 10 seconds, 50°C for 5 seconds, and 60°C for 4 minutes. The amplified products were then purified using Centri-Sep columns (Princeton Separations, Adelphia, NJ). Sequencing was done on an ABI 3100 sequencer (Applied Biosystems). The pair of sequences was then analyzed on CLUSTALW.22 Mutation analysis of the V region sequencing was performed according to the method of Giudicelli et al.23 IMGT/V-QUEST (ImMunoGeneTics Information System, Montpellier, France), an integrated software program for immunoglobulin and T-cell receptor V-J and V-D-J rearrangement analysis, was used.
Results
Clinical features
Clinical features of the 8 patients are presented in Table 1. There were 5 men and 3 women. Age ranged from 30 to 67 years (median, 59 years). In patients 4 and 7, both FL and the H/DC sarcoma presented synchronously in the same site. In patient 6, the tumors were detected synchronously but in different sites, and in patient 5, the H/DC diagnosis followed the FL by only 2 months, in a different site. In patients 1, 2, and 3, the H/DC was diagnosed from 2 to 12 years following the original FL. Therapy was variable following diagnosis of H/DC, with survival ranging from 8 months to 10 years, for those patients in whom follow up could be obtained. Only patient 1 received rituximab therapy, which was given in combination with cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP), leading to clinical remission for his follicular lymphoma, 2 years prior to the development of a histiocytic sarcoma.
Histologic and immunophenotypic features
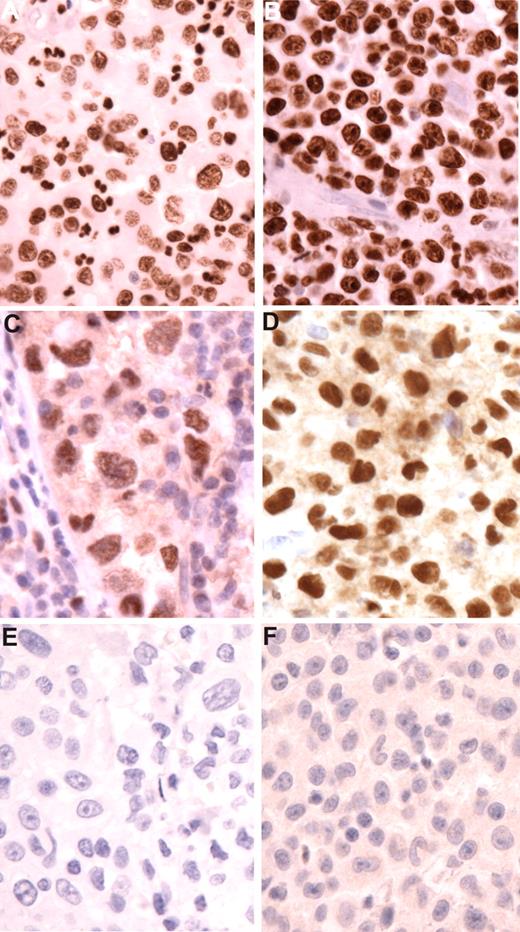
The FL in all cases had typical follicular architecture. Six cases were grade 1, one was grade 2, and one was grade 3a (Table 1). All were positive for Bcl2 protein by immunohistochemistry. The H/DC neoplasms were classified by WHO criteria (Table 2). All tumors exhibited cytologic features of malignancy. Seven were classified as histiocytic sarcomas, 5 of which exhibited evidence of dendritic cell differentiation based on expression of S100 or CD1a expression (2 cases). One tumor was classified as an interdigitating cell sarcoma based on strong S100 positivity and absence of CD163. Immunophenotypic features are summarized in Table 2. None showed expression of CD20, PAX5, or other B-cell markers. However, both CEBPβ and PU.1 were positive in all of the H/DC tumors tested. (Figure 1) Bcl2 expression was seen in 2 of 5 tumors tested (data not shown).
Transcription factor expression in histiocytic sarcomas. (A,B) Staining for CEBP beta in cases 5 and 1. Tumor cells show strong nuclear expression. Note negative blood vessel in panel B. (C,D) Staining for PU.1 in cases 4 and 1. Neoplastic cells (C) show sinusoidal localization. Note negative adjacent lymphocytes. Histiocytic cells have pleomorphic nuclei and prominent nucleoli. (E,F) Staining for PAX5 in cases 8 and 1. No nuclear staining is observed. Follicular lymphomas demonstrated normal expression of PAX5 (not shown). Original magnification for all figures ×400 (hematoxylin counterstain).
Transcription factor expression in histiocytic sarcomas. (A,B) Staining for CEBP beta in cases 5 and 1. Tumor cells show strong nuclear expression. Note negative blood vessel in panel B. (C,D) Staining for PU.1 in cases 4 and 1. Neoplastic cells (C) show sinusoidal localization. Note negative adjacent lymphocytes. Histiocytic cells have pleomorphic nuclei and prominent nucleoli. (E,F) Staining for PAX5 in cases 8 and 1. No nuclear staining is observed. Follicular lymphomas demonstrated normal expression of PAX5 (not shown). Original magnification for all figures ×400 (hematoxylin counterstain).
In the 6 patients in whom the 2 tumors involved different sites, there was no morphologic or immunophenotypic evidence of FL cells intermixed in the H/DC tumors. However, in 1 case (case 3), the original FL was accompanied by an unusual histiocytic reaction without overt malignant features; 3 years later, the patient developed a histiocytic sarcoma at a different lymph node site without histologic or immunophenotypic evidence of FL. In 2 cases, the 2 tumors involved the same site(s). One case showed a H/DC sarcoma, which initially surrounded the neoplastic follicles of the FL (case 4; Figure 2). This tumor subsequently recurred without histologic evidence of FL. In case 7, both tumors were present in the same tongue biopsy, but were architecturally separated (Figure 3). The histiocytic sarcoma was intimately associated with the tongue epithelium, while the FL was noted in adjacent lymphoid tissue. Both tumors also were noted in an ipsilateral cervical lymph node. In the patients who recurred with H/DC after 2 or more years, evidence of FL was not identified clinically at the same time.
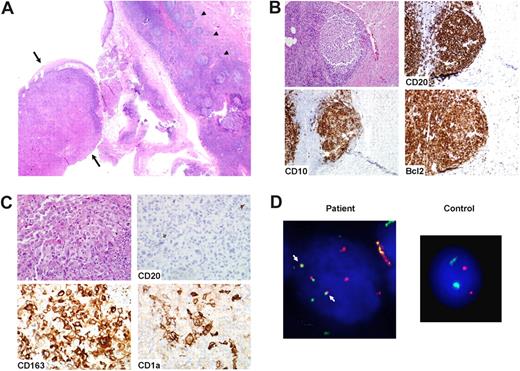
Case 4: synchronous FL and interdigitating cell sarcoma (IDCS). (A) An inguinal lymph node biopsy showed both FL, grade 1, and IDCS. The latter process occupied the majority of the lymph node biopsy, with only focal areas of FL. Magnification ×200. (B) The IDCS showed strong staining for S100 protein. Magnification ×200. (C) Cells of IDCS showed nuclear staining for CEBP beta, negative in FL lymphocytes. Magnification ×400. (D) The IDCS showed splitting of the BCL2 signals (arrow), consistent with presence of the t(14;18), using a breakapart fluorescence in situ hybridization (FISH) probe. A control sample shows no splitting of the BCL2 signals. Magnification ×1250.
Case 4: synchronous FL and interdigitating cell sarcoma (IDCS). (A) An inguinal lymph node biopsy showed both FL, grade 1, and IDCS. The latter process occupied the majority of the lymph node biopsy, with only focal areas of FL. Magnification ×200. (B) The IDCS showed strong staining for S100 protein. Magnification ×200. (C) Cells of IDCS showed nuclear staining for CEBP beta, negative in FL lymphocytes. Magnification ×400. (D) The IDCS showed splitting of the BCL2 signals (arrow), consistent with presence of the t(14;18), using a breakapart fluorescence in situ hybridization (FISH) probe. A control sample shows no splitting of the BCL2 signals. Magnification ×1250.
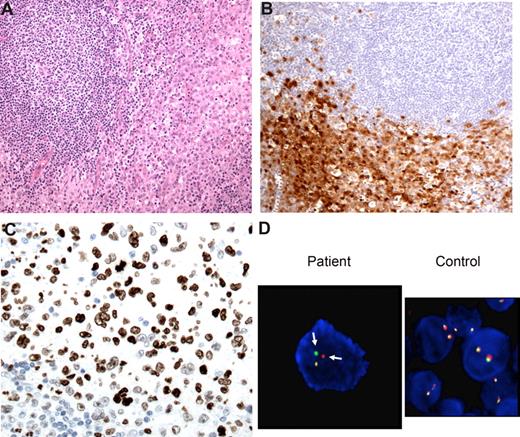
Case 7: synchronous FL and histiocytic sarcoma with dendritic cell differentiation. (A) A mass in the tongue and neck showed both FL, grade 1, and H/DC sarcoma. The FL was associated with lingual lymphoid tissue (arrowheads), while the H/DC neoplasm was associated with the epithelium (arrows). Magnification ×20. (B) The FL was positive for CD20, CD10, and Bcl2. Magnification ×200. (C) The H/DC was positive for CD163 and focally for CD1a, as well as CD68 and S100 (not shown). There was no evidence of infiltrating FL cells, as shown in the CD20 stain. Magnification ×400. (D) The H/DC neoplasm showed fusion of the BCL2 and IGH signals (arrows), consistent with presence of the t(14;18), using dual-fusion FISH (D-FISH) probes. A control sample shows no fusion. Magnification ×1250.
Case 7: synchronous FL and histiocytic sarcoma with dendritic cell differentiation. (A) A mass in the tongue and neck showed both FL, grade 1, and H/DC sarcoma. The FL was associated with lingual lymphoid tissue (arrowheads), while the H/DC neoplasm was associated with the epithelium (arrows). Magnification ×20. (B) The FL was positive for CD20, CD10, and Bcl2. Magnification ×200. (C) The H/DC was positive for CD163 and focally for CD1a, as well as CD68 and S100 (not shown). There was no evidence of infiltrating FL cells, as shown in the CD20 stain. Magnification ×400. (D) The H/DC neoplasm showed fusion of the BCL2 and IGH signals (arrows), consistent with presence of the t(14;18), using dual-fusion FISH (D-FISH) probes. A control sample shows no fusion. Magnification ×1250.
Comparative genetic and cytogenetic studies
Several techniques were used to compare the genetic and cytogenetic characteristics of the tumors (Table 3), including FISH, PCR, and comparative sequence analysis. FISH for evidence of t(14;18) was successfully performed in 6 of 8 cases. Pronounced differences in nuclear morphology between the H/DC sarcoma cells and the FL cells allowed analysis even in the 2 cases where both tumors were present in the same specimen. Furthermore, evaluation of the tissue architecture allowed isolation of nuclei for FISH from portions of the block corresponding to areas of FL or H/DC. In the 6 patients in whom the H/DC sarcoma was not in the same specimen as the FL, the presence of FL was excluded by both morphology and immunohistochemistry.
FISH demonstrated the presence of t(14;18) in 6 H/DC tumors (Figures 2,3), and in the FL cells in patients 1 to 5 (in patient 7, FISH was not interpretable in the FL cells). FISH studies were unsuccessful in cases 6 and 8. In 4 cases in which nuclear counting yielded evaluable data, positive signals were detected in 21% to 44% of FL cells (mean, 29%) and in 12% to 54% of HD/C sarcoma cells (mean, 28%), compared with 4% of cells in reactive lymph node. Six cases demonstrated clonal IGH gene rearrangements, which were the same size in the corresponding follicular lymphomas and H/DC sarcomas. Comparative sequence analysis was successfully performed in 4 of 6 cases; in 2 cases (nos. 3 and 6) the DNA fragment size obtained from the paraffin-embedded tissue was such that adequate sequencing could not be performed. In all successfully analyzed cases, sequencing detected sequence similarity of the rearranged IGH gene (Figure 4). In one case (case 8), sequence identity between the 2 tumors was confirmed by analysis of the BCL2/JH breakpoints. Mutation analysis of the amplified IGH V regions of paired samples from 3 patients showed a mutated phenotype in all cases. In case 2, while the paired samples taken 12 years apart were similar, the H/DC demonstrated more mutations than the earlier FL specimen.
Results of sequencing of IGH gene rearrangements or JH/BCL2 breakpoint regions. All of the sequences are arranged with the specimen sequence at the top, compared with the lower IMGT gene bank sequence. The dashes indicate the sequence is identical to the gene bank sequence, and the nucleotides indicate areas where they differ. Illustrated are cases 1 and 4 (Fr2) and cases 2 and 5 (Fr 3). While the FL and H/DC paired samples from case 2 are similar, the H/DC tumor biopsied 12 years after the FL demonstrates additional mutations. Sequencing of BCL2/JH breakpoint region obtained with MBR probe is illustrated in case 8.
Results of sequencing of IGH gene rearrangements or JH/BCL2 breakpoint regions. All of the sequences are arranged with the specimen sequence at the top, compared with the lower IMGT gene bank sequence. The dashes indicate the sequence is identical to the gene bank sequence, and the nucleotides indicate areas where they differ. Illustrated are cases 1 and 4 (Fr2) and cases 2 and 5 (Fr 3). While the FL and H/DC paired samples from case 2 are similar, the H/DC tumor biopsied 12 years after the FL demonstrates additional mutations. Sequencing of BCL2/JH breakpoint region obtained with MBR probe is illustrated in case 8.
Discussion
We report a series of 8 patients with both FL and H/DC tumors, and provide evidence for the first time of the same underlying molecular oncogenetic alteration in both neoplasms. FISH demonstrated the presence of t(14;18), considered the genetic hallmark of FL, in 6 of 8 H/DC tumors. In the 2 cases in which FISH was not successfully performed, a clonal relationship between the FL and H/DC tumors was indicated by identical BCL2/JH or clonal IGH gene rearrangements or both. Sequence identity by examination of identical IGH or BCL2- MBR breakpoints was confirmed in 5 paired samples, in one patient occurring 12 years apart. Although the H/DC tumors in this series varied in phenotype, the majority demonstrated a mixture of histiocytic and dendritic cell features by morphology and immunohistochemistry. Also of note was the finding that in no case did the H/DC tumor precede the diagnosis of FL.
Recent data suggest there is more plasticity between the B-cell and myeloid lineages than previously thought.24 This plasticity is associated with changes in transcription factor expression and/or activity. Xie et al demonstrated transdifferentiation of mature B cells into macrophages in vitro induced by enforced expressed of the transcription factors C/EBP α and β, and associated with inhibition of the B-cell commitment transcription factor PAX5.25 PU.1 played a synergistic role in inducing macrophage differentiation. Moreover, enforced expression of PAX5 has been shown to prevent myeloid transdifferentiation of B cells.26 Most recently Cobaleda et al showed that conditional deletion of Pax5 causes mature B cells to dedifferentiate into uncommitted precursors in the bone marrow.27 Moreover, they showed that Bcl2-mediated survival cooperates with PAX5 loss in a murine model, leading to the development of “lymphomas” composed of progenitor cells. PAX5-deficient progenitors carrying rearranged Ig genes could differentiate into myeloid cells and T cells, but not into B cells. Evidence for transdifferentiation has been shown in murine models, but has not previously been documented to occur in the clinical setting.
We postulate that changes in transcription factor expression may have led to transdifferentiation from a lymphoid phenotype (either precursor or mature FL) to a H/DC phenotype. For example, epigenetic silencing of the c-fms (csf1r) gene occurs during B-cell development, in part mediated by Pax5. However, c-fms expression can be reactivated by conditional deletion of Pax5, indicating that these steps are reversible.28 All evaluable H/DC tumors were negative for PAX5, despite genotypic evidence of a B-cell derivation. In contrast, CEBPβ and PU.1 were highly expressed in the same tumors; both of these transcription factors play a major role in mediating macrophage and myeloid differentiation.25 Based on the model provided by Cobaleda et al, continued expression of Bcl2 in the absence of PAX5 would be expected to lead to hematolymphoid tumors of non–B-cell lineage, as seen in our report.27 We cannot exclude that these tumors manifested an intermediate step, undergoing dedifferentiation to a stem cell stage, prior to differentiation to a histiocytic lineage. However, no evidence of an immature hematolymphoid neoplasm was identified clinically, either in peripheral lymphoid organs or in bone marrow or peripheral blood.
We note that rituximab does not appear to have played any role in altering the lineage of the malignant clone, since 6 of 8 patients were treated prior to 2000, prior to the rituximab era, and only 1 of the more recent cases received any rituximab therapy prior to being diagnosed with a histiocytic sarcoma. Other therapeutic manipulations are also probably not involved, since 4 of the 8 cases were diagnosed prior to any therapy for the primary follicular lymphoma.
Alternative mechanisms have been proposed to explain the presence of lymphoma-associated genetic aberrations in nonlymphoid cells that are not, strictly speaking, clonally related (ie, do not derive from the same common precursor).29 Spontaneous fusion is one mechanism by which cells of one lineage may contain genetic material from cells of a different lineage.30 Fusion of H/DC cells and FL cells would result in a tetraploid hybrid that contains FL-specific genetic changes. However, no evidence of tetraploidy was evident by FISH in our cases (data not shown). In a similar vein, H/DC tumors or their precursor cells might have the ability to phagocytose genetic material from apoptotic bodies derived from FL cells, so-called horizontal transfer of DNA.31 This mechanism is unlikely in the present series of cases since HD/C tumors contained both t(14;18) and entire, sequence-identical rearranged immunoglobulin genes.
The presence of a common precursor has been suggested as a possible mechanism to explain the finding of a clonal relationship between B-cell and H/DC neoplasms.5,19 Bipotential B-macrophage progenitors have been demonstrated in adult bone marrow from mice.32 Hou et al reported the identification in healthy human bone marrow specimens of a small fraction of primitive CD34+/CD19+/CXCL4− cells termed B-cell/myeloid progenitors (BMPs).33 When placed in B-lymphoid culture conditions, BMPs acquired characteristics of mature B cells, including surface light chain expression. In myeloid culture conditions, however, the BMPs formed myeloid colonies. Our data indicate that the HD/C tumors contained both clonal IGH gene rearrangements and t(14;18), both which are believed to occur as recombinase-mediated events in the bone marrow in the pre-B stage of differentiation (by normal or illegitimate VDJ rearrangement, respectively).34,35 Thus, a postulated common precursor would be differentiated at least to the point of recombination activating gene (RAG) complex activity. Spontaneously occurring H/DC tumors have not shown IGH gene rearrangements.3
Understanding the regulation of differentiation in lymphomas provides insight into rare “secondary neoplasms” and indicates that such tumors represent part of the spectrum of clonal evolution, which is a common event in FL. While our clinical data are limited, the development of a H/DC tumor was associated with progressive disease in most cases.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
This work was supported by the Center for Cancer Research, National Cancer Institute.
National Institutes of Health
Authorship
Contribution: A.L.F. wrote the paper and provided and analyzed data; D.A.A. and S.P. provided and analyzed data; J.S.B., A.M., M.C., M.R., and R.W. provided data; E.S.J. designed the research, wrote the paper, and provided data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Elaine S. Jaffe, Bldg 10, Rm 2B42, 10 Center Dr MSC 1500, National Cancer Institute, Bethesda, MD 20892-1500; e-mail: elainejaffe@nih.gov.