Interferons (IFNs) have been shown to negatively regulate osteoclastogenesis. In a proteomic study to assess protein expression during osteoclastogenesis, we discovered that the expression level of Jak1 was significantly decreased during the early stage of osteoclast differentiation from mouse bone marrow macrophages (BMMs) upon stimulation with receptor activator of nuclear factor κB ligand (RANKL). RANKL induced Jak1 ubiquitination, and a proteasome inhibitor MG132 efficiently blocked the RANKL-induced degradation of Jak1. The expression level of Jak1 correlated with the susceptibility of osteoclast precursors to the negative regulatory effects of IFN-β on osteoclastogenesis, since preosteoclasts (pOCs) in which Jak1 expression is significantly reduced could proceed with osteoclastogenesis in the presence of IFN-β. Forced down-regulation of Jak1 by small interfering RNA (siRNA) resulted in the efficient osteoclast differentiation of BMMs in the presence of inhibitory IFN-β, while overexpression of Jak1 in pOCs elicited IFN-β–dependent inhibition of osteoclastogenesis. Furthermore, we found that the IFN-β–induced inhibition of osteoclastogenesis required STAT3 downstream of Jak1. These data suggest that the regulation of Jak1 expression during osteoclast differentiation might serve as an intrinsic mechanism that determines osteoclast lineage commitment by modulating the negative regulation by IFN-β.
Introduction
Osteoclasts (OCs) are multinucleated cells for bone resorption originated from the monocyte/macrophage lineage of hematopoietic cells.1,2 OC differentiation from its precursor is a multistep process requiring both macrophage colony-stimulating factor (M-CSF) and receptor activator of nuclear factor κB ligand (RANKL).3 The precursor cells first differentiate into mononuclear preosteoclasts (pOCs) and fuse to form multinucleated OCs. M-CSF induces expression of RANKL receptor (RANK) as well as supports survival and proliferation of OC precursors.3 RANKL stimulates differentiation of precursors into OCs by inducing the expression of OC-specific genes, including tartrate-resistant acid phosphatase (TRAP), cathepsin K, and calcitonin receptor.1,2,4 RANKL is also required for the survival and activity of mature OCs.1,2 Although more detailed mechanism is still expected to be unveiled, RANKL signaling includes the recruitment of TNF receptor–associated factors (TRAFs), the activation of transcription factors NF-κB, c-Fos, and NFATc1. It has been also demonstrated that the mitogen-activated protein kinase (MAPK) signaling pathways of ERK, JNK, and p38 as well as the phosphatidylinositol 3-kinase (PI3K)–Akt pathway are involved in RANKL-dependent OC differentiation and function.5,–7
Considering the balance between the activities of osteoblast and OC for bone homeostasis, it is conceivable that the negative regulators of osteoclastogenesis might be equally important as pro-osteoclastogenic factors for maintaining normal bone mass.2 In this context, type I (IFN-α/β) and type II (IFN-γ) IFNs have been shown to strongly inhibit OC differentiation and function both in vivo and in vitro. IFN-γ accelerates proteasomal degradation of TRAF6, thereby abrogating RANKL-dependent signaling.8 On the other hand, it has been shown that IFN-β inhibits the expression of c-Fos.9 Recent findings that the members of suppressors of cytokine signaling (SOCS), which oppose Janus kinase (JAK)/STAT signaling induced by IFN-γ and IFN-β, reversed the inhibitory actions of the IFNs on osteoclastogenesis suggest the involvement of Jak signaling in the IFN-mediated inhibition of osteoclastogenesis.10,–12 However, the role of Jak kinases during osteoclastogenesis remains to be fully understood.
Many of the skeletal diseases involve unregulated osteoclastogenesis leading to excessive bone resorption, which are frequently observed in osteoporosis, rheumatoid arthritis, periodontal diseases, cancer metastasis, and multiple myeloma.2,5 With increasing body of evidence that the IFN/Jak/STAT signaling pathway is involved in osteoclastogenesis, a more detailed understanding of the role for the components of this pathway during OC differentiation may also provide insights into strategies for the intervention of bone destructive diseases.
In this study, using a proteomic approach, we discovered that the expression level of Jak1 is significantly decreased during the early differentiation of bone marrow macrophages (BMMs) into pOCs upon RANKL stimulation via a ubiquitin-mediated proteasomal degradation mechanism. Thus, the inhibitory activity of IFN-β on OC differentiation was limited to the early stage of osteoclastogenesis due to the lack of Jak1 in the later stages of differentiation. These results indicate the existence of a mode of regulation in OC precursors that negates the negative regulatory effect of IFN on osteoclastogenesis by reducing Jak1 upon RANKL stimulation, which may in part explain the excessive osteoclastogenesis in rheumatoid arthritis despite high levels of IFN-β in synovium.
Methods
Animals and reagents
ICR mice at 5 weeks old were obtained from Charles River Laboratories (Wilmington, MA). Wild-type (WT) and STAT1-deficient mice (129S6/SvEv background) aged 5 weeks were from Taconic (Aramachi, Japan). Animal experimental protocols were approved by the committee on the care and use of animals in research at Seoul National University. Recombinant human soluble RANKL and human M-CSF were purchased from PeproTech (Rocky Hill, NJ). Leukocyte acid phosphatase assay kit was from Sigma (St Louis, MO). Lipofectamine 2000 was from Invitrogen Life Technologies (Carlsbad, CA). Antibodies (Abs) against ubiquitin (FL-76) and Jak1 (HR-785) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti–β-actin (AC-74) was from Sigma. Phosphospecific Abs for STAT1 (Tyr701), STAT3 (Tyr705), STAT3 (Ser727), and STAT5 (Tyr694) were from Cell Signaling Technology (Beverly, MA). Anti-STAT1, anti-STAT3, and anti-STAT5 Abs were also purchased from Cell Signaling Technology. Anti-Jak1(73) and anti–phospho-Jak1 (Tyr1022/Tyr1023) Abs were from BD Biosciences Pharmingen (San Diego, CA) and Abcam (Cambridge, United Kingdom), respectively. Proteasome inhibitor MG132 was obtained from Calbiochem (La Jolla, CA). All other chemicals were purchased from Sigma.
Cell-surface biotinylation and purification of biotinylated proteins
BMMs (4 × 105 cells/well in 6-well plates) were cultured with 30 ng/mL M-CSF alone or M-CSF plus 200 ng/mL RANKL for 36 hours. A total of 2 × 107 cells were surface biotinylated using the Pinpoint cell-surface protein isolation kit (Pierce, Rockford, IL) following the manufacturer's instructions, lysed, and subjected to an immobilized NeutrAvidin gel column (Pierce). The biotinylated proteins were eluted with SDS-PAGE sample buffer.
Gel electrophoresis and protein digestion
Biotinylated proteins were loaded onto a 16 × 16-cm 8% to 16% gradient SDS-PAGE gel (Bio-Rad, Hercules, CA). After separation, gel was stained with SYPRO Ruby dye (Invitrogen) and visualized on a UV illuminator. After cutting protein bands showing differential expression between macrophages and pOCs, proteins were in-gel digested and eluted out of the gel for subsequent liquid chromatography/tandem mass spectrometry (LC/MS/MS) analyses.
LC/MS/MS experiments and data analyses
Digested protein bands were separately analyzed by nano-LC/MS/MS equipped with an LTQ-FT mass spectrometer (Thermo Electron, San Jose, CA) and an Agilent 1100 Series NanoLC pump (Agilent Technologies, Palo Alto, CA). Data from the mass spectrometer were analyzed by using the Bioworks 3.2 software (Thermo Electron) using a database constructed by combining the mouse International Protein Index database (ipi.MOUSE. v.3.10, ftp://ftp.ebi.ac.uk, downloaded 2005.9.24)13 and common contaminant proteins such as trypsin and keratin. The search results were subsequently evaluated by PeptideProphet14 and a home-built software.15 A detailed description of LC/MS/MS experiments is included in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Immunoprecipitation and immunoblotting
Immunoprecipitation and Western blotting were performed as described previously.16 For experiments requiring multiple Western blots using several phosphospecific antibodies, lysates were divided into same amounts and subjected to SDS-PAGE and immunoblotting individually. Each membrane was stripped no more than 3 times. Although actin blots for loading control were done for each membrane, only 1 representative blot was shown. For the quantification of band intensities, developed films were scanned (PowerLook1100; Umax, Taipei, Taiwan) and analyzed by TotalLab gel image analysis software (Nonlinear Dynamics, Newcastle, United Kingdom). The Jak1 protein expression was normalized using the actin band density of same lane to compensate for the protein loading.
Quantitative PCR analysis
Total RNAs were isolated using the RNeasy kit (Qiagen, Hilden, Germany), and 1 μg RNA was reverse transcribed with the AccuPower kit (Bioneer, Daejon, Korea) according to the manufacturer's instructions. For quantitative real-time polymerase chain reaction (PCR) analyses, 4 μg cDNA was amplified with SYBR green PCR master mix (Applied Biosystems, Warrington, United Kingdom) in a MicroAmp optical tube using an AB 7500 instrument (Applied Biosystems) for 30 cycles of 15-second denaturation at 95°C and 1-minute amplification at 60°C. The PCR primer sequences were as follows: Jak1, 5′-GTCCCTGAAGCCTGAGAGTG-3′ (forward) and 5′-CTTGATACCATTGCCTCCGT-3′ (reverse); and HPRT, 5′-CCTAAGATGAGCGCAAGTTG-3′ (forward) and 5′-CCACAGGGACTAGAACACCTGCTAA-3′ (reverse). Data were analyzed by 7500 system sequence detection software (version 1.3; Applied Biosystems), and the mRNA expression level of Jak1 was normalized using the level of HPRT to be presented as relative expression.
OC differentiation
Mouse bone marrow cells, isolated by flushing the marrow space of femora and tibiae, were incubated overnight on culture dishes in α–modified Eagle medium (MEM) supplemented with 10% fetal bovine serum (FBS). After discarding adherent cells, cells were further incubated with 20 ng/mL M-CSF on Petri dishes. BMMs became adherent after a 3-day culture and were used as OC precursor cells.17 These cells were differentiated into OCs by incubating 3 × 104 cells/well in 48-well plates with 30 ng/mL M-CSF and 100 ng/mL RANKL. At the end of incubation, cells were stained for TRAP activity using a leukocyte acid phosphatase assay kit (Sigma). Upon observation under light microscope (100× magnification with an Olympus BX51 microscope [Olympus, Center Valley, PA], with 10×/0.30 Uplan FL N objective equipped with a Spot Insight digital camera system [Diagnostics Instruments, Sterling Heights, MI]), cells positive for TRAP activity with 3 or more nuclei were counted as multinuclear OCs, while TRAP+ cells with less than 3 nuclei were regarded as pOCs. Although it usually took 2 days in our experimental conditions for the cells to differentiate into pOCs and 3 days to differentiate into OCs, the exact time required for differentiation slightly varied between experiments (eg, the osteoclastogenesis was slower in virus-infected cells).
Plasmids
pMX-IRES-GFP mouse Jak1WT and Jak1V658F18 were previously described, and human Tyk2WT was cloned into pREX-IRES-CD4 vector. For Jak1 knock-down experiments, oligonucleotides for siRNA were generated by targeting a 21-base sequence starting from the number 701 of mouse Jak1 (GeneBank accession number NM_146145). The resulting oligonucleotide sequences of 5′-GATCCCGTTATTCGCATCCTGGTAAGAAT-TGATATCCGTTCTTACCAGGATGCGAATAATTTTTTCCAAA-3′ and 5′-AGCTTTTGGAAAAAATTATTCGCATCCTGGTAAGAACGGATA-TCAATTCTTACCAGGATGCGAATAACGG-3′ were annealed and ligated into pSuper-retro vector (Oligoengine, Seattle, WA) using BamHI and HindIII sites. For STAT3 knock-down experiments, oligonucleotides for siRNA were generated by targeting a 21-base sequence starting from the number 510 of mouse STAT3 (GeneBank accession number NM_213659). The resulting oligonucleotide sequences were 5′-GATCCCGTTGTAGTTGAAATCAAAGTCGTTGATATCCGGGACTTT-3′ and 5′-AGCTTTTGGAAAAAATTGTAGTTGAAATCAAAATCAACGACTTTGATTTCAACTACAACGG-3′.
Retroviral gene transfer
Retroviruses were packaged by transfecting pMX-IRES-GFP-Jak1WT, -Jak1V658F, pSuper-retro-Jak1siRNA, and pSuper-retro-STAT3siRNA plasmids into Plat-E cells as described previously.16 Retroviral infection was performed by incubating cells with virus-containing media with the addition of 10 μg/mL polybrene. After overnight incubation, cells were either induced for OC differentiation or used for signaling experiments.16 For the siRNA experiments, successful down-regulation of proteins required 24 to 48 hours of incubation after viral infection depending on the target.
STAT3 DNA binding assay
The DNA binding activity of STAT3 was determined in nuclear extracts (200 μg protein) from IFN-β–stimulated cells using the Duoset IC human/mouse active STAT3 activity assay kit (R&D Systems, Minneapolis, MN) following the manufacturer's protocol.
Statistical analysis
The Student t test was used to determine the significance of differences between results.
Results
Jak1 expression is decreased during BMM differentiation into pOCs
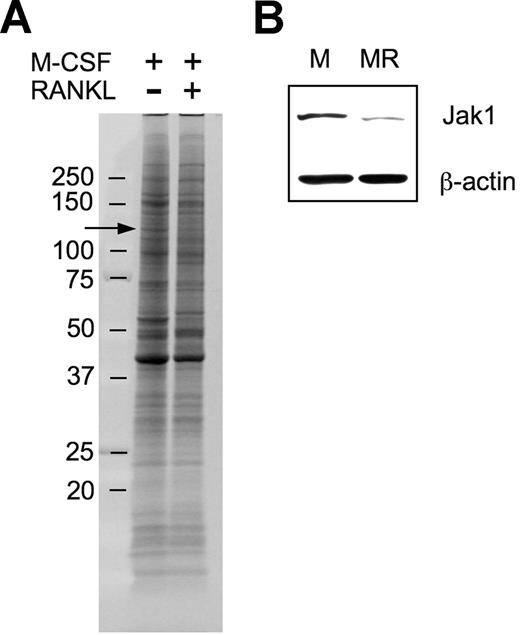
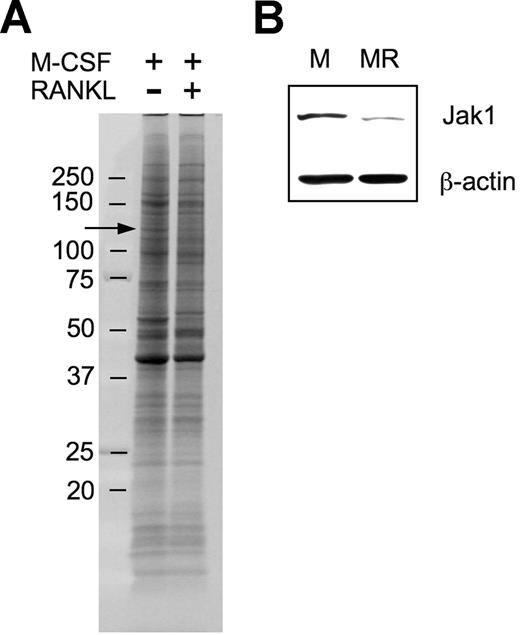
In an attempt to explore molecular mechanisms underlying macrophage differentiation into OCs, we used a proteomic approach to ascertain differentially expressed proteins during the early stage of OC differentiation under the assumption that those differentially regulated proteins may have a role in OC differentiation. After culturing BMMs in the presence of 30 ng/mL M-CSF alone or in combination with 200 ng/mL RANKL for 36 hours, surface proteins were biotinylated, subjected to SDS-PAGE, and visualized (Figure 1A). There were 4 protein bands of which the intensity was distinctively increased, and 8 protein bands of which the intensity was decreased. Those protein bands were cut off of the gel and were analyzed by LC/MS/MS experiments. Positively identified proteins from the approximately 130-kDa band showing decreased expression upon RANKL treatment (Figure 1A arrow) included mouse Jak1 tyrosine kinase. A total of 3 unique Jak1 peptide sequences were identified (Table 1). To confirm the decrease of Jak1 protein by RANKL treatment, total-cell lysates of BMMs treated with M-CSF alone or M-CSF plus RANKL for 36 hours were subjected to Western blot analysis. Figure 1B confirms that the expression of Jak1 is significantly decreased by treatment with RANKL for 36 hours.
Proteomic identification of Jak1 as a protein whose expression was decreased during early osteoclast differentiation. (A) BMMs were incubated with 30 ng/mL M-CSF in combination with 200 ng/mL RANKL for 36 hours. At the end of incubation, cell-surface proteins were biotinylated and isolated. The biotinylated proteins were separated on an 8% to 16% SDS gel and visualized by SYPRO Ruby staining. Protein bands showing differential expression in RANKL-stimulated cells were subjected to LC/MS/MS analysis for protein identification. Arrow indicates the protein band in which Jak1 was included. (B) The decreased expression of Jak1 in RANKL-stimulated cells was confirmed by Western blotting. BMMs were cultured with 30 ng/mL M-CSF alone (M) or M-CSF plus 200 ng/mL RANKL (MR) for 36 hours, and total-cell lysates were subjected to SDS-PAGE followed by immunoblotting using anti-Jak1 antibody. Antiactin blot was shown to ensure same loading between the 2 lanes.
Proteomic identification of Jak1 as a protein whose expression was decreased during early osteoclast differentiation. (A) BMMs were incubated with 30 ng/mL M-CSF in combination with 200 ng/mL RANKL for 36 hours. At the end of incubation, cell-surface proteins were biotinylated and isolated. The biotinylated proteins were separated on an 8% to 16% SDS gel and visualized by SYPRO Ruby staining. Protein bands showing differential expression in RANKL-stimulated cells were subjected to LC/MS/MS analysis for protein identification. Arrow indicates the protein band in which Jak1 was included. (B) The decreased expression of Jak1 in RANKL-stimulated cells was confirmed by Western blotting. BMMs were cultured with 30 ng/mL M-CSF alone (M) or M-CSF plus 200 ng/mL RANKL (MR) for 36 hours, and total-cell lysates were subjected to SDS-PAGE followed by immunoblotting using anti-Jak1 antibody. Antiactin blot was shown to ensure same loading between the 2 lanes.
Jak1 protein but not the mRNA expression level is affected by RANKL treatment
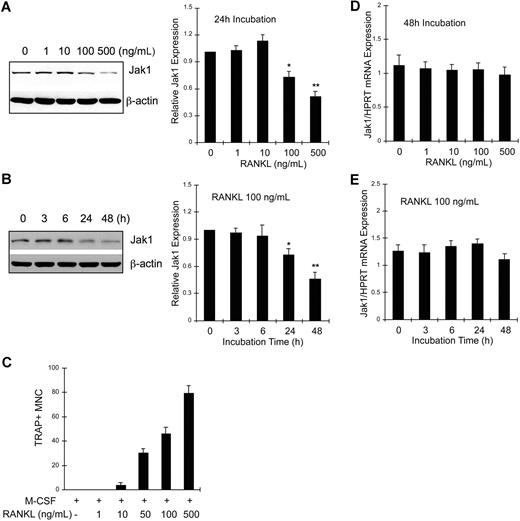
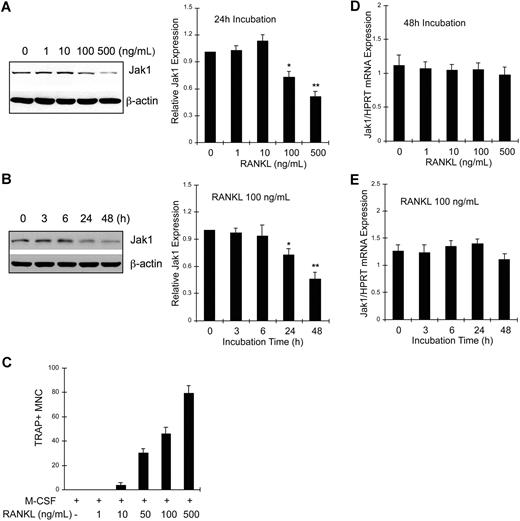
BMMs were treated with M-CSF and 0 to 500 ng/mL RANKL for 24 hours, and cell lysates were analyzed for Jak1 expression by Western blotting (Figure 2A). Jak1 expression was not altered by incubation with RANKL up to 10 ng/mL. However, 100 ng/mL or greater concentrations of RANKL significantly reduced Jak1 expression (Figure 2A left panel). Densitometry results from 3 independent experiments showed that Jak1 expression was reduced to 72.3% plus or minus 6.1% and 50.7% plus or minus 5.2% of control after 100 and 500 ng/mL RANKL treatment, respectively (Figure 2A right panel). At 48 hours after stimulation, more profound reduction of Jak1 expression was observed in cells treated with 100 or 500 ng/mL RANKL, but not in BMMs treated with 10 ng/mL or less RANKL (data not shown). We next investigated the time-course of Jak1 expression levels upon RANKL stimulation. Results in Figure 2B show that the protein expression of Jak1 started to significantly decrease at 24 hours after RANKL stimulation (Figure 2B left panel). When Jak1 band densities were analyzed by densitometry from 3 independent experiments (Figure 2B right panel), the mean Jak1 density at 24 hours and 48 hours after RANKL stimulation was 72.6% plus or minus 6.7% and 45.6% plus or minus 7.4% of control (0 hours), respectively. A similar pattern of reduction in Jak1 expression level was also observed during the RANKL-induced osteoclastogenesis in RAW264.7 cells (Figure S1). However, Jak2 and Jak3 proteins were not down-regulated by RANKL in BMMs (Figure S2). The reduced expression of Jak1 and the formation of TRAP+ multinuclear OCs seemed to correlate. Figure 2C shows that the formation of OCs was minimal after the treatment of BMMs with 10 ng/mL RANKL for 72 hours. However, 100 or 500 ng/mL RANKL, which reduced Jak1 expression significantly, induced efficient formation of OCs. Similarly, the formation of mononuclear pOCs measured at 48 hours after stimulation also correlated with the RANKL-dependent reduction of Jak1 expression (data not shown). Next, we tested if Jak1 expression was regulated at the transcription level. BMMs were treated with 30 ng/mL M-CSF and 0 to 500 ng/mL RANKL for 48 hours, and total RNAs were isolated. Quantitative real-time PCR analysis resulted in no significant difference in Jak1 mRNA levels during a 48-hour treatment of cells with 0 to 500 ng/mL RANKL (Figure 2D) and during the course of 48 hours of incubation with 100 ng/mL RANKL (Figure 2E). Similarly, reverse transcription (RT)–PCR analysis showed no significant change in the Jak1 mRNA level (Figure S3).
Jak1 protein but not mRNA expression level was decreased during RANKL-induced osteoclast differentiation. (A) BMMs were cultured with 30 ng/mL M-CSF in the presence of indicated concentrations of RANKL for 24 hours. Lysates were subjected to Western blotting, and Jak1 expression was determined by enhanced chemiluminescence. Jak1 band densities were quantified as described in “Immunoprecipitation and immunoblotting” (right panel). (B) BMMs were cultured with 30 ng/mL M-CSF in the presence of 100 ng/mL RANKL for indicated times, and total-cell lysates were subjected to Western blotting using anti-Jak1 antibody (left panel). Jak1 expression was quantified as in panel A (right). (C) BMMs were cultured in the presence of 30 ng/mL M-CSF and 1 to 500 ng/mL RANKL for 3 days, and the formation of TRAP+ multinuclear OCs was observed. Cells were counted in 3 random fields under the microscope and the average number plus or minus SD was shown. Similar results were obtained from 2 independent experiments. (D) BMMs were cultured for 48 hours in the presence of 30 ng/mL M-CSF and 0 to 500 ng/mL RANKL, and total RNAs were isolated, reverse-transcribed, and subjected to quantitative real-time PCR analysis for Jak1. Jak1 expression levels were normalized by the levels of HPRT in each sample. (E) RNA samples from the cells cultured as in panel B were subjected to quantitative real-time PCR analysis. Western blot images are representative of at least 3 experiments with similar results. Values in graphs are expressed as means (± SD) from 3 (A,D) or 4 (B,E) experiments. *P < .05; **P < .01 versus control values.
Jak1 protein but not mRNA expression level was decreased during RANKL-induced osteoclast differentiation. (A) BMMs were cultured with 30 ng/mL M-CSF in the presence of indicated concentrations of RANKL for 24 hours. Lysates were subjected to Western blotting, and Jak1 expression was determined by enhanced chemiluminescence. Jak1 band densities were quantified as described in “Immunoprecipitation and immunoblotting” (right panel). (B) BMMs were cultured with 30 ng/mL M-CSF in the presence of 100 ng/mL RANKL for indicated times, and total-cell lysates were subjected to Western blotting using anti-Jak1 antibody (left panel). Jak1 expression was quantified as in panel A (right). (C) BMMs were cultured in the presence of 30 ng/mL M-CSF and 1 to 500 ng/mL RANKL for 3 days, and the formation of TRAP+ multinuclear OCs was observed. Cells were counted in 3 random fields under the microscope and the average number plus or minus SD was shown. Similar results were obtained from 2 independent experiments. (D) BMMs were cultured for 48 hours in the presence of 30 ng/mL M-CSF and 0 to 500 ng/mL RANKL, and total RNAs were isolated, reverse-transcribed, and subjected to quantitative real-time PCR analysis for Jak1. Jak1 expression levels were normalized by the levels of HPRT in each sample. (E) RNA samples from the cells cultured as in panel B were subjected to quantitative real-time PCR analysis. Western blot images are representative of at least 3 experiments with similar results. Values in graphs are expressed as means (± SD) from 3 (A,D) or 4 (B,E) experiments. *P < .05; **P < .01 versus control values.
RANKL induces ubiquitination of Jak1
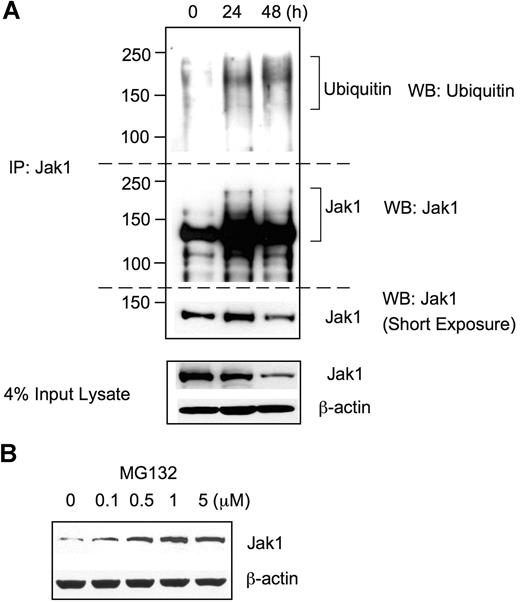
Since Jak1 protein expression level was substantially reduced after RANKL stimulation without any significant change in the mRNA level, we hypothesized that Jak1 protein expression might be regulated by ubiquitin-mediated proteasomal degradation. To test this possibility, BMMs were grown in the presence of 30 ng/mL M-CSF plus 100 ng/mL RANKL, and cell lysates were subjected to immunoprecipitation using Jak1-specific Ab followed by Western blotting with antiubiquitin Ab (Figure 3A). Upon stimulation with RANKL for 24 hours and 48 hours, extensively ubiquitinated protein bands having 130 kDa or higher molecular weights were immunoprecipitated with anti-Jak1 Ab, whereas Jak1 immunoprecipitates from control lysates (0 hours) showed only slight levels of basal ubiquitination. When the same membrane was stripped and reprobed with anti-Jak1 Ab, protein bands having higher molecular weight than that of the unmodified Jak1 (approximately 130 kDa), corresponding to the ubiquitinated Jak1, appeared in 24- and 48-hour stimulation samples, which were absent in control. To further confirm the Jak1 degradation via a ubiquitin-mediated proteasome pathway, BMMs were stimulated with RANKL for 48 hours, and a proteasome inhibitor MG132 was added during the final 4 hours of incubation (Figure 3B). Western blotting revealed that the incubation of BMMs with the increasing concentrations of MG132 resulted in the increased level of Jak1 in cell lysates, suggesting that the proteasome-mediated degradation of Jak1 was blocked by MG132. Taken together, these results suggested that the decreased level of Jak1 following RANKL stimulation resulted from the RANKL-induced ubiquitination of Jak1 and subsequent proteasome-mediated degradation.
Jak1 was ubiquitinated upon RANKL stimulation. (A) BMMs were incubated with 30 ng/mL M-CSF and 100 ng/mL RANKL for the indicated times, and total-cell lysates were subjected to immunoprecipitation using anti-Jak1 antibody. Immunoprecipitates were separated by SDS-PAGE and immunoblotted using antiubiquitin antibody. The blot was stripped and reprobed for Jak1. Jak1 and actin expression was also examined from a small portion of input lysates. (B) BMMs were cultured with M-CSF (30 ng/mL) and RANKL (100 ng/mL) for 48 hours. MG132 was added for the final 4 hours of incubation. Total-cell lysates were subjected to Western blotting for the examination of Jak1 expression. (A,B) Blots are representative of at least 3 experiments with similar results.
Jak1 was ubiquitinated upon RANKL stimulation. (A) BMMs were incubated with 30 ng/mL M-CSF and 100 ng/mL RANKL for the indicated times, and total-cell lysates were subjected to immunoprecipitation using anti-Jak1 antibody. Immunoprecipitates were separated by SDS-PAGE and immunoblotted using antiubiquitin antibody. The blot was stripped and reprobed for Jak1. Jak1 and actin expression was also examined from a small portion of input lysates. (B) BMMs were cultured with M-CSF (30 ng/mL) and RANKL (100 ng/mL) for 48 hours. MG132 was added for the final 4 hours of incubation. Total-cell lysates were subjected to Western blotting for the examination of Jak1 expression. (A,B) Blots are representative of at least 3 experiments with similar results.
Jak1 expression on OC precursors correlates with the IFN-β susceptibility during OC differentiation
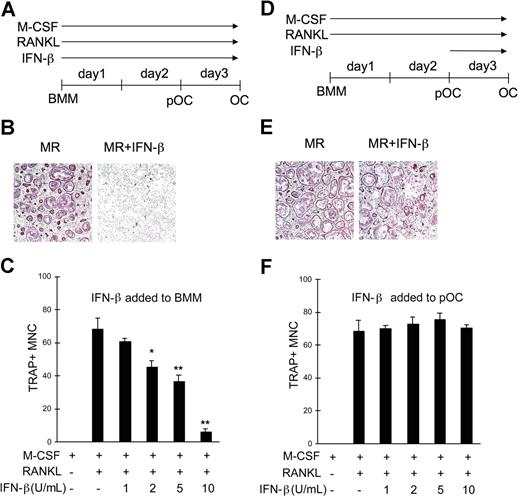
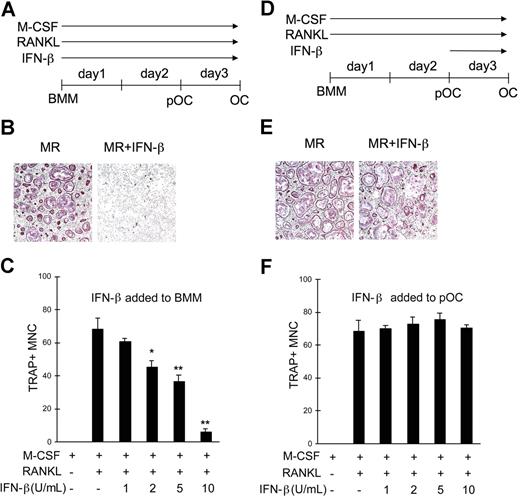
We next examined the physiologic significance of decreased Jak1 expression in OC precursor cells during RANKL-induced osteoclastogenesis. Recently, it has been demonstrated that IFN-β exerts negative effects on RANKL-induced OC formation,9 which was confirmed in our study by culturing BMMs with RANKL in the presence of IFN-β (Figure 4A). The presence of IFN-β significantly reduced the formation of OCs as determined by the dramatically reduced formation of TRAP+ large multinuclear cells after 3-day culture with RANKL (Figure 4B). When an increasing concentration of IFN-β was used to inhibit OC formation, as little as 2 U/mL IFN-β was sufficient to significantly inhibit osteoclastogenesis induced by 100 ng/mL RANKL (Figure 4C). The number of OCs formed under the presence of 10 U/mL IFN-β for 3 days was less than 10% of those in the absence of IFN-β. Since the lack of Jak1 was known to be responsible for the unresponsiveness of cells to IFNs,19 we postulated that pOCs, in which Jak1 expression is significantly reduced (Figure 2), might be resistant to the inhibition of osteoclastogenesis by IFN-β. To test this, BMMs were first cultured with RANKL for 2 days, treated with 10 U/mL IFN-β, and stained for TRAP activity at day 3 (Figure 4D). As predicted, IFN-β had no negative effect on OC differentiation when added at 48 hours after RANKL stimulation (Figure 4E). There was no significant difference in the OC numbers with the tested concentrations of IFN-β (1-10 U/mL; Figure 4F). The expression of type I IFN receptor was not affected by RANKL up to 500 ng/mL at 48 hours after stimulation (data not shown). To further dissect the time point when BMMs cultured with RANKL become resistant to the negative effect of IFN-β, 10 U/mL IFN-β was added at different times after the addition of RANKL (Figure S4). When IFN-β was added at the start of culture, there was significant reduction in the formation of OCs. However, when IFN-β was added at 24 hours or 48 hours after RANKL addition, no significant decrease in the OC number was observed. Furthermore, presence of IFN-β only for the first 24 hours after RANKL stimulation was enough to inhibit osteoclastogenesis. Thus, it seemed that the inhibitory action of IFN-β on osteoclastogenesis was limited to the early stage of OC differentiation, when Jak1 expression is still intact.
pOCs were not susceptible to the IFN-β–mediated inhibition of osteoclastogenesis. (A) BMMs were cultured with M-CSF (30 ng/mL) and RANKL (100 ng/mL) for 3 days. IFN-β was added at the start of culture for experiments described in panels B and C. (B) At the end of culture, cells were fixed and stained for TRAP activity. Osteoclasts were identified as TRAP+ (purple) multinuclear cells. MR represents M-CSF plus RANKL. Results are representative of at least 3 experiments using 10 U/mL IFN-β. (C) OC numbers were counted after a 3-day culture of BMMs with M-CSF and RANKL in combination with IFN-β. Cells were counted in 3 random fields. Results are presented as means plus or minus SD and are representative of at least 3 experiments with similar results. Asterisks represent statistical difference between the values obtained from cells cultured with or without IFN-β in the presence of M-CSF and RANKL (*P < .05; **P < .01). (D) BMMs were first differentiated into pOCs by incubating cells with M-CSF and RANKL for 2 days, and IFN-β was added at the end of day 2 for experiments in panels E and F. (E) After a 3-day culture, cells were stained for TRAP activity. (F) OC numbers were counted after a 3-day culture of BMMs with M-CSF and RANKL. Varying concentrations of IFN-β were added for the final 24 hours. Data are presented as in panel C.
pOCs were not susceptible to the IFN-β–mediated inhibition of osteoclastogenesis. (A) BMMs were cultured with M-CSF (30 ng/mL) and RANKL (100 ng/mL) for 3 days. IFN-β was added at the start of culture for experiments described in panels B and C. (B) At the end of culture, cells were fixed and stained for TRAP activity. Osteoclasts were identified as TRAP+ (purple) multinuclear cells. MR represents M-CSF plus RANKL. Results are representative of at least 3 experiments using 10 U/mL IFN-β. (C) OC numbers were counted after a 3-day culture of BMMs with M-CSF and RANKL in combination with IFN-β. Cells were counted in 3 random fields. Results are presented as means plus or minus SD and are representative of at least 3 experiments with similar results. Asterisks represent statistical difference between the values obtained from cells cultured with or without IFN-β in the presence of M-CSF and RANKL (*P < .05; **P < .01). (D) BMMs were first differentiated into pOCs by incubating cells with M-CSF and RANKL for 2 days, and IFN-β was added at the end of day 2 for experiments in panels E and F. (E) After a 3-day culture, cells were stained for TRAP activity. (F) OC numbers were counted after a 3-day culture of BMMs with M-CSF and RANKL. Varying concentrations of IFN-β were added for the final 24 hours. Data are presented as in panel C.
Jak1 overexpression in pOCs elicits IFN-β–mediated inhibition of osteoclastogenesis
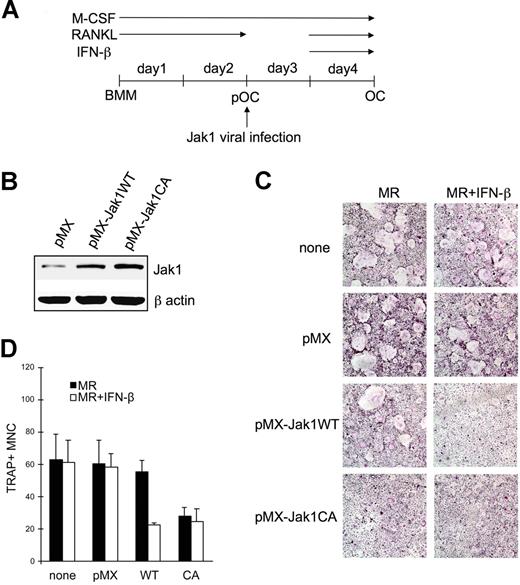
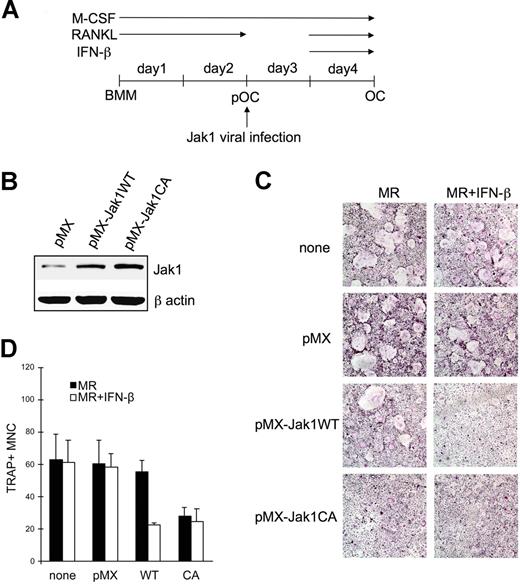
Next, we hypothesized that the forced expression of Jak1 into pOCs would induce these cells to be sensitive to the inhibitory effect of IFN-β. To test this, BMMs were first cultured in the presence of RANKL for 2 days, and cells were infected with retroviruses harboring WT Jak1 or constitutively active Jak1 (Figure 5A). Figure 5B shows that the infection of cells with viruses harboring WT Jak1 (pMX-Jak1WT) or constitutively active Jak1 (pMX-Jak1CA; Jak1V658F)18 successfully increased Jak1 protein expression. After viral infection, pOCs were further differentiated into OCs using RANKL in the presence of IFN-β. Uninfected and control virus–infected pOCs showed no difference in the numbers of differentiated OCs in the presence or absence of IFN-β (Figure 5C). However, the overexpression of WT Jak1 in pOCs sensitized pOCs to the antiosteoclastogenic effect of IFN-β, leading to the significantly reduced formation of OCs in the presence of IFN-β. An approximately 60% reduction in the numbers of TRAP+ OCs was observed (Figure 5D). Finally, overexpression of constitutively active Jak1 inhibited OC differentiation from pOCs independent of IFN treatment.
Jak1 overexpression sensitized pOCs to the IFN-β-mediated inhibition of osteoclastogenesis. (A) BMMs were first differentiated into pOCs by culturing with M-CSF (30 ng/mL) and RANKL (100 ng/mL). After the infection with viruses harboring empty vector (pMX), WT Jak1 (pMX-Jak1WT), or constitutively active Jak1 (pMX-Jak1CA), cells were further differentiated into OCs with RANKL in the presence or absence of IFN-β (10 U/mL). (B) At 1 day after Jak1 viral infection, total-cell lysates were investigated for Jak1 expression by Western blotting. (C) At the end of culture, cells were stained for TRAP activity. MR represents M-CSF plus RANKL. Results are representative of at least 3 experiments with similar results. (D) OC numbers in panel C were counted. Data are means (± SD) from 3 random fields in representative experiment.
Jak1 overexpression sensitized pOCs to the IFN-β-mediated inhibition of osteoclastogenesis. (A) BMMs were first differentiated into pOCs by culturing with M-CSF (30 ng/mL) and RANKL (100 ng/mL). After the infection with viruses harboring empty vector (pMX), WT Jak1 (pMX-Jak1WT), or constitutively active Jak1 (pMX-Jak1CA), cells were further differentiated into OCs with RANKL in the presence or absence of IFN-β (10 U/mL). (B) At 1 day after Jak1 viral infection, total-cell lysates were investigated for Jak1 expression by Western blotting. (C) At the end of culture, cells were stained for TRAP activity. MR represents M-CSF plus RANKL. Results are representative of at least 3 experiments with similar results. (D) OC numbers in panel C were counted. Data are means (± SD) from 3 random fields in representative experiment.
Since the addition of the proteasome inhibitor MG132 significantly increased Jak1 expression (Figure 3B), the effect of IFN-β on osteoclastogenesis in the presence of MG132 was tested. BMMs were first cultured for 2 days with 100 ng/mL RANKL, and 1 μM MG132 was added during the final 4 hours of incubation (Figure 5A). Although the addition of MG132 alone slightly decreased the OC numbers, treatment of pOCs with MG132 resensitized cells to the inhibitory effect of IFN-β (Figure S5B). While IFN-β at a 10 U/mL concentration was ineffective to inhibit OC formation when added at 48 hours after RANKL stimulation, as little as 1 U/mL IFN-β significantly reduced the numbers of OCs formed from the precursors pretreated with MG132. Treatment of cells with MG132 in our experimental scheme showed no cytotoxicity as tested by a tetrazolium-based assay (data not shown).
Jak1 knock-down protects BMMs from the IFN-β–mediated inhibition of osteoclastogenesis
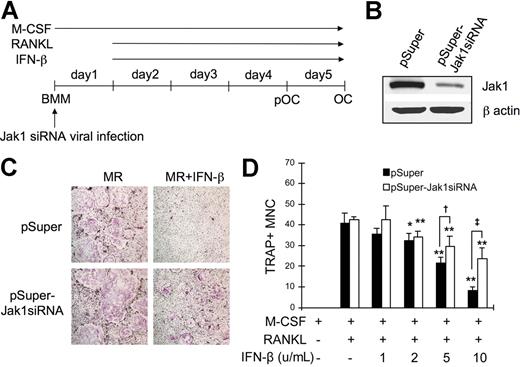
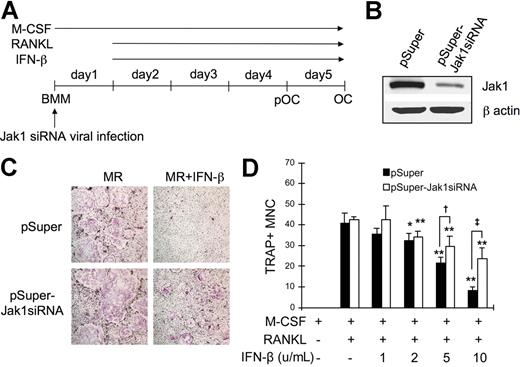
Since the overexpression of Jak1 in pOCs conferred IFN-β sensitivity during RANKL-induced osteoclastogenesis, it was conversely expected that the knock-down of Jak1 in BMMs would protect these OC precursors from the IFN-β–mediated inhibition of OC differentiation. To test this, the expression of Jak1 in BMMs were down-regulated by introducing Jak1-specific siRNA (Jak1siRNA) via retroviral infection (Figure 6A). Compared with the cells infected with control viruses, cells cultured with viruses harboring Jak1siRNA showed significantly reduced Jak1 expression (Figure 6B). These BMMs were then differentiated into OCs using RANKL, and the inhibitory effect of IFN-β on the osteoclastogenesis was examined. As shown in Figure 6C, introduction of siRNA specific for Jak1 protected BMMs from the inhibitory effect of IFN-β on the RANKL-induced OC differentiation, while cells infected with control viruses failed to differentiate into OCs in the presence of IFN-β. When the increasing concentrations of IFN-β were used, 2 U/mL IFN-β was sufficient to significantly inhibit osteoclastogenesis in BMMs infected with control virus (Figure 6D). Similarly, IFN-β at 2 U/mL or greater concentrations significantly reduced the formation of TRAP+ OCs upon RANKL stimulation in Jak1siRNA virus–infected BMMs. However, consistently higher number of OCs was observed in Jak1 knocked-down cells after IFN-β addition when compared with control virus-infected cells. Specifically, 1.4-fold and 2.8-fold more OCs were observed in Jak1 knocked-down cells than in control virus-infected cells in the presence of 5 U/mL and 10 U/mL IFN-β, respectively.
Jak1 knock-down protected BMMs from the IFN-β-mediated inhibition of osteoclastogenesis. (A) BMMs were first infected with viruses harboring empty vector (pSuper) or Jak1 siRNA construct (pSuper-Jak1siRNA). After 24 hours, cells were stimulated with RANKL (100 ng/mL) to induce OC differentiation in the presence or absence of IFN-β. (B) At 1 day after Jak1 siRNA viral infection, total-cell lysates were examined for Jak1 expression by Western blotting. (C) At the end of culture, cells were stained for TRAP activity. MR represents M-CSF plus RANKL. IFN-β was used at 10 U/mL. Results are representative of at least 3 experiments with similar results. (D) OC numbers in experiments using varying concentrations of IFN-β were counted. Data are means (± SD) from 3 random fields in representative experiment. Asterisks represent statistical difference between the values obtained from cells cultured in the presence or absence of IFN-β with M-CSF and RANKL (*P < .05; **P < .01). Daggers represent significant difference between pSuper- and pSuper-Jak1siRNA–infected cells at the same IFN-β concentration (†P < .05; ‡P < .01).
Jak1 knock-down protected BMMs from the IFN-β-mediated inhibition of osteoclastogenesis. (A) BMMs were first infected with viruses harboring empty vector (pSuper) or Jak1 siRNA construct (pSuper-Jak1siRNA). After 24 hours, cells were stimulated with RANKL (100 ng/mL) to induce OC differentiation in the presence or absence of IFN-β. (B) At 1 day after Jak1 siRNA viral infection, total-cell lysates were examined for Jak1 expression by Western blotting. (C) At the end of culture, cells were stained for TRAP activity. MR represents M-CSF plus RANKL. IFN-β was used at 10 U/mL. Results are representative of at least 3 experiments with similar results. (D) OC numbers in experiments using varying concentrations of IFN-β were counted. Data are means (± SD) from 3 random fields in representative experiment. Asterisks represent statistical difference between the values obtained from cells cultured in the presence or absence of IFN-β with M-CSF and RANKL (*P < .05; **P < .01). Daggers represent significant difference between pSuper- and pSuper-Jak1siRNA–infected cells at the same IFN-β concentration (†P < .05; ‡P < .01).
The Jak1-dependent negative regulation of osteoclastogenesis by IFN-β is mediated through STAT3
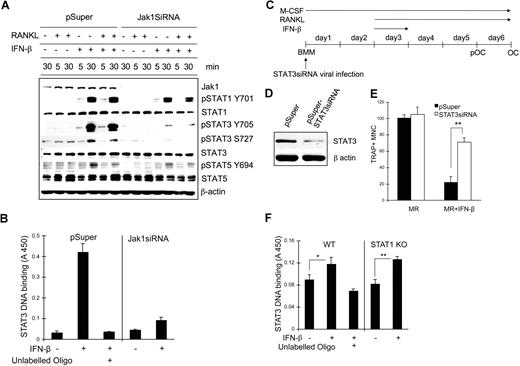
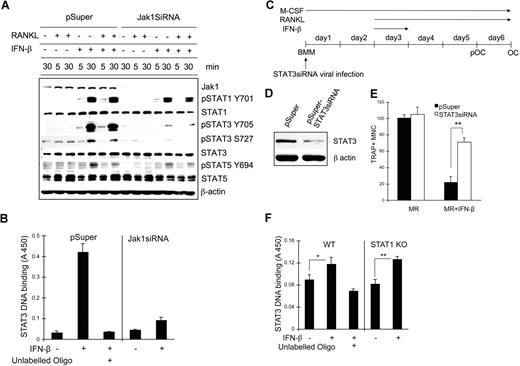
We next examined the signaling pathways affected upon IFN-β stimulation when Jak1 expression is decreased. BMMs were infected with control viruses or viruses harboring Jak1siRNA plasmid and stimulated with RANKL, IFN-β, or RANKL plus IFN-β for 5 minutes or 30 minutes; cell lysates were subjected to Western blotting using phosphospecific antibodies (Figure 7A). Treatment of cells with IFN-β or RANKL in combination with IFN-β significantly induced STAT1 and STAT3 tyrosine phosphorylation at 30 minutes after stimulation while inducing slight phosphorylation in STAT5. However, in Jak1 knocked-down cells, IFN-β–induced STAT3 tyrosine phosphorylation was clearly reduced, although STAT1 phosphorylation was nearly unaffected. STAT5 tyrosine phosphorylation was only slightly reduced at 30 minutes after IFN-β stimulation in Jak1 knocked-down cells compared with control BMMs. There was similar reduction of STAT3 serine phosphorylation after IFN-β stimulation in Jak1 knocked-down cells. Similar selective reduction in the IFN-β–induced phosphorylation of STAT3 was also observed in pOCs (Figure S6A). To confirm the reduced STAT3 activity in Jak1 knocked-down cells, STAT3 binding to the STAT3 consensus binding site was examined in nuclear extracts from control and Jak1siRNA virus–infected BMMs (Figure 7B). In control virus-infected BMMs, stimulation with 100 U/mL IFN-β for 30 minutes significantly induced STAT3 DNA binding activity. However, in Jak1 knocked-down cells, STAT3 DNA binding upon IFN-β stimulation was greatly reduced. Since Jak1 knock-down seemed to prominently hamper IFN-β–dependent STAT3 activation, the role of STAT3 during IFN-β–mediated inhibition of osteoclastogenesis was investigated. BMMs were infected with STAT3 siRNA viruses (Figure 7C), and the reduction of STAT3 expression was confirmed by Western blotting in control and STAT3 knocked-down BMMs (Figure 7D). When these BMMs were cultured with RANKL in the presence of IFN-β, STAT3 knocked-down BMMs were clearly protected from the inhibition of osteoclastogenesis by IFN-β (Figures 7E, S6B). While incubating cells with IFN-β for the first 24 hours following RANKL stimulation was enough to significantly inhibit osteoclastogenesis, more than 3 times the number of OCs were observed in STAT3 knocked-down cells compared with control in the presence of 10 U/mL IFN-β. The STAT3 activity in BMMs appeared to be independent on STAT1 expression, as we observed similar STAT3 DNA binding activity upon IFN-β stimulation in STAT1 knock-out cells (Figure 7F) and STAT1 siRNA–infected cells (Figure S7) compared with the respective control cells. These results suggested that the decreased expression of Jak1 and subsequently impeded STAT3 signaling correlates with the lack of IFN-mediated inhibition of osteoclastogenesis of already committed pOCs.
STAT3 signaling pathway was involved in the IFN-β–mediated inhibition of osteoclastogenesis. (A) BMMs were infected with viruses harboring empty vector (pSuper) or Jak1 siRNA construct (Jak1siRNA). At 1 day after viral infection, cells were stimulated with IFN-β (100 U/mL) in combination with RANKL (500 ng/mL) for the indicated times. Total-cell lysates were subjected to Western blotting using phosphospecific antibodies. The expression of each protein was examined by directly stripping and reprobing the same membrane used to detect the phosphoform of the protein with respective antibody. (B) Vector or Jak1siRNA-infected BMMs were stimulated with 100 U/mL IFN-β for 30 minutes. Nuclear extracts were prepared and assayed for the DNA binding activities of STAT3 as described in “Methods.” Excess unlabeled oligo DNA was added to confirm the specificity of assay. (C) BMMs were first infected with viruses harboring empty vector (pSuper) or STAT3 siRNA construct (pSuper-STAT3siRNA). (D) After 48 hours, total-cell lysates were examined for STAT3 expression by Western blotting. Antiactin blot was shown to ensure same loading between 2 lanes. (E) Cells in panel D were incubated with 10 U/mL IFN-β for 24 hours and further incubated with 100 ng/mL RANKL for 3 days in the presence of 30 ng/mL M-CSF. At the end of culture, cells were stained for TRAP activity. The number of TRAP+ multinuclear OCs was counted from 3 random fields under microscope. Data are means plus or minus SD and are representative of 3 independent experiments. Asterisks represent statistical difference (**P < .01). MR represents M-CSF plus RANKL. (F) WT or STAT1-deficient BMMs were stimulated with 100 U/mL IFN-β for 30 minutes. Nuclear extracts were prepared and assayed for the DNA binding activities of STAT3 as described in “STAT3 DNA binding assay.” Excess unlabeled oligo DNA was added to confirm the specificity of assay. Data are means (± SEM) of 5 independent experiments. Asterisks represent statistical difference (*P < .05; **P < .01).
STAT3 signaling pathway was involved in the IFN-β–mediated inhibition of osteoclastogenesis. (A) BMMs were infected with viruses harboring empty vector (pSuper) or Jak1 siRNA construct (Jak1siRNA). At 1 day after viral infection, cells were stimulated with IFN-β (100 U/mL) in combination with RANKL (500 ng/mL) for the indicated times. Total-cell lysates were subjected to Western blotting using phosphospecific antibodies. The expression of each protein was examined by directly stripping and reprobing the same membrane used to detect the phosphoform of the protein with respective antibody. (B) Vector or Jak1siRNA-infected BMMs were stimulated with 100 U/mL IFN-β for 30 minutes. Nuclear extracts were prepared and assayed for the DNA binding activities of STAT3 as described in “Methods.” Excess unlabeled oligo DNA was added to confirm the specificity of assay. (C) BMMs were first infected with viruses harboring empty vector (pSuper) or STAT3 siRNA construct (pSuper-STAT3siRNA). (D) After 48 hours, total-cell lysates were examined for STAT3 expression by Western blotting. Antiactin blot was shown to ensure same loading between 2 lanes. (E) Cells in panel D were incubated with 10 U/mL IFN-β for 24 hours and further incubated with 100 ng/mL RANKL for 3 days in the presence of 30 ng/mL M-CSF. At the end of culture, cells were stained for TRAP activity. The number of TRAP+ multinuclear OCs was counted from 3 random fields under microscope. Data are means plus or minus SD and are representative of 3 independent experiments. Asterisks represent statistical difference (**P < .01). MR represents M-CSF plus RANKL. (F) WT or STAT1-deficient BMMs were stimulated with 100 U/mL IFN-β for 30 minutes. Nuclear extracts were prepared and assayed for the DNA binding activities of STAT3 as described in “STAT3 DNA binding assay.” Excess unlabeled oligo DNA was added to confirm the specificity of assay. Data are means (± SEM) of 5 independent experiments. Asterisks represent statistical difference (*P < .05; **P < .01).
Discussion
In the present study, we discovered that the expression of Jak1 is significantly reduced during the early stage of OC differentiation by using unbiased proteomic approach. Our results suggested that Jak1 expression is regulated via ubiquitin-mediated proteasomal degradation upon RANKL stimulation. To our knowledge, this is the first evidence that directly shows the ubiquitination of Jak1 during any physiologic process. Recently, it was shown that the ubiquitin-mediated proteasomal degradation regulates the members of the Jak/STAT pathway. Jak2 was reported to be down-regulated via the ubiquitin-proteasome pathway upon stimulation of cells by hormones and cytokines such as prolactin, IL-3, and IFN-γ.20,21 It was also demonstrated that STAT1 and STAT4 were ubiquitinated by the ubiquitin E3 ligase SLIM.22 Thus, our results add to a growing body of evidence that the ubiquitin-mediated degradation of the Jak/STAT pathway proteins constitutes a novel mechanism regulating cellular events. At present, several questions remain to be answered regarding the RANKL-induced ubiquitination of Jak1, such as signaling pathways resulting in the Jak1 ubiquitination and the identity of ubiquitin E3 ligase.
The role of Jak1 is crucial in signaling events initiated by cytokines, including IFNs.19,23 However, in spite of increasing evidence that IFN-mediated signaling is one of the major mechanisms involved in the inhibition of osteoclastogenesis, the role of Jak1 during RANKL-induced OC differentiation has never been investigated. In this study, we showed that the decreased expression of Jak1 during early OC differentiation correlated with the IFN-responsiveness of OC precursors. The “commitment” of OC precursors (ie, resistance to the IFN-mediated inhibition of osteoclastogenesis) seemed to occur within first 24 hours following RANKL stimulation, since the existence of IFN-β in the culture media during this 24-hour period significantly inhibited osteoclastogenesis, while OC differentiation occurred independent of IFN-β thereafter. Furthermore, once BMMs were cultured with IFN-β for the first 24 hours and washed out, BMMs could not differentiate into mature OCs up to 1 week (data not shown), while it usually took 3 days in our culture system for BMMs to differentiate into OCs. The importance of the Jak1 in this commitment step was confirmed by showing that IFN-β could inhibit osteoclastogenesis at 48 hours after RANKL stimulation, far beyond the normal commitment stage, when Jak1 was overexpressed. Forced expression of constitutive active Jak1 resulted in the IFN-independent suppression of osteoclastogenesis. Furthermore, inhibition of the ubiquitin-mediated degradation of Jak1 by proteasome inhibitor also rendered IFN responsiveness beyond this commitment stage (48 hours after RANKL stimulation). On the other hand, down-regulation of Jak1 by siRNA protected OC precursors from the negative regulation by IFN-β, confirming the crucial role of Jak1 during OC differentiation. Our results are in accordance with those reported by Mochizuki et al, which showed that the OC lineage commitment occurred within the first 24 hours after RANKL stimulation.24 In their report, those committed (treated with RANKL for 24 hours) cells became resistant to the inhibition of osteoclastogenesis by GM-CSF, IFN-γ, and IL-4. From our perspective, it is likely that the regulation of Jak1 expression demonstrated in this study is involved in the OC lineage commitment at least in the protection from the inhibitory action of IFN-β.
Takayanagi et al suggested that IFN-β might inhibit RANKL-induced osteoclastogenesis via a negative feedback mechanism, since RANKL stimulated generation of endogenous IFN-β.9 We could observe no significant increase in the RANKL-induced osteoclastogenesis in Jak1 knocked-down BMMs compared with control cells (Figure 6). Theses results can be at least partially explained by the fact that RANKL-induced expression of SOCS1 and SOCS3 were reduced in Jak1 knocked-down BMMs (Figure S8). Since SOCS proteins were reported to protect osteoclastogenesis from the inhibition by RANKL-induced endogenous IFN-β,11,12 it is possible that the effects of decreased Jak1 expression and decrease in the RANKL-induced SOCS proteins were balanced out.
STAT proteins have been suggested as mediators of negative signaling by IFNs during osteoclastogenesis. STAT1 played a crucial role in IFN-mediated inhibition of osteoclastogenesis since STAT1-deficient precursor cells were resistant to the suppression of OC differentiation by IFN-β and IFN-γ.8,9 Hematopoietic cell–specific STAT3-deficient mice were described to show osteoporotic phenotype with increased number of OCs, suggesting an involvement of this member of STAT family proteins in the negative regulation of osteoclastogenesis.25 We observed significantly impaired STAT3 phosphorylation, and to a lesser extent, reduced STAT5 phosphorylation upon stimulation with IFN-β in Jak1 knocked-down BMMs as well as in pOCs compared with control BMMs (Figure 7). Furthermore, STAT3 knocked-down BMMs could differentiate into OCs in the presence of IFN-β. On the other hand, there was no significant difference in the IFN-β–induced tyrosine phosphorylation of STAT1 in Jak1 knocked-down BMMs and in pOCs compared with control BMMs (Figures 7A, S6A). The overexpression of Tyk2 in BMMs resulted in more potent phosphorylation of STAT1 upon IFN-β stimulation (Figure S9), suggesting that Tyk2 might be playing a compensatory role to activate STAT1 when Jak1 is down-regulated. Thus, it is likely that STAT3 is playing important role downstream of Jak1 during IFN-β–mediated inhibition of osteoclastogenesis described in the present study. Although the exact mechanism by which STAT3 modulates osteoclastogenesis still needs to be unveiled, it was demonstrated that the expression of c-Fos was significantly increased in STAT3-deficient BMMs.25 Since IFN-β is known to inhibit osteoclastogenesis by down-regulating c-Fos,9 one possibility is that the reduced Jak1 expression and concomitant reduction in the STAT3 activity shown in the current study facilitates osteoclastogenesis by impeding the IFN-β–mediated decrease in c-Fos expression.
Our results may also have implications for skeletal diseases involving uncontrolled osteoclastogenesis even in the presence of negative regulators. For example, rheumatoid arthritis (RA) is characterized by the destruction of cartilage and bone in joints with greatly increased OC number and activity.26,–28 Surprisingly, markedly increased expression of IFN-β was reported in synovial tissue from patients with RA,29 in spite of the current understanding that IFN-β suppresses osteoclastogenesis. Furthermore, controlled clinical studies found no significant beneficial effect of IFN-β for the treatment of patients with RA,30 despite some promising results with animal models.31 Taken together, it is possible that the reduction of Jak1 expression in RANKL-primed OC precursors shown in the present study might be playing a role in protection from the negative effect of high levels of IFN-β in RA joints.
To summarize, we observed the down-regulation of Jak1 resulted from the ubiquitin-mediated degradation during early OC differentiation of BMMs induced by RANKL. The reduction in the Jak1 expression level correlated with the resistance to the antiosteoclastogenic effect of IFN-β. The mechanism described in this study may be a part of the differentiation program that determines OC lineage commitment.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the 21C Frontier Functional Proteomics Project (FPR05C2-280) and the Research Program for New Drug Target Discovery (M10748000257-07N4800-25710), and grants from the Korea Science & Engineering Foundation, Ministry of Science and Technology, Korea.
Authorship
Contribution: Y.L. designed and performed the research, collected and analyzed the data, and wrote the paper. S.-W.H. H.J.J., and H.-J.K.performed the research and collected the data. Z.H.L., J.S., and S.N.C contributed vital new reagents. E.-J.C. and S.-W.L. analyzed the data. H.-H.K. designed the research, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hong-Hee Kim, Department of Cell and Developmental Biology, BK21 Program, DRI, Seoul National University, 28 Yeongon-Dong, Chongno-Gu, Seoul 110-749, Korea; e-mail: hhbkim@snu.ac.kr.