Abstract
CD1d-restricted T cells have been implicated in the pathogenesis of several chronic inflammatory states. However, the nature of the specific ligands recognized by these cells in vivo in patients with inflammatory or malignant diseases remains unknown. We took a biochemical approach to directly isolate and characterize the nature of CD1d-binding ligands from the plasma of myeloma patients. Characterization of these ligands revealed several lysophosphatidylcholine (LPC) species. Human LPC-CD1d dimer binding cells are T-cell receptorαβ+ T cells but predominantly Vα24−Vβ11−. Cytokine secretion by LPC-specific T cells is skewed toward IL-13 secretion, and the frequencies of these cells are increased in myeloma patients relative to healthy donors. These data identify a distinct population of human CD1d-restricted T cells specific for inflammation-associated lysolipids and suggest a novel mechanism for inflammation mediated immune regulation in human cancer.
Introduction
In contrast to CD4 and CD8+ T cells that recognize peptide ligands in the context of major histocompatibility complex (MHC) class I and II molecules, a distinct subset of T cells recognize lipid antigens in the context of CD1d molecules on antigen presenting cells.1,2 Broadly, 2 distinct types of CD1d-restricted T cells have been identified.3 One subset of CD1d-restricted T cells, also known as invariant or type I natural killer T (NKT) cells, express an invariant T-cell receptor (iTCR: Vα24/Vβ11 in humans and Vα14 in mice) and NK markers, such as CD161.2,4 However, particularly in humans, many CD1d-restricted T cells lack iTCR and have been termed type II NKT cells.5,6 Most of the early work on CD1d-restricted T cells was based on the recognition of a synthetic ligand α-galactosyl ceramide (α-GalCer), by iNKT cells.4 Recent studies have shown that type I NKT cells can recognize some microbial glycolipids and self-antigens, such as isoglobotrihexosylceramide (iGb3; reviewed by Brutkiewicz7 ). The nature of antigens specifically recognized by type II NKT cells is less clear and limited to sulfatide and nonlipidic small molecules.8,9 Binding of CD1d molecules to phospholipids has also been demonstrated.10-13 However, whether these molecules are commonly recognized by populations of human T cells is not known.
Several studies have suggested an important role for CD1d-restricted T cells in the regulation of chronic inflammatory states. For example, type I NKT cells can promote allergen-induced asthma and atherosclerosis and can mediate protection against some autoimmune states in mice.1 Cancer is intricately linked to inflammation. NKT cells have been implicated in both immune surveillance and immune regulation in cancer, attributed to type I and II NKT cells, respectively.1,14 In some settings, GD3 and tumor-derived glycosphingolipids have been implicated as ligands for iNKT cells.15,16 However, the nature of the specific ligands recognized by either type I or II NKT cells in inflammation or cancer in humans remains unclear.
Multiple myeloma (MM) is a plasma cell tumor wherein both the innate and adaptive limbs of the immune response are able to recognize cancer cells.17 In prior studies, we have shown that clinical progression in myeloma is associated with dysfunction of type I NKT cells.18 To identify the nature of CD1d-binding ligands in human myeloma, we took a biochemical approach to directly isolate and characterize the CD1d-binding lipids from the plasma of these patients. Here we show that CD1d-binding ligands isolated from myeloma patients who are recognized by human T cells are inflammation-associated lysophospholipids.
Methods
Plasma and cells from patients and healthy donors
Peripheral blood mononuclear cells (PBMCs) were obtained from healthy donors (buffy coats from New York Blood Center) and from myeloma patients after informed consent was obtained in accordance with the Declaration of Helsinki and approved by the Institutional Review Boards of the Rockefeller University and St Vincent's Cancer Center. Blood mononuclear cells were isolated by density gradient centrifugation using Ficoll-Paque (GE Healthcare, Little Chalfont, United Kingdom). Plasma was harvested from the top of the gradient and cryopreserved until used for lipid isolation. Samples from myeloma patients were collected under the auspices of a protocol for collection of samples for research approved by the Institutional Review Board at St Vincent's Cancer Center and at Rockefeller University.
Reagents
All lipid products were purchased from Avanti Polar Lipids (Alabaster, AL), dissolved in chloroform at 10 mg/mL, and stored at −20°C as storage stock. To prepare 1 mg/mL of working stock, phospholipids were first diluted 1:5 with 100% dimethyl sulfoxide, then 1:2 with water. All phospholipid compounds were stored in either amber glass vials or siliconized Eppendorf tubes covered with aluminum foil (to minimized light exposure). Anti–Vα24-fluorescein isothiocyanate and anti–Vβ11-phycoerythrin (PE) antibodies were purchased from Immunotech (Fullerton, CA). CD1d-DimerX and most fluorochrome-conjugated antibodies were purchased from BD Biosciences (San Jose, CA). Anti–CD8-PE-Texas-Red, anti–CD19-PE-Cy5.5, and anti–CD4-PacificBlue antibodies were purchased from Invitrogen (Carlsbad, CA). The α-galactosyl-ceramide (α-GalCer) was kindly provided by Kirin Breweries (Tokyo, Japan).
Isolation of bulk lipids from plasma
Extraction of bulk lipids was performed based on the method of Bligh and Dyer.19 Plasma (50 mL) was mixed 1:1 with phosphate-buffered saline (PBS), stirred in a beaker while adding ammonium sulfate (Fisher Scientific, Pittsburgh, PA) slowly until saturation at room temperature, and transferred and stirred continuously overnight at 4°C for protein precipitation. The mixture was centrifuged at 1000g for 1 hour at 4°C to pellet the solid precipitates, and liquid mixture was then collected, frozen, and lyophilized using a freeze-dry system (Labconco, Kansas City, MO). The lyophilized powder was then extracted sequentially with 3 combinations of organic solvents (chloroform:methanol:water at 2:1:0, 1:2:0, 10:10:1). The 3 extracts were pooled and organic solvents removed using rotary evaporator. The extract was then redissolved in 5 mL water and stored frozen in aliquots at −20°C until use.
Mass spectrometry analysis
Lipid samples from extracts and elutes were desalted, cleaned and concentrated using micro-C18 ZipTip (Millipore, Billerica, MA). The lipids were eluted from ZipTip with 2 μL aqueous acetonitrile solution containing 50% acetonitrile (vol/vol) and 0.1% formic acid, and loaded into a GlassTip (New Objective, Woburn, MA). The analysis was performed using an Applied Biosystems QSTAR QqTOF mass spectrometer (Foster City, CA) operated in both positive and negative modes. Ions of interest were selected and subsequently dissociated by collision for analysis of the ionic fragments.
Cross-linking CD1d-DimerX to beads and elution of lipids
The crosslinking of CD1d-DimerX to magnetic beads was done based per the manufacturer's protocol. In brief, Dynabeads M-280 (sheep antimouse IgG from Invitrogen) were washed twice in PBS, resuspended in PBS/bovine serum albumin (BSA) at 1 μg dimer to 1 to 2 × 107 beads ratio, and incubated overnight at 4°C with constant rotation. Then, dimer beads were washed twice with 0.2 M triethanolamine, pH 8.2, and crosslinked with freshly prepared 5.4 mg/mL dimethyl pimelimidate dihydrochloride (Sigma-Aldrich, St Louis, MO) at room temperature with constant rotation for 30 minutes. The crosslinking was stopped by resuspending in 1 mL 50 mM Tris, pH 7.5, and incubated for 15 minutes with rotational mixing. Beads were then washed 3 times with PBS/BSA before storage in the original volume of beads. As a control, some Dynabeads were processed through the same procedure as above but without CD1d-DimerX. For ligand loading, 200 μL crosslinked beads was washed 3 times with PBS while the bulk extract was diluted 60:40 with dimethyl sulfoxide. At the end of the wash, the beads were mixed with 125 μL bulk lipid extract and incubated overnight at 37°C. For elution, ligand-loaded beads were washed once with ice-cold PBS, then mixed with 0.1 M citrate at pH 2.0, incubated for 10 minutes at 37°C. CD1d-conjugated beads not loaded with lipid ligands were used as additional controls. The lipid elute was then collected after magnetic separation and saved for analysis. The beads were washed 3 times in PBS and saved for the next use.
Loading CD1d dimers and detection of CD1d-lipid–reactive T cells
Loading and staining using CD1d-DimerX was done based on the manufacturer's protocol. For loading with α-GalCer (as positive control), 1 μL DimerX (0.5 mg/mL) was mixed with 1 μL of a 10-molar excess of α-GalCer (20 μg/mL). For loading with lysophosphatidylcholine (LPC), 1 mg/mL LPC was first diluted to 80:20 with water (0.8 mg/mL), then mixed 1:1 with CD1d-DimerX for the final concentration of 50 to 400 μM. The loading of all other phospholipids was done in equimolar ratio to LPC. The unloaded dimer control was prepared by mixing CD1d-DimerX with the carrier only. The dimer mix was incubated overnight at 37°C and stored at 4°C before use. For some experiments, mixtures of defined lipids at different ratios (eg, LPC and phosphatidylcholine [PC]) were concurrently loaded onto CD1d. For some experiments, bulk lipids from myeloma or healthy donor plasma were directly used to load CD1d dimers. For some experiments, CD1d dimerX was preincubated with defined lipid ligands (1:1 ratio) for 4 hours at 37°C as described under “Loading CD1d dimers and detection of CD1d-lipid–reactive T cells.” After 4 hours, a 10-molar excess of α-GalCer or carrier control was added and mixed before incubating overnight at 37°C.
For staining human T cells with loaded CD1d dimer, T cells were first washed twice in PBS and then stained with dead cell exclusion dye (Live/Dead Aqua dead cell stain kit; Invitrogen) for 15 minutes as recommended by the manufacturer. Cells were then washed twice in wash buffer (PBS with 0.5% BSA), blocked with blocking buffer (wash buffer with 0.1 mg/mL human IgG whole molecules; Jackson ImmunoResearch Laboratories, West Grove, PA) at a concentration of approximately 1 to 3 million cells per 40 μL for 10 minutes at room temperature. Dimer staining mix was then prepared by first diluting CD1d-Dimer mix at 0.5 μg of CD1d-DimerX to 10 μL blocking buffer and then added to blocked cells at a 1:5 ratio. Cells were then stained for approximately 30 minutes to 1 hour at room temperature in the dark. After dimer staining, cells were washed twice with wash buffer, blocked with 10% rat serum in PBS for 5 minutes at room temperature (1-3 million in 25 μL), and then stained with antimouse-IgG1-allophycocyanin (APC) or PE (clone X56 from BD Biosciences) at 1:1 ratio for the final dilution of 1:500 of antibody for 15 to 30 minutes on ice. Cells were then washed twice with wash buffer and stained with additional markers. Typically, the following antibodies were used for staining: anti–Vα24-fluorescein isothiocyanate, anti–Vβ11-PE, anti–CD3-PE, anti–CD3-Alexa700, anti–CD56-PECy7, anti–CD8-PE-TexasRed, anti–CD4-PacBlue, anti–CD19-PECy5.5. For some experiments, cells were also stained with anti–TCRαβ-PE and anti–TCRγδ-PE. For acquisition using BD-LSR II, the cells were resuspended in wash buffer and data acquired directly. For acquisition using a BD-Calibur (BD Biosciences), staining with 7-amino-actinomycin D was used for excluding dying cells.
Expansion of Lyso-PC–specific T cells
PBMCs were separated into CD14+ and CD14− fractions using CD14 magnetic beads (Miltenyi Biotec, Auburn, CA). CD14+ monocytes were used for dendritic cell (DC) culture and cultured in either 1% plasma or in serum-free Aim-V media (Invitrogen) in the presence of granulocyte-macrophage colony-stimulating factor and IL-4 as previously described.20 CD14− PBMCs were similarly cultured in either RPMI 1640 (Cellgro-Mediatech, Manassas, VA) with 5% pooled human serum (Labquip, Niagara Falls, NY) or in Aim-V medium. For some experiments, expansion of invariant NKT cells using α-GalCer–loaded DCs was performed as previously described.21 For loading LysoPC, immature DCs on day 5 of culture were cultured for 48 hours in the presence of 20 μg/mL of LPC (LPC-C18-1). For the expansion of LPC specific T cells, DCs were cultured with responder CD14− cells (DC-responder ratio of 1:10 to 1:20) in media supplemented weekly with 50 U/mL of IL-2 (Chiron, Emeryville, CA). For some experiments, the cocultures were setup in serum free conditions, without or with supplementation with 20 μg/mL of LPC. After 1 to 2 weeks of coculture, the presence of LPC specific T cells was monitored by flow cytometry, as described under “Loading DC1d dimers and detection of CD1d-lipid–reactive T cells.”
Activation of human invariant NKT (iNKT) cells by immobilized CD1d-DimerX
The loading of CD1d dimers with defined lipids or α-GalCer was performed as specified under “Loading CD1d dimers and detection of CD1d-lipid–reactive T cells.” After overnight incubation, lipid-loaded CD1d dimer, or unloaded CD1d dimer as control, was diluted in PBS at a ratio of 1 μg of CD1d dimer to 100 μL of PBS, added to a 96-well U-bottom plate, centrifuged at 300g at room temperature, and incubated at 37°C for approximately 4 to 6 hours. The dimer-PBS mixture was removed just before adding purified iNKT cells (0.5-1 × 106 cells/well). Culture supernatants were harvested after 40 to 48 hours of culture and analyzed for the presence of interferon-γ by enzyme-linked immunosorbent assay (ELISA) following the manufacturer's protocol (BioSource Invitrogen, Carlsbad, CA).
Detection of cytokine production by CD1d-LysoPC+ T cells
CD3+ CD1d-LPC+Va24− T cells (> 95% purity) were obtained from cultures of human T cells described in “Expansion of Lyso-PC–specific T cells” by flow sorting using a BD FACSAria cell-sorting system (BD Biosciences, San Jose, CA). As a control, CD3+ CD1d-LysoPC− Vα24− T cells were also sorted. For stimulation with phorbol myristate acetate (PMA) and ionomycin, sorted cells (CD1d-LysoPC+ or CD1d-LysoPC−) were plated at concentration of 5 to 10 × 104 cells in 100 μL fresh culture media (RPMI with 5% pooled human serum), and stimulated with PMA (500 ng/mL) and ionomycin (1 μM) in a 96-well U-bottom plate. For stimulation with immobilized anti-CD3, CD1d-LPC+ sorted cells were plated at 104 cells/100 μL fresh culture media in anti–human-CD3 antibody-coated or control plate (BD Biosciences) as suggested by the manufacturer. For some experiments, T cells were depleted of CD1d-LPC+ cells or mock-depleted by sorting, before plating at 106 cells per 200 μL in anti–human-CD3 antibody-coated or control plates. For some experiments, sorted 5 × 104 LysoPC-CD1d dimer+ T cells were cultured with CD1d expressing cell line (C1rd cells; kindly provided by Dr Mark Exley, Boston), or mock-transfected control cells at a T cell:antigen-presenting cell ratio of 1:10, as described,6 with or without prior pulsing with 40 μg/mL LPC-C181, with 5 ng/mL PMA and 5 U/mL IL-2 in Aim-V media. After 40 to 48 hours of coculture, supernatants were harvested and the presence of cytokines was measured by Luminex analysis with human cytokine detection kits from Upstate Biotechnology (Charlottesville, VA), as described.22 In some experiments, IL-17 was detected by an ELISA kit following the manufacturer's protocol (eBioscience, San Diego, CA).
Statistics
Comparison of data between groups was performed using the Mann Whitney test. Significance was analyzed with 2-sided P values less than .05.
Results
To identify the nature of CD1d-binding lipids in the plasma of myeloma patients, bulk lipids from these samples were obtained by chloroform/methanol extraction.19 Analysis of bulk lipids by electrospray ionization mass spectrometry revealed the presence of many species in the extracts from both patients tested (Figure 1A). To identify the CD1d-binding fraction, we first loaded the bulk lipids onto CD1d dimer conjugated to magnetic beads and then eluted the bound fragments as described in “Cross-linking CD1d-DimerX to beads and elution of lipids.” Magnetic beads not conjugated with CD1d were used as a control. Another control was CD1d-conjugated beads not treated with lipids that did not yield any contaminating lipids in the beads themselves (data not shown). Although the mass spectra in bulk lipids from both patients differed and included several lipid species as expected, the eluates from CD1d-conjugated beads in both patients were remarkably similar (Figure 1B). The most abundant ions were observed at m/z 496, 520, 522, 524, and 544. To further validate the enrichment of these ions, the samples eluted from beads alone or CD1d-conjugated beads were spiked with C14-LysoPC (m/z 468) as an internal standard and analyzed by electrospray ionization mass spectrometry. These data again confirmed the relative enrichment of previously identified species in the elutes from CD1d-conjugated beads (Figure 1C). The tandem mass spectra (MS/MS) of ions at m/z 496.33 are displayed in Figure 1D. It shows a major fragment at m/z 184. The m/z of this fragment corresponds to the head group of phosphatidylcholine. Indeed, the other fragments observed at m/z 60, 86, and 104 are also typical dissociation products of phosphatidylcholine.23,24 The same samples were also analyzed in electrospray ionization (ESI)-negative mode. An ion at m/z 480.28 was observed that corresponds to the [M-15]− species observed at m/z 496.33 in positive mode. The [M-15]− species was formed by the loss of the methyl group from phosphatidylcholine. The MS/MS of 480.28 is displayed in Figure 1E. A major fragment was observed at m/z 255, which matches to one of the fatty acid anions in phosphatidylcholine according to the general rules of fragmentation of phopholipid in negative mode. The molecular weight of palmitic acid is 256, suggesting that one side chain of phosphatidylcholine at m/z 496.3 for [M + H]+ or 480.28 for [M-15]− is palmitic acid. Therefore, we conclude that the major ion binding to CD1d molecule is hexadecanoly-sn-glycero-3-phosphocholine (Figure 1F). To determine the regioisomer of this lysophospholipid species, we examined the MS/MS of the sodiated hexadecanoly-sn-glycero-3-phosphocholine at m/z 518.32 and found that the intensity of a fragment at m/z 104 is more abundant than that at m/z 147, demonstrating that species we identified in this work is 1-hexadecanoly-2-hydroxy-sn-glycero-3-phosphocholine (LPC).24 The analysis of other species identified in these studies revealed that they were all LPC species with different acyl chains (16:0, 18:0, 18:1, 18:2, 20:4; Table 1). To further confirm our results, authentic 1-hexadecanoly-2-hydroxy-sn-glycero-3-phosphocholine was purchased and analyzed using tandem mass spectrometry. Fragmentation patterns of authentic 1-hexadecanoly-2-hydroxy-sn-glycero-3-phosphocholine were identical to what we observed in the present work (not shown). Therefore, the great majority of CD1d-binding lipids isolated from the plasma of myeloma patients were LPC species, which are structurally distinct from ceramides. The isolation of LPC from myeloma plasma is consistent with prior studies showing that levels of this lipid in myeloma is much higher than in healthy donors.25 Consistent with this, when bulk lipids from myeloma patients were loaded onto CD1d dimer, the staining for human T cells was better than that seen with lipids from healthy donors (Figure 1G).
Isolation and identification of CD1d-binding lipids from myeloma patient plasma. (A,B) ESI-mass spectra of lipid species from bulk extracts (A) and CD1d-conjugated bead eluents (B) from plasma of 2 myeloma patients (MM1 and MM2). (C) ESI-mass spectra of eluents from beads only control and CD1d beads, spiked with an LPC species (LPC-C14, m/z 368, arrow) as an internal reference. Data shown are representative of findings on 4 separate patients. (D,E) Tandem mass spectra (MS/MS) of major species observed for eluents of myeloma patient studied in panel C. (F) MS/MS of 496.3 of [M + H]+. (G) MS/MS of 480.3 of [M-15]. (F) Structure of 1-hexadecanoly-2-hydroxy-sn-glycero-3-phosphocholine, or lysophosphatidylcholine (LPC)-C16:0. (G) Bulk lipids isolated from myeloma patients or healthy donors were used to load CD1d dimers and stain cultures of human T cells as described in “Loading CD1d dimers and detection of CD1d-lipid–reactive T cells.” Numbers represent percentage of cells in the quadrant.
Isolation and identification of CD1d-binding lipids from myeloma patient plasma. (A,B) ESI-mass spectra of lipid species from bulk extracts (A) and CD1d-conjugated bead eluents (B) from plasma of 2 myeloma patients (MM1 and MM2). (C) ESI-mass spectra of eluents from beads only control and CD1d beads, spiked with an LPC species (LPC-C14, m/z 368, arrow) as an internal reference. Data shown are representative of findings on 4 separate patients. (D,E) Tandem mass spectra (MS/MS) of major species observed for eluents of myeloma patient studied in panel C. (F) MS/MS of 496.3 of [M + H]+. (G) MS/MS of 480.3 of [M-15]. (F) Structure of 1-hexadecanoly-2-hydroxy-sn-glycero-3-phosphocholine, or lysophosphatidylcholine (LPC)-C16:0. (G) Bulk lipids isolated from myeloma patients or healthy donors were used to load CD1d dimers and stain cultures of human T cells as described in “Loading CD1d dimers and detection of CD1d-lipid–reactive T cells.” Numbers represent percentage of cells in the quadrant.
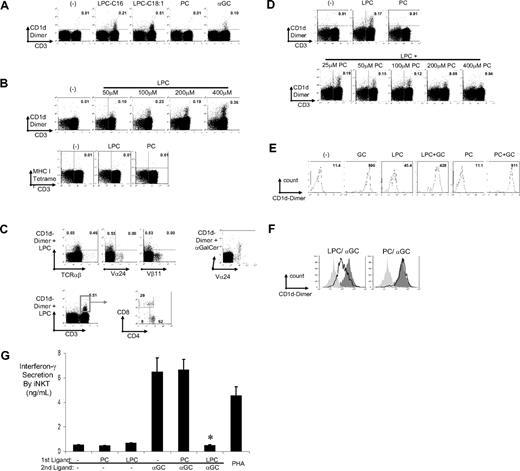
Several species of LPC exist in vivo depending on the properties of the acyl chain.26 We focused on 2 of the species (C16:0 and C18:1) identified in our experiments. To confirm the findings with elution experiments, we loaded equimolar quantities of LPC (C16:0 and C18:1) and PC on CD1d dimer and compared their subsequent binding to cultures of human T cells (Figure 2A). CD1d dimers loaded with LPC showed clear staining for human T cells. Staining with C18:1 LPC was generally superior to that with C16:0 (Figure 2A), and this species was used in subsequent experiments. The efficiency of staining with LPC-loaded CD1d dimers was dependent on the dose of LPC used to load the dimer and evident at a dose of 50 to 100 μM (Figure 2B). As an irrelevant MHC molecule, MHCI tetramer coincubated with LPC or PC did not stain these T cells. In cultures of human T cells, the predominant population stained with LPC-CD1d was TCRαβ+ and a subset of Vα24−Vβ11− CD3+ T cells, which consisted predominantly of CD4+, but also CD8+ T cells (Figure 2C). In contrast to CD1d-LPC, CD1d dimer loaded with α-GalCer clearly stained Vα24+ T cells in these cultures. Equimolar loading of CD1d with the parent lipid, PC, led to minimal staining. However, the addition of increasing concentration of PC to LPC led to dose-dependent inhibition of staining of T cells, which is consistent with the capacity of PC to bind CD1d12,13 and/or interfere with the capacity of LPC to load CD1d and stain type II NKT cells (Figure 2D). Together, these data show that LPC loaded onto CD1d can bind a distinct population of human T cells.
Binding of CD1d dimer molecules loaded with lysophosphatidylcholine (LysoPC) to a subpopulation of CD3+ T cells. (A) CD1d dimers loaded with 2 species of LPC (LPC-C16 and LPC-C18:1) stained a unique population of CD3+ T cells: FACS dot plots of CD1d dimers loaded with vehicle control (−), 400 μM of 2 species of LPC (C16 and C18:1) or PC, or 20 μM α-GalCer, and used to stain cultured human PBMCs. Numbers in the quadrant correspond to percentage of cells of total CD3+ cells, and data shown are gated for live cells. The staining is representative of 4 individual donors. (B) CD1d dimers were loaded with different concentrations of C18:1 LPC before staining of human T cell cultures as described in “Loading CD1d dimers and detection of CD1d-lipid–reactive T cells.” As controls, T cells were also stained with MHC-I tetramer cultured with 400 μM of LPC or PC. Numbers in the quadrant correspond to percentage cells of total CD3+ cells, and data shown are gated for live cells. (C) Phenotype of CD1d-LPC-C18:1+ cells: FACS dot plots of staining of CD1d-LPC-C181 (as in panel A) but plotted against different markers, TCRαβ, Vα24, and Vβ11 as indicated. Data for the staining of Vα24+ cells in these cultures with CD1d-αGalCer are shown as a control. Bottom panel shows FACS dot plot gated for CD1d-LPC-C18:1+ T-cell population for the proportion of CD4+/CD8+/double-negative subpopulations. (D) CD1d dimers were loaded with either 400 μM of LysoPC or PC alone, or with a mixture of LysoPC and PC as indicated, before use for staining T cells as in panel A. (E) Histogram plots of staining of iNKT cells by CD1d dimers loaded with α-GalCer (20 μM), LPC, PC, or the combination of α-GalCer with PC or LPC. Numbers represent mean fluorescent intensity of staining. (F) Competition between LPC and α-GalCer for binding to iNKT cells: Histogram overlay plots comparing mean fluorescence shifts between CD1d dimer loaded with vehicle control (light gray solid), positive control (α-GalCer, dark gray solid), and LPC-C18:1, or PC (open curves). Competition was set up by first loading CD1d dimer with vehicle control, LPC-C18:1, or PC, for 4 hours, then loading with vehicle control or α-GalCer. Ligand-loaded CD1d dimers were then used to stain iNKT cells. Data are representative of 4 similar experiments. (G) Differential iNKT stimulation by ligand-loaded and immobilized CD1d dimer. Polyclonal populations of Vα24+ T cells were isolated using magnetic beads from cultures expanded in vitro using α-GalCer. Purified Vα24+ T cells were cultured for 40 to 48 hours in plates with immobilized CD1d dimer, loaded with vehicle control or α-GalCer with or without preincubation with control, PC, or LPC as competing ligands as described in “Loading CD1d dimers and detection of CD1d-lipid–reactive T cells.” PHA was used as positive control. The level of IFN-γ in the supernatant was monitored using ELISA. Data are representative of 3 similar experiments (*P < .05). Error bars represent SD.
Binding of CD1d dimer molecules loaded with lysophosphatidylcholine (LysoPC) to a subpopulation of CD3+ T cells. (A) CD1d dimers loaded with 2 species of LPC (LPC-C16 and LPC-C18:1) stained a unique population of CD3+ T cells: FACS dot plots of CD1d dimers loaded with vehicle control (−), 400 μM of 2 species of LPC (C16 and C18:1) or PC, or 20 μM α-GalCer, and used to stain cultured human PBMCs. Numbers in the quadrant correspond to percentage of cells of total CD3+ cells, and data shown are gated for live cells. The staining is representative of 4 individual donors. (B) CD1d dimers were loaded with different concentrations of C18:1 LPC before staining of human T cell cultures as described in “Loading CD1d dimers and detection of CD1d-lipid–reactive T cells.” As controls, T cells were also stained with MHC-I tetramer cultured with 400 μM of LPC or PC. Numbers in the quadrant correspond to percentage cells of total CD3+ cells, and data shown are gated for live cells. (C) Phenotype of CD1d-LPC-C18:1+ cells: FACS dot plots of staining of CD1d-LPC-C181 (as in panel A) but plotted against different markers, TCRαβ, Vα24, and Vβ11 as indicated. Data for the staining of Vα24+ cells in these cultures with CD1d-αGalCer are shown as a control. Bottom panel shows FACS dot plot gated for CD1d-LPC-C18:1+ T-cell population for the proportion of CD4+/CD8+/double-negative subpopulations. (D) CD1d dimers were loaded with either 400 μM of LysoPC or PC alone, or with a mixture of LysoPC and PC as indicated, before use for staining T cells as in panel A. (E) Histogram plots of staining of iNKT cells by CD1d dimers loaded with α-GalCer (20 μM), LPC, PC, or the combination of α-GalCer with PC or LPC. Numbers represent mean fluorescent intensity of staining. (F) Competition between LPC and α-GalCer for binding to iNKT cells: Histogram overlay plots comparing mean fluorescence shifts between CD1d dimer loaded with vehicle control (light gray solid), positive control (α-GalCer, dark gray solid), and LPC-C18:1, or PC (open curves). Competition was set up by first loading CD1d dimer with vehicle control, LPC-C18:1, or PC, for 4 hours, then loading with vehicle control or α-GalCer. Ligand-loaded CD1d dimers were then used to stain iNKT cells. Data are representative of 4 similar experiments. (G) Differential iNKT stimulation by ligand-loaded and immobilized CD1d dimer. Polyclonal populations of Vα24+ T cells were isolated using magnetic beads from cultures expanded in vitro using α-GalCer. Purified Vα24+ T cells were cultured for 40 to 48 hours in plates with immobilized CD1d dimer, loaded with vehicle control or α-GalCer with or without preincubation with control, PC, or LPC as competing ligands as described in “Loading CD1d dimers and detection of CD1d-lipid–reactive T cells.” PHA was used as positive control. The level of IFN-γ in the supernatant was monitored using ELISA. Data are representative of 3 similar experiments (*P < .05). Error bars represent SD.
To further evaluate the ability of LPC to bind CD1d, we took advantage of the ability of α-GalCer to load CD1d and of the loaded CD1d dimer to stain Vα24+Vβ11+ T cells (also termed type I or iNKT cells) with high mean fluorescence intensity (Figure 2E). When LPC (400 μM) was coincubated with α-GalCer (20 μM) for loading CD1d dimer, there was mild reduction in the intensity of staining, but iNKT cells could still be stained (Figure 2E). However, when CD1d dimers were preincubated with LPC before loading α-GalCer, it clearly reduced the intensity of staining for iNKT cells, suggesting that, under these conditions, LPC could competitively inhibit the loading of α-GalCer on CD1d (Figure 2F). Culture of purified human iNKT cells with immobilized CD1d dimer-α-GalCer normally leads to a rapid release of cytokines, such as interferon-γ.21 However, when these CD1d molecules were preincubated with LPC before α-GalCer, their ability to induce interferon-γ from purified iNKT cells was inhibited, which further supported the observed capacity of preloading with LPC to inhibit α-GalCer–dependent activation of iNKT cells (Figure 2G). Notably, immobilized CD1d dimer loaded with LPC alone elicited little IFN-γ from Vα24+Vβ11+ T cells, which is consistent with Vα24−Vβ11− T cells being the predominant population stained with these reagents. Interestingly, preloading with PC under these conditions did not alter GalCer-mediated iNKT staining and activation. This is somewhat surprising in view of the interference of LysoPC-dependent staining of a different type of NKT cells (type II NKT cells) by concurrent addition of PC (Figure 2D). However, it is consistent with the possibility that the recognition of CD1d by type I vs type II NKT cells may differ and/or that the structural aspects of LPC loading into CD1d (which has not yet been examined) may differ from that of PC.27
Next we examined whether human LPC specific T cells could be expanded in culture. Culture of human T cells in 5% pooled human serum with both unpulsed DCs, as well as those pulsed with LPC led to expansion of LPC specific T cells in culture (Figure 3A,B). Although this was initially surprising, we realized that several studies have shown the presence of LPC in normal human serum.25,28,29 The presence of LPC in lipids from 5% pooled human serum was confirmed by matrix-assisted laser-desorption/ionization (data not shown), which is consistent with prior studies.25,28,29 Therefore, we repeated these cocultures in serum-free media. In this setting, the expansion of LPC specific T cells in the presence of unpulsed DCs was abrogated and specific expansion of LPC specific T cells could be demonstrated only in the presence of the specific ligand (Figure 3C,D). These data demonstrate the feasibility of ligand-dependent expansion of human LPC specific T cells in culture.
Expansion of human CD1d-LPC+ T cells in culture. (A) Expansion of CD1d-LPC+ cells in serum-containing media: representative FACS plot showing the presence of LPC specific T cells in freshly isolated PBMCs or after coculture with DC pulsed with or without LPC in culture media with 5% pooled human serum. Numbers in the quadrant correspond to percentage cells of total CD3+ cells, and data shown are gated for live cells. (B) Summary of experiments (n = 8) with unpulsed-DC or LPC-pulsed DC in serum-containing culture media. Each dot corresponds to percentage of CD1d-LPC+ cells per total CD3+ cells, and each line corresponds to data before and after 2-week expansion. (C) Expansion of CD1d-LPC+ cells in serum-free media: representative FACS plot showing the presence of LPC specific T cells in freshly isolated PBMCs or after coculture with DC pulsed with or without LPC in serum free media. Numbers in the quadrant correspond to percentage of cells of total CD3+ cells, and data shown are gated for live cells. (D) Summary of experiments (n = 4) with unpulsed DC or LPC pulsed DCs in serum-free media. Each dot corresponds to percentage of CD1d-LPC+ cells per total CD3+ cells, and each line corresponds to data before and after 1-week expansion.
Expansion of human CD1d-LPC+ T cells in culture. (A) Expansion of CD1d-LPC+ cells in serum-containing media: representative FACS plot showing the presence of LPC specific T cells in freshly isolated PBMCs or after coculture with DC pulsed with or without LPC in culture media with 5% pooled human serum. Numbers in the quadrant correspond to percentage cells of total CD3+ cells, and data shown are gated for live cells. (B) Summary of experiments (n = 8) with unpulsed-DC or LPC-pulsed DC in serum-containing culture media. Each dot corresponds to percentage of CD1d-LPC+ cells per total CD3+ cells, and each line corresponds to data before and after 2-week expansion. (C) Expansion of CD1d-LPC+ cells in serum-free media: representative FACS plot showing the presence of LPC specific T cells in freshly isolated PBMCs or after coculture with DC pulsed with or without LPC in serum free media. Numbers in the quadrant correspond to percentage of cells of total CD3+ cells, and data shown are gated for live cells. (D) Summary of experiments (n = 4) with unpulsed DC or LPC pulsed DCs in serum-free media. Each dot corresponds to percentage of CD1d-LPC+ cells per total CD3+ cells, and each line corresponds to data before and after 1-week expansion.
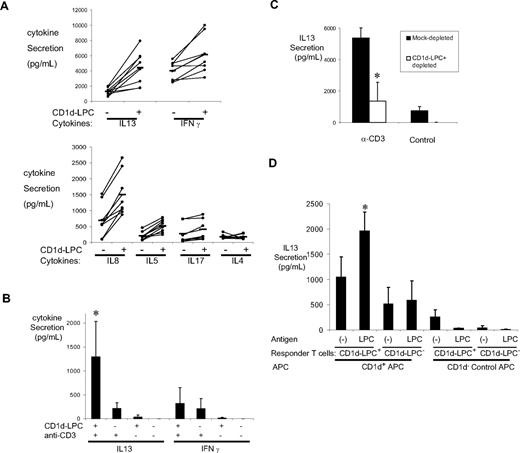
To characterize the functional properties of these cells, we sorted CD1d-LPC+ CD3+ T cells and compared their cytokine profile in response to stimulation by PMA/ionomycin, relative to CD1d-LPC− CD3+ T cells (Figure 4A). Vα24− CD1d-LPC+ T cells secreted higher levels of several cytokines, including IL-13, IL-5, IL-8, and IFN-γ in response to PMA/ionomycin, relative to CD1d-LPC− control CD3+ T cells (Figure 4A). Next, we compared the capacity of these sorted T cells to secrete IL-13 and IFN-γ in response to plate bound anti-CD3. Vα24− CD1d-LPC+ T cells secreted more IL-13 relative to CD1d-LPC− control T cells under these conditions (Figure 4B). Interestingly, depletion of this subpopulation from these cultures led to significant decline in the anti–CD3-mediated secretion of IL-13, compared with mock sorted or undepleted cells, suggesting that this subpopulation of T cells may be a major producer of IL-13 in these cultures (Figure 4C). LPC specific T cells also secreted IL-13 in response to LPC-loaded CD1d expressing APCs, suggesting that they can respond to ligand-specific stimulation by secretion of this cytokine (Figure 4D).
Cytokine secretion by CD1d-LPC+ T cells. (A) T-cell subsets were flow-sorted as indicated for CD1d-LPC+ and CD1d-LPC− cell population and stimulated with PMA and ionomycin. The supernatant was harvested and analyzed for the presence of IL-13, IL-5, IL-8, IL-17, IFN-γ, and IL-4 by Luminex, IL-17 by ELISA. The data shown are for each donor (n = 8) for each cytokine. Significant P values: IL-13, .001; IL-5, .003; IL-8, .02; IFN-γ, .049. (B) Differential expression of IL-13 and IFN-γ by CD1d-LysoPC+ T cells after anti-CD3 stimulation: CD3+CD1d-LysoPC+ and CD3+CD1d-LPC− were flow-sorted as indicated in panel A and stimulated by plate immobilized anti-CD3 antibodies or control. Secretion of IL-13 and IFN-γ was monitored by Luminex after 40 to 48 hours of culture. Data shown are means (± SD) of 4 experiments (*P < .05). (C) Decline in anti-CD3–stimulated IL-13 secretion in human PBMCs after the depletion of CD1d-LysoPC+ cells: cultured human PBMCs were either mock-depleted or depleted of CD1d-LysoPC+ cells by flow sorting. The cells were then stimulated with anti-CD3, and the production of IL-13 monitored by Luminex. Data shown are means (± SD) of 3 experiments (*P < .05). (D) Secretion of IL-13 in response to LysoPC stimulation. Cultured PBMCs were flow-sorted as indicated in panel A. CD1d-LysoPC+ and CD1d-LysoPC− T cells were then stimulated with LysoPC pulsed CD1d-expressing cell line (or unpulsed cells, as well as CD1d− control cells as a control) in serum-free media for 40 to 48 hours. Secretion of IL-13 was monitored by Luminex. Data shown are means (± SD) of 3 similar experiments (*P < .05).
Cytokine secretion by CD1d-LPC+ T cells. (A) T-cell subsets were flow-sorted as indicated for CD1d-LPC+ and CD1d-LPC− cell population and stimulated with PMA and ionomycin. The supernatant was harvested and analyzed for the presence of IL-13, IL-5, IL-8, IL-17, IFN-γ, and IL-4 by Luminex, IL-17 by ELISA. The data shown are for each donor (n = 8) for each cytokine. Significant P values: IL-13, .001; IL-5, .003; IL-8, .02; IFN-γ, .049. (B) Differential expression of IL-13 and IFN-γ by CD1d-LysoPC+ T cells after anti-CD3 stimulation: CD3+CD1d-LysoPC+ and CD3+CD1d-LPC− were flow-sorted as indicated in panel A and stimulated by plate immobilized anti-CD3 antibodies or control. Secretion of IL-13 and IFN-γ was monitored by Luminex after 40 to 48 hours of culture. Data shown are means (± SD) of 4 experiments (*P < .05). (C) Decline in anti-CD3–stimulated IL-13 secretion in human PBMCs after the depletion of CD1d-LysoPC+ cells: cultured human PBMCs were either mock-depleted or depleted of CD1d-LysoPC+ cells by flow sorting. The cells were then stimulated with anti-CD3, and the production of IL-13 monitored by Luminex. Data shown are means (± SD) of 3 experiments (*P < .05). (D) Secretion of IL-13 in response to LysoPC stimulation. Cultured PBMCs were flow-sorted as indicated in panel A. CD1d-LysoPC+ and CD1d-LysoPC− T cells were then stimulated with LysoPC pulsed CD1d-expressing cell line (or unpulsed cells, as well as CD1d− control cells as a control) in serum-free media for 40 to 48 hours. Secretion of IL-13 was monitored by Luminex. Data shown are means (± SD) of 3 similar experiments (*P < .05).
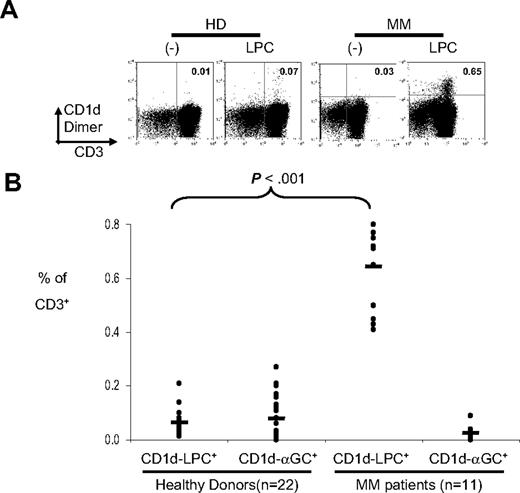
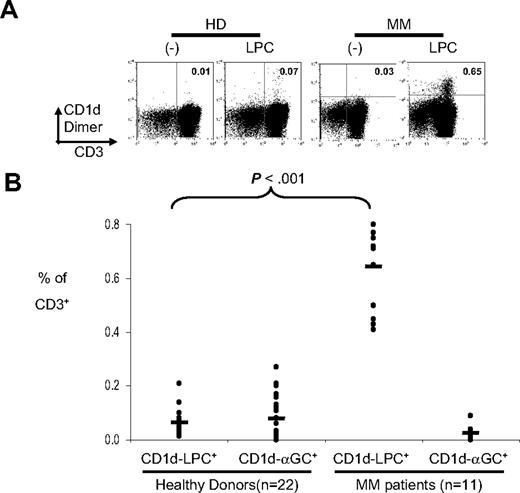
Prior studies have documented that sera from myeloma patients contain much higher levels of LPC, as well as higher palmitic/linoleic acid ratio, relative to healthy controls.25 This may help explain the facile loading of bulk lipids from myeloma patients, but not healthy donors, on CD1d dimer that we observed. Therefore, we tested whether the frequency of LPC specific T cells was altered in human myeloma. CD1d multimers loaded with LPC (vs PC or unloaded CD1d dimer as a control) were used to stain fresh human PBMCs from healthy donors or myeloma patients (Figure 5A,B). The frequency of CD1d-LPC+ T cells was increased in myeloma patients relative to healthy controls. These cells lack the expression of invariant T-cell receptor. Therefore, MM patients have an increase in a distinct population of CD1d-restricted T cells relative to healthy controls.
Detection of CD1d-LPC+ T cells in multiple myeloma patients. (A) Representative FACS dot plots of a healthy donor and a myeloma patient stained with CD1d dimer loaded with vehicle control or LPC. The number in the quadrant corresponds with the percentage of cells of total CD3+ cells. Data shown are gated for live cells. (B) Summary of percentage of CD1d-LPC+ cells of total CD3+ for healthy donors (n = 22) and multiple myeloma patients (n = 11). Each dot corresponds to one donor. Horizontal bars represent means (*P < .05).
Detection of CD1d-LPC+ T cells in multiple myeloma patients. (A) Representative FACS dot plots of a healthy donor and a myeloma patient stained with CD1d dimer loaded with vehicle control or LPC. The number in the quadrant corresponds with the percentage of cells of total CD3+ cells. Data shown are gated for live cells. (B) Summary of percentage of CD1d-LPC+ cells of total CD3+ for healthy donors (n = 22) and multiple myeloma patients (n = 11). Each dot corresponds to one donor. Horizontal bars represent means (*P < .05).
Discussion
In this study, we have identified a novel subset of human CD1d-restricted T cells that recognize LPC. LPC is generated by the action of phospholipase A2 on PC. It is a major constituent of oxidized low density lipoprotein in atherosclerotic plaques, and elevated levels of lysophospholipids have been observed in allergic and autoimmune inflammation, asthma, and human cancer, including in MM.25,26,30,31 Therefore, the findings made in this study may apply to the putative role for CD1d-restricted T cells in diverse inflammatory states.
Staining human T cells with LPC-loaded CD1d dimers was inhibited in the presence of PC. However, in contrast to LPC, CD1d dimers loaded with PC did not stain human T cells. Although binding some T-cell lines to murine CD1d molecules with PC or PI species has been observed, these ligands have not been shown to be recognized by bulk populations of human or murine T cells.10-13,32-34 These lipids may, however, play important chaperone-like roles in intracellular assembly of CD1 molecules.13 Our data are consistent with a model wherein the recognition of CD1d by these T cells may depend on the balance between PC and LPC, suggested to be a sensitive indicator of inflammation.35 These studies therefore support the hypothesis that some CD1d-restricted cells may serve as a cellular sensor system for the detection of inflammation in tissues. In this regard, it is notable that several species of LPC exist in vivo,26 and the differences in acyl chains may lead to different functional consequences.36 Inflammation is a common feature of many human cancers and can lead to high levels of several species of LPC or related ligands in the malignant tissue.26 Therefore, the generation of LPC may be a mechanism by which inflammation might paralyze innate immunity against cancer.18
The majority of cells that bind LPC-loaded CD1d dimers in humans were Vα24−Vβ11− T cells.3 Such CD1d-restricted T cells have been termed type II NKT cells. It is of interest that the cytokine profile of these cells after TCR-based stimulation is somewhat skewed toward IL-13. Indeed, IL-13 was the only cytokine detected after ligand-specific stimulation in culture. Type II NKT cells have been implicated in suppressing tumor immune surveillance in mice in an IL-13–dependent manner in settings where the release of such ligands (eg, release of platelet activating factor, a structurally similar lipid, after UV-induced injury) might occur in vivo.14,37 IL-13–producing type II NKT cells have also been observed in the involved tissues in patients with ulcerative colitis.38 The preferential production of IL-13 by LPC specific T cells is also of interest because these cytokines have been implicated in promoting tumor growth, fibrosis, and angiogenesis in malignant tissues and inflammation.39,40 Specific targeting of these cells may therefore provide novel approaches to regulate inflammation and innate immunity in the clinic.
LPC is also known to be a major target of naturally occurring antibodies that recognize oxidized low-density lipoprotein and molecular motifs on dying cells.41 Interestingly, recent studies have begun to implicate such autoreactive B cells as preferred targets of transformation in human B-cell tumors.42 Therefore, the detection of increased levels of LPC specific T cells in myeloma patients may be of additional and more direct relevance to the pathogenesis of myeloma. Further studies are needed to evaluate the potential role of lipid antigens in driving the origin of B-cell tumors and to evaluate lipid reactive T cells in related B-cell tumors, particularly chronic lymphocytic leukemia.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ralph M. Steinman for his mentorship, Philip Livingston and Kavita M. Dhodapkar for discussions about this paper, Judy Adams for help with figures, Arlene Hurley, RN, for help with clinical aspects, John Gathuru for technical assistance, and members of the Dhodapkar laboratory for many helpful discussions.
This work was supported in part by funds from the National Institutes of Health (CA106802, CA109465, Rockefeller Clinical and Translational Science Award), Damon Runyon Cancer Research Fund, and Dana Foundation. D.H.C. is a Human Immunology Fellow of the Dana Foundation/CRI-Irvington Institute.
National Institutes of Health
Authorship
Contribution: D.H.C. performed research, analyzed data, and helped write and prepare the manuscript; H.D., P.M., J.K., G.R., and R.S. performed some aspects of research; A.M., D.H.V., and S.J. performed clinical research; and M.V.D. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Madhav V. Dhodapkar, Section of Hematology, Yale University, 333 Cedar Street, New Haven, CT 06510; e-mail: madhav.dhodapkar@yale.edu.

![Figure 1. Isolation and identification of CD1d-binding lipids from myeloma patient plasma. (A,B) ESI-mass spectra of lipid species from bulk extracts (A) and CD1d-conjugated bead eluents (B) from plasma of 2 myeloma patients (MM1 and MM2). (C) ESI-mass spectra of eluents from beads only control and CD1d beads, spiked with an LPC species (LPC-C14, m/z 368, arrow) as an internal reference. Data shown are representative of findings on 4 separate patients. (D,E) Tandem mass spectra (MS/MS) of major species observed for eluents of myeloma patient studied in panel C. (F) MS/MS of 496.3 of [M + H]+. (G) MS/MS of 480.3 of [M-15]. (F) Structure of 1-hexadecanoly-2-hydroxy-sn-glycero-3-phosphocholine, or lysophosphatidylcholine (LPC)-C16:0. (G) Bulk lipids isolated from myeloma patients or healthy donors were used to load CD1d dimers and stain cultures of human T cells as described in “Loading CD1d dimers and detection of CD1d-lipid–reactive T cells.” Numbers represent percentage of cells in the quadrant.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/112/4/10.1182_blood-2008-04-149831/6/m_zh80160822650001.jpeg?Expires=1764985938&Signature=2i-wkOA9vKEOKfPnPbP~jMrIDOj2GI2rR7MgwCxTgmdf~FZdsZo6PJrVMY4N6EnK8DVcexUMG43-wqo7Did9qiu1xWGxZEOLiMKUcShNBCZagDNj-Q2vtc8CgE7l12zopB681pQoqBJEpy0aBkP9E66ksvbER42jw8Q~wD34FiI5kGD0IF5jHtNQqB2UH~D9di~7t~fIW-om-6LdTcsAOtNvD-UxxRcW-4YjDw6Fy5uSb0w3fv92~Lj5kBrAPBnGE-SggSiXDIRfRjJdN~Sf0syZ2otaZ3cuBRx5IfM7f77JZJqXUIlYzgtVpGYYeWu09OGLlyAyWOjgMTjjDOTZRg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




![Figure 1. Isolation and identification of CD1d-binding lipids from myeloma patient plasma. (A,B) ESI-mass spectra of lipid species from bulk extracts (A) and CD1d-conjugated bead eluents (B) from plasma of 2 myeloma patients (MM1 and MM2). (C) ESI-mass spectra of eluents from beads only control and CD1d beads, spiked with an LPC species (LPC-C14, m/z 368, arrow) as an internal reference. Data shown are representative of findings on 4 separate patients. (D,E) Tandem mass spectra (MS/MS) of major species observed for eluents of myeloma patient studied in panel C. (F) MS/MS of 496.3 of [M + H]+. (G) MS/MS of 480.3 of [M-15]. (F) Structure of 1-hexadecanoly-2-hydroxy-sn-glycero-3-phosphocholine, or lysophosphatidylcholine (LPC)-C16:0. (G) Bulk lipids isolated from myeloma patients or healthy donors were used to load CD1d dimers and stain cultures of human T cells as described in “Loading CD1d dimers and detection of CD1d-lipid–reactive T cells.” Numbers represent percentage of cells in the quadrant.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/112/4/10.1182_blood-2008-04-149831/6/m_zh80160822650001.jpeg?Expires=1765298107&Signature=buz7tXue-QXWEXO3-3WJQXFm3BJsA2S3DZUBDxi9f5PaOBMIbG1UCyKHK8Qz0fHTnR7A6cfYxKSqzvs4lcE6QKe~BnETcSf-f-e-ZDqsQy5rUMnt5kg3H-8n2nSE7q2N2-OYAcKTmc0uhjZavcwST6~ZrOI6z0RySE61-Wanz3e4AdcUbDTWo7IZGr8jw4FjuGwpOzNXxAkgPtOiyGlQdB-hAjByVhro2Y0Ju6B8znzj3S8XzWA2QCsR7EKRSzJzhoG6u5N4zGnN33~T3THOk~dZOPJFMDcKsoLsrkiBZvtm8TcXQeBO7LhkyNqk4-0doTfyOgkvD4i7PramBTmD2w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)