Abstract
DNA methylation of CpG islands around gene transcription start sites results in gene silencing and plays a role in leukemia pathophysiology. Its impact in leukemia progression is not fully understood. We performed genomewide screening for methylated CpG islands and identified 8 genes frequently methylated in leukemia cell lines and in patients with acute myeloid leukemia (AML): NOR1, CDH13, p15, NPM2, OLIG2, PGR, HIN1, and SLC26A4. We assessed the methylation status of these genes and of the repetitive element LINE-1 in 30 patients with AML, both at diagnosis and relapse. Abnormal methylation was found in 23% to 83% of patients at diagnosis and in 47% to 93% at relapse, with CDH13 being the most frequently methylated. We observed concordance in methylation of several genes, confirming the presence of a hypermethylator pathway in AML. DNA methylation levels increased at relapse in 25 of 30 (83%) patients with AML. These changes represent much larger epigenetic dysregulation, since methylation microarray analysis of 9008 autosomal genes in 4 patients showed hypermethylation ranging from 5.9% to 13.6% (median 8.3%) genes at diagnosis and 8.0% to 15.2% (median 10.6%) genes in relapse (P < .001). Our data suggest that DNA methylation is involved in AML progression and provide a rationale for the use of epigenetic agents in remission maintenance.
Introduction
Acute myeloid leukemia (AML) is a hematologic malignancy with frequent nonrandom singular or multiple genetic changes. Various gene translocations and mutations are involved in specific AML subtypes and structurally altered genes play a role in AML development and affect its prognosis.1 The importance of epigenetics in the pathogenesis of leukemia is gaining recognition.2,3 The methylation of cytosines in the palindromic CpG sites clustered in gene promoter regions is important in epigenetic silencing; methylation also plays a role in the aging process and acts as an alternative mechanism of tumor suppressor inactivation in cancer.4,5 DNA methylation affects not only the development of leukemia but also its progression and relapse. Methylation of the tumor suppressor gene p15 is the most frequently reported epigenetic event in myeloid malignancies and is frequently used to assess treatment success.6,7 Numerous other genes are methylated in AML,2,8-17 but the effects of these changes on the development, progression, and relapse of disease are not completely understood.
In this study, we used bisulfite pyrosequencing to determine the DNA methylation status of CpG islands of genes previously cloned by methylated CpG island amplification coupled with representational difference analysis (MCA/RDA).18-21 We confirmed that aberrant methylation was frequent in AML and also showed that methylation levels of all genes except of the LINE-1 repetitive element were higher at relapse than at diagnosis. This phenomenon was more apparent in patients with stable karyotypes compared with patients who developed additional genetic abnormalities at relapse. Our results indicate that DNA methylation changes are important factors in disease progression and/or chemotherapy resistance in AML.
Methods
Patients
We examined bone marrow or peripheral blood cells from 30 patients with AML who had been treated at the University of Texas M. D. Anderson Cancer Center (Houston, TX) from 1998 to 2004. All patients received high-dose chemotherapy containing Ara-C combined with idarubicin (11 patients), daunorubicin (6 patients), fludarabine (5 patients), cyclophosphamide and topotecan (7 patients), and troxacitabine (1 patient). None of the patients received DNA-demethylating drugs. Available samples from patients who had achieved complete remission but had later relapsed were identified through a database search. The samples were obtained from established tissue banks at M. D. Anderson Cancer Center and included Ficoll-separated mononuclear cells that had been stored at −80°C (14 samples) and bone marrow that had been embedded in paraffin (46 samples). The patients were divided into 3 cytogenetic risk groups: good (inv 16, del 16q), intermediate (normal karyotype, +4, +6, +8, +11, +13, +14, +22, including 1 or 2 of these abnormalities), and poor (inv 9 or inv 17, t(6,9), t(1,6), −5/del 5q, −7, and complex karyotype with 3 or more chromosomal abnormalities).1 Alterations of 11q23 were not detected in any patient. For normal controls, peripheral blood mononuclear cells were obtained from 26 healthy volunteers (18 to 53 years of age). DNA was isolated from cells and from paraffin-embedded tissue as previously described.22 The institutional review board at M.D. Anderson approved all protocols, and all patients gave informed consent for the collection of residual tissues as per institutional guidelines and in accordance with the Declaration of Helsinki.
Cell lines
The leukemia cell lines used in this study (BV173R, HL60, HEL, K562, KG-1, KG-1a, ML-1, MV4:11, KBM5R, OCI-AML3, TF-1, CEM, JTAg, Jurkat, MOLT-4, T-ALL, Peer, ALL1, BJAB, Raji, and RS4;11) were obtained from the ATCC (Manassas, VA) and M. D. Anderson repositories.
DNA methylation analysis
DNA was extracted and treated with bisulfite, as reported previously.21,23 Genomic DNA (2 μg) was denatured by 0.2 M NaOH at 37°C for 10 minutes followed by incubation with freshly prepared 30 μL of 10 mM hydroquinone and 520 μL of 3 M sodium bisulfite (pH 5.0) at 50°C for 16 hours. DNA was purified with a Wizard Miniprep Column (Promega, Madison, WI), desulfonated with 0.3 M NaOH at 25°C for 5 minutes, precipitated with ammonium acetate and ethanol, and dissolved in 50 μL of TE buffer (Tris-HCl 10 mM, EDTA 1 mM, pH 8.0). Bisulfite-treated DNA (40-80 ng) was amplified with gene-specific primers in a 2-step polymerase chain reaction (PCR). The second step of PCR was used to label single DNA strand with biotin using a universal primer tag24 or gene-specific primers biotinylated at the 5′ end. Each PCR step was performed in a total volume of 20 μL of 67 mM Tris-HCl (pH = 8.8), 16 mM ammonium sulfate, 2 mM MgCl2, 0.125 mM dNTPs, 1 U Taq polymerase, and 100 nM PCR primers. TQ30 oligonucleotide (50 nM) was used as a reversible inhibitor of Taqpolymerase in the first step of the PCR.25
The PCR primer sequences and annealing temperatures used are listed in Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article. The following PCR conditions were used: initial denaturation at 95°C for 5 minutes, followed by 40 cycles comprising denaturation at 94°C for 15 seconds, annealing at the appropriate temperature for 30 seconds, and extension at 72°C for 15 seconds. We used 45 cycles for the second step to completely exhaust the biotinylated primer. We measured levels of DNA methylation as the percentage of bisulfite-resistant cytosines at CpG sites by pyrosequencing with the PSQ HS 96 Pyrosequencing System (Biotage, Charlottesville, VA) and Pyro Gold CDT Reagents (Biotage) as previously described.24 A pyrosequencing assay typically interrogates 2 to 8 adjacent CpG sites. We found high concordance in methylation between adjacent sites and we therefore used mean values from all pyrosequenced CpG sites as a measure of methylation of a given gene. For each assay, we determined 95% confidence intervals of normal values by measuring the DNA methylation levels in the controls. Methylation values above this interval were considered abnormal. Single assays close to transcription start sites were used for NOR1, CDH13, p15, NPM2, OLIG2, and HIN1 genes. We used 2 assays for SLC26A4: (−15), which was positioned at CpGs located −32 to −15 bases from transcription start site, and (+355), which was in the region cloned by MCA/RDA with CpG sites 355 to 365 bases downstream from transcription start site. The progesterone receptor gene has 2 CpG islands corresponding to its 2 isoforms: a short A and a long B isoform (PGRA and PGRB); thus, we tested both CpG islands. We also measured DNA methylation at the 5′ end of the LINE-1 repeat (GenBank accession number X58075) to assess global methylation levels.26,27 Maps of the investigated CpG islands are shown in Figure S1. Typical pyrograms are shown in Figure S2.
Methylated CpG island microarray (MCAM) analysis
We used gDNA from the bone marrow samples of 4 patients with AML, obtained at the time of the initial diagnosis and at the time of the first relapse. A gDNA pool made from blood cells of 4 healthy donors was used as a control. Methylated CpG island amplification was performed as described.18 Amplicons from patients with AML were labeled with the Cy5 dye and cohybridized against amplicons from pooled blood cells from 4 healthy donors labeled with the Cy3 dye on Agilent Technologies 4 × 44K custom DNA microarrays (Agilent, Santa Clara, CA) as described previously.28,29 This method allows parallel analysis of 42 222 probes corresponding to 9008 autosomal genes. The probes on the array were selected to recognize SmaI/XmaIfragments, mostly around gene transcription start sites.
Statistical analysis
We used the Wilcoxon matched pairs test and χ2 test to compare methylation levels at diagnosis and relapse. The Spearman nonparametric correlation test was used to compare methylation and clinical data. Survival was computed using the Kaplan-Meier analysis. A 2-tailed P value of .05 or less was considered statistically significant.
To determine the combined significance of all genes studied for methylation, we calculated the methylation z score. The z score was derived by subtracting the population mean from an individual raw value and then dividing the difference by the population standard deviation. It allowed us to compare observations from different genes as well as of all studied genes in individual patients at AML diagnosis or relapse.
Results
DNA methylation of CpG islands cloned by MCA/RDA in leukemia cell lines
Methylated CpG island amplification (MCA) coupled with representational difference analysis (RDA) is an unbiased method to identify CG-rich DNA fragments hypermethylated in cancer.18 We previously used MCA/RDA to identify more than 40 genes as hypermethylated in hematologic malignancies19-21 and selected 14 genes along with the p15 tumor suppressor gene and LINE-1 repetitive element for detailed analysis by bisulfite pyrosequencing.
We assessed the methylation status of these 15 genes in 21 leukemia cell lines. The results are summarized in Figure S3. We found abnormal methylation in a minimum of 9 of 15 genes in the leukemia cell lines KG-1 and OCI-AML3 and in a maximum of 13 of 15 genes in the KG-1a, ML-1, T-ALL, and Raji leukemia cell lines. NPM2 gene was methylated in all cell lines, whereas FADS2 and TYSND1 genes were methylated in only 2 cell lines each (K562 and Raji, and K562 and TF-1, respectively). The tumor suppressor gene p15 was deleted in the myeloid leukemia cell lines K562 and KBM5R and the lymphoid leukemia cell lines JTAg and RS4;11.
Next, we selected 9 loci representing the 7 genes most frequently methylated in leukemia cell lines (NOR1, CDH13, NPM2, OLIG2, PGRA, PGRB, HIN1, SLC26A4 [−15], SLC26A4 [+355], and p15) to determine DNA methylation in patients with AML at diagnosis and relapse (Table 1). We also determined the methylation of the LINE-1 repetitive element, which is present in the human genome in multiple copies and is a surrogate marker of global genomic methylation.26,30
DNA methylation in AML at diagnosis and relapse
The clinical characteristics of the studied group of 30 patients with AML are shown in Table 2. The median age of patients was 56 years, M2 was the most common subtype on the basis of the French-American-British (FAB) classification, and the majority of patients had poor cytogenetics (57%). The median time from diagnosis to relapse was 11 months, and the median overall survival duration from diagnosis was 18 months.
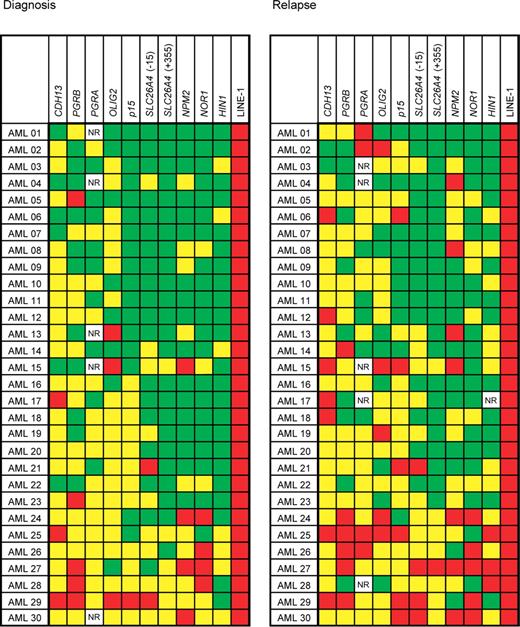
We measured levels of DNA methylation in the CpG islands or CpG-rich regions of 9 loci and the LINE-1 repetitive element by bisulfite pyrosequencing. The results are summarized in Figure 1. We observed abnormal methylation values in varying proportions of AML patients for every gene studied. At diagnosis, abnormally increased methylation was most common in CDH13 (25 of 30 patients [83%]), followed by OLIG2 (23 of 30 patients [77%]), PGRA (18 of 25 patients [72%]; no result was obtained in 5 patients), PGRB (21 of 30 patients [70%]), NOR1 (19 of 30 patients [63%]), NPM2 (19 of 30 patients [63%]), p15 (14 of 30 patients [47%]), SLC26A4(−15) (12 of 30 patients [40%]), SLC26A4(+355) (9 of 30 patients [30%]), and HIN1 (7 of 30 patients [23%]). The frequencies of abnormal methylation further increased at relapse. The most common hypermethylated gene was again CDH13 (28 of 30 patients [93%]), followed by PGRA (23 of 25 patients [92%]; no result was obtained in 5 patients), p15 (24 of 30 patients [80%]), NPM2 (22 of 30 patients [73%]), OLIG2 (21 of 30 patients [70%]), PGRB (20 of 30 patients [67%]), NOR1 (19 of 30 patients [63%]), SLC26A4(−15) (19 of 30 patients [63%]), SLC26A4(+355) (14 of 30 patients [47%]), and HIN1 (14 of 30 patients [47%]).
DNA methylation in AML at diagnosis and relapse. Methylation in normal peripheral blood controls is shown as box plots with bars indicating 5% to 95% confidence intervals. Methylation densities in individual patients with AML and their changes are shown in the second and third columns. PGRA, PGRB, CDH13, and OLIG2 genes were methylated above the cutoff at diagnosis in most patients and their methylation densities further increased at relapse. LINE-1 repetitive element showed a higher methylation in AML compared with normal peripheral blood controls and a slight decrease in methylation at relapse compared with diagnosis.
DNA methylation in AML at diagnosis and relapse. Methylation in normal peripheral blood controls is shown as box plots with bars indicating 5% to 95% confidence intervals. Methylation densities in individual patients with AML and their changes are shown in the second and third columns. PGRA, PGRB, CDH13, and OLIG2 genes were methylated above the cutoff at diagnosis in most patients and their methylation densities further increased at relapse. LINE-1 repetitive element showed a higher methylation in AML compared with normal peripheral blood controls and a slight decrease in methylation at relapse compared with diagnosis.
We assessed the degree of concordance in DNA methylation of individual genes at diagnosis. A positive correlation was observed in 11 of 55 (20%) gene pairs. The highest correlation was observed between methylation of NOR1 and NPM2 (r = .693, P < .001). The only negative correlation of 55 gene pairs was observed between p15 and LINE-1 (r = −.551, P = .002). Correlation of methylation between individual genes is summarized in Table S2.
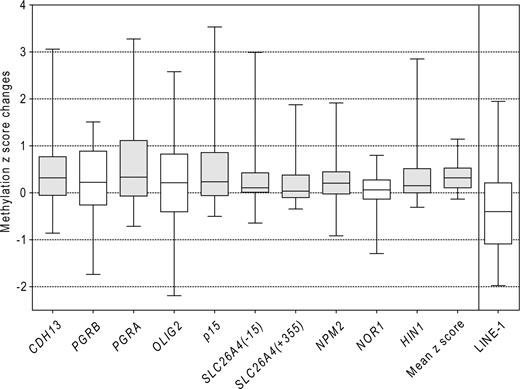
All patients had abnormal methylation in at least 1 of the 9 genes at diagnosis and in at least 2 genes at relapse. The median number of genes methylated at diagnosis was 4, which increased to 6 at relapse, P < .001 (Figure 2). In addition to the methylation frequency (ie, the number of abnormally methylated genes) the quantitative aspect of pyrosequencing allowed us to study methylation density (ie, the percentage of CpG methylation). The increase in methylation density at relapse was statistically significant (P < .05) for CDH13 (mean increase of 10%), p15 (15%), NPM2 (7%), PGRA (14%), HIN1 (9%), and SLC26A4 (7%) genes. We also calculated the composite mean methylation z score for each patient, which included all genes studied. The methylation z score increased by +0.340 at relapse. This finding was statistically significant (P < .001). These results are shown in Figure 3.
DNA methylation in patients with AML at diagnosis and relapse. Patients are arranged horizontally, genes vertically. Average methylation densities of 0% to 10% are shown in green (0%-15% for PGRB), 10% to 50% methylation in yellow (15%-50% for PGRB); and methylation over 50% in red. NR, no result. We observed increased methylation in AML at diagnosis in 144 of 295 data points and in 175 of 294 data points at relapse.
DNA methylation in patients with AML at diagnosis and relapse. Patients are arranged horizontally, genes vertically. Average methylation densities of 0% to 10% are shown in green (0%-15% for PGRB), 10% to 50% methylation in yellow (15%-50% for PGRB); and methylation over 50% in red. NR, no result. We observed increased methylation in AML at diagnosis in 144 of 295 data points and in 175 of 294 data points at relapse.
Changes in methylation between relapse and diagnosis. Changes in methylation z scores between relapse and diagnosis are shown as box plots. The boxes represent the 25% to 75% interquartile range, horizontal lines inside the boxes show the medians, and vertical bars show the 5% to 95% range. Gray boxes denote a significant increase in methylation in relapse (P < .05, Wilcoxon signed rank test). Mean z score was calculated as the mean of individual gene z scores. All genes showed increase in methylation at relapse with the exception of the LINE-1 repetitive element.
Changes in methylation between relapse and diagnosis. Changes in methylation z scores between relapse and diagnosis are shown as box plots. The boxes represent the 25% to 75% interquartile range, horizontal lines inside the boxes show the medians, and vertical bars show the 5% to 95% range. Gray boxes denote a significant increase in methylation in relapse (P < .05, Wilcoxon signed rank test). Mean z score was calculated as the mean of individual gene z scores. All genes showed increase in methylation at relapse with the exception of the LINE-1 repetitive element.
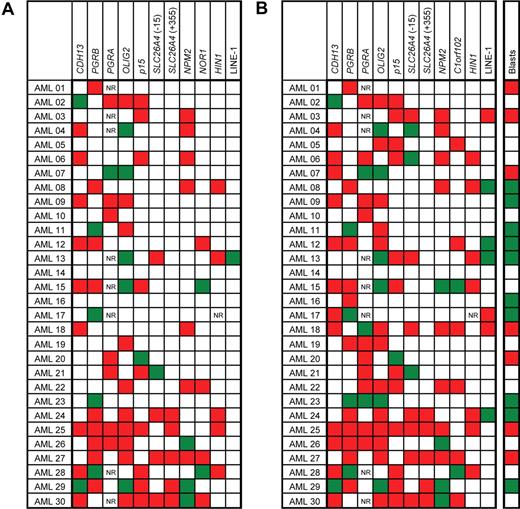
The changes in DNA methylation status in individual patients between diagnosis and relapse are shown in Figure 4. We observed a new appearance of abnormal methylation over the 95% confidence control limit in 51 of 291 measurements (ie, the total number of paired methylation values compared), the disappearance of methylation in 20 of 291 measurements and no change in the methylation status in 220 of 291 measurements (P = .07, χ2 test; Figure 4A). The highest increase in newly appearing methylation was observed in HIN1 (8 of 29 measurements), followed by p15 and NPM2, with 7 of 30 measurements for each gene. The disappearance of methylation at relapse was never observed in p15 gene and SLC26A4(+355) locus. Similarly, HIN1 and SLC26A4 (−15) showed a loss of aberrant methylation at relapse in only 1 patient each (Figure 4A).
Changes in methylation between diagnosis and relapse in patients with AML. (A) Categorical changes. Red boxes denote changes from unmethylated at diagnosis to methylated at relapse. Green boxes show genes methylated at diagnosis and unmethylated at relapse. White boxes denote no change in methylation status between diagnosis and relapse. NR, no result available. Genes were considered methylated if their methylation density exceeded the 95% confidence interval established in normal controls. (B) Differences in methylation densities. Red boxes denote an increase in methylation density at relapse of more than 10% when compared with diagnosis; green boxes show decrease of methylation density at relapse over 10%, and white boxes denote methylation changes within the 10% interval. Blasts, bone marrow blast count changes between the diagnosis and relapse. Red boxes denote an increase in blasts at relapse of more than 10% when compared with diagnosis; green boxes show a decrease of blasts at relapse over 10%; and white boxes denote changes of blasts within the 10% interval. Changes in blast counts did not correlate with methylation changes (Spearman nonparametric test not significant).
Changes in methylation between diagnosis and relapse in patients with AML. (A) Categorical changes. Red boxes denote changes from unmethylated at diagnosis to methylated at relapse. Green boxes show genes methylated at diagnosis and unmethylated at relapse. White boxes denote no change in methylation status between diagnosis and relapse. NR, no result available. Genes were considered methylated if their methylation density exceeded the 95% confidence interval established in normal controls. (B) Differences in methylation densities. Red boxes denote an increase in methylation density at relapse of more than 10% when compared with diagnosis; green boxes show decrease of methylation density at relapse over 10%, and white boxes denote methylation changes within the 10% interval. Blasts, bone marrow blast count changes between the diagnosis and relapse. Red boxes denote an increase in blasts at relapse of more than 10% when compared with diagnosis; green boxes show a decrease of blasts at relapse over 10%; and white boxes denote changes of blasts within the 10% interval. Changes in blast counts did not correlate with methylation changes (Spearman nonparametric test not significant).
We also analyzed the changes in methylation between the diagnosis and relapse using a cutoff of 10% for the difference in methylation densities (Figure 4B). This level is approximately 3 times higher than the value of standard deviation in our assays and is thus likely significant. Increased methylation at relapse was found in 91 of 291 (31%) measurements, and a decrease was seen in 25 of 291 (9%) measurements. No changes outside the 10% difference interval were found in the remaining 175 of 291 (60%) measurements (P = .02, χ2 test; Figure 4B). There was no correlation between changes in bone marrow blast counts and changes in methylation.
Methylation of the LINE-1 repetitive element
We did not find significant differences in methylation of the LINE-1 repetitive element in patients at diagnosis compared with healthy controls (median 73% vs 67%, respectively; P = .15, Mann-Whitney test; Figure 1), suggesting that AML is not characterized by global hypomethylation of repetitive sequences. Mean LINE-1 methylation levels at diagnosis (median 74%) were similar to the levels at relapse (median 72%, P =.15, Figure 4).
Association between clinicopathologic features and DNA methylation
No differences were found in clinicopathologic features between diagnosis and relapse, including leukemic blast counts. Bone marrow and peripheral blood blast counts were positively associated with increased NOR1 and NPM2 methylation levels, both at diagnosis and relapse (Spearman nonparametric test, P < .05). Methylation of these genes was also associated with decreased platelet counts at diagnosis (P < .05). Higher methylation of p15 at diagnosis was associated with younger age (P = .02). We observed no association between methylation at diagnosis or at relapse and survival in our group of patients who where all selected based on having relapsed.
Cytogenetics and DNA methylation
DNA methylation status at diagnosis or relapse and methylation changes between the diagnosis and relapse were independent of the 3 cytogenetic risk groups: good, intermediate, and poor (data not shown). To compare the changes of DNA methylation as an epigenetic phenomenon with genetic changes manifested as karyotype instability, we divided the patients into 2 groups: those with a stable karyotype (no cytogenetic changes at relapse) and those with an unstable karyotype (a gain of chromosomal abnormalities between diagnosis and relapse). Unexpectedly, the mean methylation z score at relapse was higher in patients with a stable karyotype than in patients with an unstable karyotype (median z score, 0.54 vs 0.16 [P = .07]). In the stable karyotype group, we observed a significant increase in CDH13 (median z score, −0.04 at diagnosis vs 1.14 at relapse [P = .04]) and HIN1 methylation (median z score, −0.46 at diagnosis vs 0.54 at relapse [P = .03]) with disease progression. Interestingly, an increased methylation z score of the tumor suppressor gene p15 was seen at diagnosis in patients with unstable karyotypes (median 0.19 in the unstable karyotype vs −0.52 in the stable karyotype group [P = .04]).
Methylated CpG islands microarrays (MCAMs)
We analyzed genomewide methylation pattern by MCAM in 4 patients with AML and found on average 812 genes methylated at diagnosis and 999 genes methylated in relapse (range 530-1228 genes methylated at diagnosis and 724-1373 genes methylated in relapse). These values represent a range of 5.9% to 13.6% and 8.0% to 15.2%, respectively, for diagnosis and relapse, of the total number of 9008 autosomal genes present on the microarray. Thus, the number of methylated genes significantly increased in relapse in each patient (Table 3). Next we evaluated genes that were simultaneously hypermethylated in at least 3 patients with leukemia and found that 283 of 9008 (3.1%) autosomal genes were hypermethylated at diagnosis while 562 of 9008 (5.8%) genes were hypermethylated in relapse. This 86% increase in the number of methylated genes was significant by the Chi-square test (P < .001) and supports the role of increasing methylation in leukemia progression. We performed the same analysis for the subset of amplicons located inside of CpG islands and detected by probes within the interval from −2000 to +500 bp from the transcription start site. We found that 182 of 4710 (3.9%) genes were methylated at AML diagnosis and 224 of 4710 (4.8%) genes were methylated in relapse (Figure 5). Similarly to the previous result from the whole gene set, the number of genes methylated in CpG islands near transcription start sites increased by 23% in relapse. The difference between methylation at diagnosis and in relapse was significant (P = .03). Lists of genes found consistently hypermethylated at diagnosis, relapse, or both in 3 or 4 patients with AML are in Tables S3 through S5. Five genes analyzed by bisulfite pyrosequencing were also present on our microarrays (HIN1, NOR1, NPM2, OLIG2, and PGR). Microarray data were in a good agreement with bisulfite pyrosequencing results showing 100% specificity and 69% sensitivity (Table S6). Our results suggest that methylation increase in AML progression affects in a similar fashion CpG sites that are close to transcription start sites and the more distant ones.
Microarray analysis of genes hypermethylated in AML. The Venn diagram shows the overlap and differences in genes methylated at the time of diagnosis and the first relapse in at least 3 of 4 patients with AML studied. A total number of 4710 genes was analyzed by 16 475 microarray probes recognizing CpG islands near gene transcription start sites.
Microarray analysis of genes hypermethylated in AML. The Venn diagram shows the overlap and differences in genes methylated at the time of diagnosis and the first relapse in at least 3 of 4 patients with AML studied. A total number of 4710 genes was analyzed by 16 475 microarray probes recognizing CpG islands near gene transcription start sites.
Discussion
DNA methylation and modifications of histone tails are key cooperating mechanisms in maintaining epigenetic memory in mammalian cells. Along with genetic alterations, epigenetic abnormalities play an important role in gene deregulation in cancer.5 Aberrant hypermethylation in cancer or leukemia cells may affect hundreds of promoter-associated CpG islands and cause stable epigenetic silencing of methylated genes. Many genes that are methylated in tumors are not expressed in relevant normal tissues, but silencing genes that are critically important to controlling cell proliferation contributes to the development of a malignant phenotype in the same manner as inactivating mutations of tumor suppressor genes.31 In leukemia, epigenetic silencing by DNA methylation of cyclin-dependent kinase inhibitors,6,9,32,33 DNA repair genes,15 apoptosis mediators,12,13 nuclear receptors,8,10,14 transcription factors,16 cell adhesion molecules,34 and many other genes has been reported.2,11,17
We investigated the DNA methylation status of 15 genes in 21 leukemia cell lines using bisulfite pyrosequencing. We further evaluated 9 CpG islands in primary leukemia cells and documented their methylation in 23% to 83% patients with AML at diagnosis in this small, selected group of 30 patients. The frequency and density of methylation in studied genes was further increased at relapse. We performed genomewide analysis of genes methylated at the time of diagnosis and first relapse in 4 AML patients using the MCAM microarray method. We found a similar increase (182 to 224 genes) in the number of genes consistently methylated in 3 or 4 patients when looking at CpG islands near transcription start sites of a 4710 set of autosomal genes. Functional significance of these genes in AML needs to be established; however, they can be regarded as markers associated with AML relapse and poor survival. Moreover, our microarray experiments detected methylation of candidate tumor suppressor genes previously reported as deleted or methylated in leukemia, such as ID4.35 Our findings indicate that DNA methylation is an important factor affecting hundreds of genes and likely involved in disease progression and in chemotherapy resistance.
Whether gene methylation affects the decisionmaking functions of hematopoietic stem cells or is just a signature of epigenetic instability is unknown. p15 cell-cycle inhibitor is expressed at low levels in the bone marrow and peripheral blood, and its expression has been shown to decrease with CpG hypermethylation in patients with leukemia.36 Other studied genes have low or undetectable expression in blood or bone marrow cells (data not shown). Methylation of p15, PGR, and CDH13 has been described in leukemia.6,10,34 Although PGR and CDH13 genes are not significantly expressed in mature blood cells, their expression or silencing may be important in the balance of stem cell self-renewal and differentiation. In most patients, methylation of multiple genes was observed, suggesting a general disturbance of epigenetic memory in AML and confirming the findings of the CpG island methylator phenotype37 in leukemia.2,38,39 The mechanism of concordant methylation of multiple genes is unknown. Hypermethylation of multiple genes is associated with poor prognosis in patients with acute lymphoblastic leukemia.38
We found that levels of DNA methylation increased in most patients with relapsed AML, particularly in those with the stable karyotype, suggesting that an epigenetic instability process is involved in the disease progression. Conversely, chromosomal instability may be the major driving force behind the development of additional cytogenetic changes at relapse. It is interesting that we saw differences in the degree of methylation levels between the patients with versus without chromosomal instability. This supports our hypothesis that epigenetic and chromosomal instability may be inversely correlated in AML. Our data are only suggestive and based on a small number of patients, thus requiring future validation in a large cohort of patients. Additionally, only patients with AML who subsequently relapsed were included in the study and may not accurately represent the whole spectrum of patients with AML.
Understanding the nature and extent of DNA methylation and other epigenetic alterations in AML will help us develop strategies for the use of DNA-demethylating and histone-modifying agents in the treatment of AML. A direct implication of our findings is that hypomethylating agents may be useful in AML at remission to prevent the emergence of hypermethylated (and possibly drug-resistant) clones.
The online version of this article contains a data supplement.
Presented in part at the 48th annual meeting of the American Society of Hematology, Orlando, FL, December 9-12, 2006.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Ann M. Sutton for editorial help. This work was supported by Leukemia SPORE 5P50CA100632-05 and 5P01CA108631-03 grants from the National Institutes of Health. J.P.J.I. is an American Cancer Society Clinical Research Professor.
National Institutes of Health
Authorship
Contribution: J.P.J.I., J.J., H.M.K., and H.K. designed the research; C.E.B.R. selected, reviewed, and provided paraffin-embedded bone marrow specimens; H.K., J.J., and R.H. performed the experiments; M.R.H.E., K.K., and W.C. critically contributed to microarray experiments; L.Z. and L.S. designed and annotated custom microarrays tailored to the MCAM method; and H.K., J.J., M.R.H.E., and J.P.J.I. analyzed the data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jean-Pierre J. Issa, Department of Leukemia, Unit 428, The University of Texas M.D. Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: jpissa@mdanderson.org.
References
Author notes
*H.K. and J.J. contributed equally to this study.