Abstract
T-cell responses have been implicated in the development of HPA-1a–induced neonatal alloimmune thrombocytopenia (NAIT). However, HPA-1a–specific T cells have neither been isolated nor characterized. Here, we aimed to determine whether HPA-1a–specific T cells could be isolated from HPA-1a–immunized women. In the present study, peripheral blood mononuclear cells (PBMCs) from an HPA-1a–alloimmunized woman were cultured for weeks in the presence of HPA-1a peptide, labeled with CFSE, and assayed for antigen-specific proliferation. Individual proliferating cells were isolated by fluorescence-activated cell sorting and expanded in culture. Antigen specificity and HLA restriction were determined by cytokine secretion (enzyme-linked immunospot [ELISPOT]) and proliferation assays. Several CD3+CD4+ T-cell clones were isolated that proliferated and secreted cytokines in response to HPA-1a peptide. Two of these clones have been established in long-term culture in our laboratory. Both of these recognize synthetic as well as naturally processed HPA-1a antigen, and the recognition is restricted by the MHC molecule HLA-DRB3*0101 that is strongly associated with NAIT. These HPA-1a–specific T-cell clones represent unambiguous evidence for the association of T-cell responses with NAIT, and they will serve as unique tools to elucidate the cellular immune response that may result in NAIT.
Introduction
Neonatal alloimmune thrombocytopenia (NAIT) results from maternal alloantibodies reactive with fetal platelet antigens. These antibodies are transported across the placenta via a specific receptor, FcRn, on the syncytiotrophoblast during pregnancy and can cause depletion of fetal platelets and increase the risk of bleeding. NAIT can result in intracranial hemorrhage and death, and severely affected neonates may have brain damage and lifelong severe disability.1-3 There is no safe and effective treatment of the fetus, although IvIg and steroids given to the mother during pregnancy is reported to be efficient in many cases.4,5
The most common cause of NAIT is an incompatibility in the human platelet antigen 1 (HPA-1) between the mother and the fetus: in the alloantigen system, HPA-1 is defined as a leucine/proline polymorphism at residue 33 in the β3 integrin. In homozygous HPA-1bb (Pro33) women carrying a fetus positive for HPA-1a (Leu33), maternal antibodies against HPA-1a can be formed. Alloimmunization occurs in approximately 10% of HPA-1a–negative pregnant women and is strongly associated with HLA-DRB3*0101.1,2,6,7 Severe NAIT occurs at a rate of approximately 1 in 1100 to 1200 of all pregnancies.1,6 There is currently no treatment that can prevent immunization.
Although antibodies are produced by plasma cells differentiated from activated B cells, the initiation of antibody responses usually also depends on activation of antigen-specific T cells.
However, little is known about the underlying cellular responses that results in anti–HPA-1a antibody production. Particularly, T cells specific for the HPA-1a antigen have not been positively identified. Therefore, to examine the cellular immune responses associated with NAIT, we wanted to determine whether HPA-1a–specific T cells could be isolated from a woman that had given birth to a child with severe NAIT.
Methods
The study was approved by the Regional Committee for Medical Research Ethics, North Norway. Blood samples were drawn from patients and healthy volunteers after written informed consent was obtained in accordance with the Declaration of Helsinki.
Donors
An HPA-1bb woman, who gave birth to a neonate with severe thrombocytopenia, donated peripheral blood 1 day after delivery. The woman was blood group A and typed HLA-DRB1*0101 and *0301, HLA-DRB3*0101, HLA-DQB1*0201 and *0501, HLA-DPB1*0401 and *0402. The neonate was blood group AB, HPA-1a–positive, had a low platelet count (9 × 109/L), petechiae, and intracerebral bleedings. Maternal antibodies against paternal β3 integrin were barely detectable at delivery. Seven weeks postpartum, low levels of maternal anti–HPA-1a antibody were detected (1.25 IU/mL) by a modified monoclonal antibody immobilization of platelet antigen assay.8,9 The anti–HPA-1a internal standard was the same as previously published.9 Peripheral blood was also collected from healthy volunteers from the Blood Bank at the University Hospital of North Norway.
Synthetic peptides
Three 20-mer peptide sequences were used in this study (Table 1). The HPA-1a sequence (L33; integrin β3 19-38 with a leucine at residue 33) contains the 9-residue anchor sequence that is reported to associate with the HLA-DRB3*0101 MHC molecule10,11 flanked at each end by 5 and 6 additional residues. The flanking residues were added both to extend the peptide beyond the peptide-binding groove of the HLA-DRB3*0101 molecule, which conceivably could influence T-cell recognition, and to match the HPA-1a and Lol P1 sequences in length and relative position of the anchor residues. The Lol P1 peptide sequence has been shown experimentally to bind HLA-DRB3*010111 and as such was a relevant control peptide. The HPA-1b peptide (P33), which was also used as a control, is similar to the HPA-1a sequence with a proline instead of a leucine residue at position 33. This represents the allotypic difference between HPA-1a and HPA-1b. The leucine at position 33 of HPA-1a serves as one of the anchor residues for association to HLA-DRB3*0101. HPA-1b peptides, which lack the leucine anchor, associate poorly with HLA-DRB3*0101.11 Because of the proven binding of peptide Lol P1 to HLA-DRB3*0101, this peptide was considered superior to P33 as a control peptide, and Lol P1 was consistently used as such in all experiments. The peptides (synthesized by Eurogentec, Seraing, Belgium) were dissolved to 88 μM in 60% ethanol and 40% water and stored at −20°C.
Isolation of PBMCs, platelets, and monocytes
Peripheral blood was diluted 1:1 in phosphate-buffered saline (PBS) and layered over Lymphoprep medium (Axis-Shield, Dundee, United Kingdom) followed by 15 minutes of centrifugation at 800g without brakes. The interface was collected, and 40 mL 0.2% PBSA (0.2% bovine serum albumin in PBS) was added. A centrifugation at 400g for 6 minutes pelleted the peripheral blood mononuclear cells (PBMCs), whereas the PBSA supernatant contained coisolated platelets. The platelet-rich PBSA was layered over a 1.047 g/mL gradient of Optiprep (Axis-Shield) and PBS, followed by centrifugation at 400g for 10 minutes. The platelets at the interface were collected and washed in PBSA (0.2%) and pelleted by a 5-minute centrifugation at 950g for 10 minutes. Monocytes were isolated by mixing 108 PBMCs in 1.2 mL fetal bovine serum (FBS) along with 800 μL autologous red blood cells (RBCs) (from the Lymphoprep-separated blood) and 100 μL RosetteSep Human Monocyte Enrichment Cocktail (StemCell Technologies, Vancouver, BC) and incubated for 20 minutes at room temperature. The cell suspension was diluted with 2 mL PBS, and the suspension was layered on top of 3 mL Lymphoprep and centrifuged at 800g for 15 minutes without brakes. Monocytes were collected at the interface and washed twice in PBSA (0.2%).
Cell culture and generation of HPA-1a–specific T-cell lines
PBMCs from an alloimmunized woman were cultured in complete medium (CM) consisting of Iscove modified Dulbecco medium (IMDM), 10% FBS, 4% human serum (from HPA-1a–negative donors), and penicillin-streptomycin at 37°C, in a 7.5% CO2 humidified atmosphere. The cells were stimulated with synthetic L33 peptide (5 μM) biweekly and supplemented with interleukin 2 (IL-2; 30 U/mL), IL-7 (25 ng/mL) the first 4 weeks and IL-15 (5 ng/mL) and IL-2 the next 2 weeks. All interleukins were purchased from PeproTech (London, United Kingdom). After 6 weeks in culture the cells were labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE; Invitrogen, Carlsbad, CA) and stimulated again with specific peptide L33 or control peptide Lol P1. After another 13 days in culture supplemented with IL-2 and IL-15, CD3+CD4+ lymphocytes (PE-Alexa 610–conjugated anti-CD4 and APC-conjugated anti-CD3 antibodies; Caltag, Burlingame, CA) were analyzed for proliferation by flow cytometry. Cells with reduced CFSE intensity in L33-stimulated culture were single cell sorted with a FACSAria (Becton Dickinson, Franklin Lakes, NJ) fluorescence-activated cell sorter (FACS). The FACS data were analyzed using biexponential axes, which is considered most appropriate for display of populations with low fluorescence.12 The clones were expanded using anti-CD3 mAb (30 ng/mL; Caltag), IL-2 (50 U/mL) and irradiated feeder cells (104 B-lymphoblasts [see “B-LCLs”] and 7.5 × 104 PBMCs/well).
T-cell receptor β-chain analysis
Total RNA was isolated from the T-cell clones using RNeasy Mini Kit (QIAGEN, Hilden, Germany) with DNase treatment protocol and eluted in water. cDNA was synthesized from 0.5 μg total RNA by Superscript Reverse Transcriptase II (Invitrogen) and random primers (Promega, Madison, WI) in 50-μL reactions. For determination of the T-cell receptor β-chain (TCRB) variant gene (TRBV), a set of 35 forward primers that distinguish most functional TRBV13 and a common reverse primer (BC6314 ) were used. A region within the TCRB constant gene (TRBC) was amplified using primers UpBC and LoBC,14 and it served as an internal control reaction. Polymerase chain reaction (PCR) was performed in 25-μL reactions with 2 μL cDNA and 200 nM of each primer, using QuantiTect SYBR Green PCR kit (QIAGEN), on ABI-Prism 7900-HT (Applied Biosystems, Foster City, CA). PCR cycling program was as follows: 95°C for 15 minutes followed by 50 cycles of 95°C for 15 seconds, 58°C for 1 minute, and 72°C for 15 seconds. For TCRB sequencing, a PCR with 200 nM specific TRBV primer/BC63 primer was performed with HotStarTaq (QIAGEN) in 25-μL reaction using 2.5 μL cDNA. Cycling program was as follows: initial hold of 95°C for 15 minutes followed by 35 cycles of 94°C for 30 seconds, 58° for 30 seconds and 72° for 15 seconds, and finally 72° for 10 minutes on GeneAmp 9700 (Applied Biosystems). The sequencing reaction was performed with 2 μL PCR product and 2 μL Big Dye 3.1 (Applied Biosystems) in 20-μL reactions and 160 nM specific TRBV or BC63 primer with the following cycling program: initial hold of 95°C for 15 minutes followed by 10 cycles of 94°C for 10 seconds, 58°C for 20 seconds and 72°C for 30 seconds, and 25 cycles of 94°C for 10 seconds, 55°C for 20 seconds and 72°C for 30 seconds, and a final 10 minutes at 72°C on GeneAmp 9700 (Applied Biosystems). Samples were precipitated and run on a 3130xl Genetic Analyzer (Applied Biosystems). The sequences were analyzed using Sequence Scanner 1.0 (Applied Biosystems) and IMGT/V-QUEST software15 available at http://imgt.cines.fr.
B-LCLs
B-lymphoblastoid cell lines (B-LCLs) were used as feeder cells to expand T-cell clones in numbers and as antigen-presenting cells in proliferative assays and enzyme-linked immunospot (ELISPOT). B-LCLs were established in our laboratory by transformation of B cells from HPA-1a–immunized women, with EBV (Epstein-Barr virus) as described elsewhere.16 In addition, HLA class II–defined homozygous EBV-transformed B-LCLs were obtained from International Histocompatibility Working Group (IHWG, Seattle, WA): EMJ (IHW no. 9097), STEINLIN (IHW no. 9087), and DUCAF (IHW no. 9019). All lymphoblastoid cells were cultured in IMDM, 10% FBS, and penicillin-streptomycin.
Proliferation studies
CFSE is an intracellular protein binding dye that is retained in daughter cells during cell division. Cells were washed twice in PBS and resuspended to 1 mL in PBSA (0.1%) and CFSE (50-250 ng/mL), incubated at 37°C for 10 minutes, and then cooled on wet ice and resuspended in CM. Labeled cells were washed twice in PBSA (0.2%) and once in CM. The CFSE-labeled T cells were transferred to round-bottom wells on 96-well tissue culture plates (NUNC, Roskilde, Denmark), 5 × 105 cells/well and combined with 106 HPA-1a–negative antigen-presenting cells (APCs). The APCs were either B-LCLs, peptide-pulsed by a 2-hour incubation with 2 μM peptide at 37°C and washed, or monocytes that were either peptide-pulsed as above, or cocultured overnight with HPA-1aa, HPA-1ab, or HPA-1bb platelets and washed. The cells were cultured for 5 to 15 days. PE-Alexa 610–conjugated anti-CD4 and APC-conjugated anti-CD3 antibodies (Caltag) were used to identify the T-cell population of interest, and CFSE intensity was analyzed on a FACSCalibur (Becton Dickinson) flow cytometer. Flow cytometric data were analyzed by FlowJo (TreeStar, Ashland, OR). Median CFSE fluorescence intensities (MIs) of T-cell populations were used to calculate the proliferation score (PS): MIstimulated = MInonstimulated/2PS, defined as the median number of division of stimulated T cells in the end point cultures relative to a culture of nonstimulated T cells alone (no monocytes).
ELISPOT
MultiScreen HTS 96-well microtiter plates with nitrocellulose membrane (Millipore, Billerica, MA) were wet with 35% ethanol (30 μL/well) and washed 3 times with PBS (200 μL/well), followed by overnight incubation at 4°C with 25 μL (10 μg/mL) single capture antibodies; anti–human IFNγ or IL-4 or IL-5 (BD Biosciences, San Jose, CA). Before cell culture, the membranes were blocked with CM (200 μL/well) by incubation for 2 hours at 37°C in a 7.5% CO2 humidified atmosphere. Clonal T cells and peptide-pulsed APCs were added to the wells and incubated for 30 hours at 37°C, 7.5% CO2. APCs were prepared as described for the proliferation assays. To analyze, the plates were washed 6 times with PBS/0.01% Tween 20 and once with PBS (200 μL/well), followed by an incubation with 50 μL/well biotinylated antibodies; anti–human IFNγ or IL-4 or IL-5 (BD Biosciences) at a concentration of 1 μg/mL in PBS for 2 hours at 37°C, 7.5% CO2. The plates were washed as described earlier, followed by incubation for 45 minutes at room temperature with streptavidin–alkaline phosphatase conjugate (Caltag; diluted 1:10 000 in PBS). The plates were then washed 3 times with PBS/Tween 20 (0.01%) and 4 times with PBS (200 μL/well), followed by incubation with the alkaline phosphatase substrate BCIP-NBT (5-bromo-4-chloro-3-indolyl-phosphate/nitro blue tetrazolium; Moss, Pasadena, MD) for 10 to 15 minutes. Finally, the plates were washed in tap water, dried, and scanned using Immunospot (Cellular Technology, Cleveland, OH).
Results
Isolation of HPA-1a–specific T cells from an alloimmunized woman
To determine whether HPA-1a–specific T cells are present in HPA-1a–alloimmunized women, we stimulated PBMCs from an HPA-1a–negative woman who recently had given birth to a child with severe thrombocytopenia, with a 20-mer peptide containing the HPA-1a epitope (leucine at residue 33 of the β3 integrin; L33 peptide). The responding cells were amplified in numbers by repeated biweekly stimulation in culture. The cultured cells were then labeled with CFSE dye and stimulated again in parallel cultures with either L33 or Lol P1 peptide and assayed for proliferating cells. A population of cells that had proliferated extensively was identified in the culture stimulated with the L33 peptide based on CFSE dye dilution. This population of cells, defined by a gate (Figure 1 left) constituted 1.4% of the cultured cells. In comparison, only 0.3% of the CD3+CD4+ cells in the culture stimulated with Lol P1 peptide underwent extensive proliferation (Figure 1 right), indicating successful enrichment of HPA-1a–specific cells in the culture stimulated with L33 peptide. Extensively proliferating cells were cloned directly from the L33 peptide–stimulated culture by single-cell sorting with the use of FACS. Six clonal cultures were successfully amplified by stimulation with anti-CD3 mAb.
Identification and isolation of CD4+ T cells that proliferated specifically in response to the L33 peptide. PBMCs drawn from an HPA-1a–alloimmunized woman were stimulated in vitro with L33 peptide for weeks in tissue culture, then labeled with CFSE dye, and stimulated again with L33 peptide or Lol P1 control peptide in parallel cultures. After 13 days, the cultures were assayed for proliferation by flow cytometry. A sort gate was set to define an area for optimal enrichment of HPA-1a–specific proliferating CD4+ T cells by FACS. CD3+CD4+ lymphocytes that underwent enhanced proliferation in response to L33 peptide (1.4%; left) represented a considerably enriched population compared with the control culture (0.3%; right). The displays are biexponential (see “Cell culture and generation of HPA-1a–specific T-cell lines”).
Identification and isolation of CD4+ T cells that proliferated specifically in response to the L33 peptide. PBMCs drawn from an HPA-1a–alloimmunized woman were stimulated in vitro with L33 peptide for weeks in tissue culture, then labeled with CFSE dye, and stimulated again with L33 peptide or Lol P1 control peptide in parallel cultures. After 13 days, the cultures were assayed for proliferation by flow cytometry. A sort gate was set to define an area for optimal enrichment of HPA-1a–specific proliferating CD4+ T cells by FACS. CD3+CD4+ lymphocytes that underwent enhanced proliferation in response to L33 peptide (1.4%; left) represented a considerably enriched population compared with the control culture (0.3%; right). The displays are biexponential (see “Cell culture and generation of HPA-1a–specific T-cell lines”).
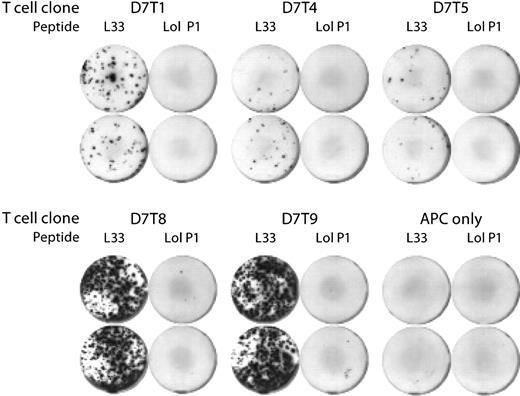
Five of the 6 T-cell clones (D7T1, D7T4, D7T5, D7T8, and D7T9) secreted IFNγ on stimulation with L33 peptide, but not with control peptide (Figure 2). Low levels of IL-4 and IL-5 secretion were also seen with some of the clones (data not shown). For analysis of D7T1 and D7T4, approximately 200 clonal T cells were assayed in each well, whereas for D7T5, D7T8, and D7T9, 20 μL was transferred directly from the expansion culture to the assay well, and the number of cells was not determined. However, on the basis of the number of spots observed in the L33-stimulated wells, there were probably more than 1000 cells/well for the 2 clones D7T8 and D7T9. The clone D7T7 secreted IFNγ when stimulated with both specific and control peptides; thus, the peptide specificity of this clone could not be determined (data not shown). The T-cell clones D7T1 and D7T4 were both successfully established as long-term cell lines and maintained by biweekly stimulation with anti-CD3.
Confirmation of peptide-specificity of isolated CD4+ T-cell clones. Clonal CD4+ T cells were cocultured for 36 hours with 50 000 peptide-pulsed B lymphoblasts and assayed for IFNγ secretion with the ELISPOT assay technique. HPA-1a–derived peptide L33, but not peptide Lol P1, induced IFNγ secretion. Duplicate assays are shown. Two hundred T cells were assayed per well of clones D7T1 and D7T4, and the number of D7T5, D7T8, and D7T9 was not determined. Wells marked “APC only” contained peptide-pulsed B lymphoblasts and no T cells. APC indicates antigen-presenting cell.
Confirmation of peptide-specificity of isolated CD4+ T-cell clones. Clonal CD4+ T cells were cocultured for 36 hours with 50 000 peptide-pulsed B lymphoblasts and assayed for IFNγ secretion with the ELISPOT assay technique. HPA-1a–derived peptide L33, but not peptide Lol P1, induced IFNγ secretion. Duplicate assays are shown. Two hundred T cells were assayed per well of clones D7T1 and D7T4, and the number of D7T5, D7T8, and D7T9 was not determined. Wells marked “APC only” contained peptide-pulsed B lymphoblasts and no T cells. APC indicates antigen-presenting cell.
HPA-1a–specific T-cell clones D7T1 and D7T4 have different T-cell receptor β-chains
The clonality of D7T1 and D7T4 was confirmed by T-cell receptor (TCR) β-chain (TCRB) analysis. Sequence analysis showed that they both have in-frame TCRB rearrangements, but different complementarity determining region 3 (CDR3) sequences, as well as different usage of TCRB variable (TRBV), TCRB joining (TRBJ), and TCRB constant (TRBC) genes. The characteristics of the TCRB rearrangements of the T-cell clones D7T1 and D7T4 are presented in Table 2. Thus, both the established HPA-1a–specific cell lines, D7T1 and D7T4, are clonal and use different T-cell receptors.
HPA-1a–specific T-cell clones D7T1 and D7T4 are HLA-DRB3*0101 restricted
Because the alloimmunization with HPA-1a is strongly associated with HLA-DRB3*0101, and the T cells were isolated from an HLA-DRB3*0101–positive woman, we hypothesized that the recognition of the L33 peptide by the isolated T-cell clones is restricted by this MHC molecule. To investigate this, the clonal T cells were stimulated with peptide-pulsed HLA class II–defined B-LCLs (Table 3) and assayed for IFNγ secretion and proliferation.
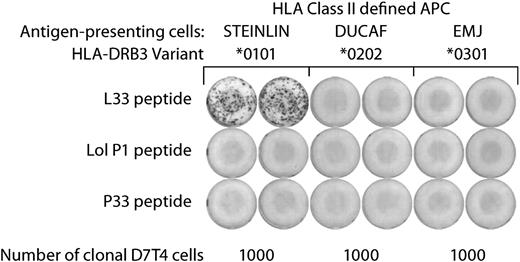
IFNγ secretion by the clonal T cells was seen only on stimulation with L33 peptide–pulsed STEINLIN cells, which carry the HLA-DRB3*0101 allelic variant (Figure 3). No response was seen with the control peptides Lol P1 and P33, or with the HLA-DRB3*0101–negative cell lines DUCAF and EMJ. Similarly, T-cell proliferation was seen only on stimulation with HLA-DRB3*0101–positive cells pulsed with specific peptide (Table 3).
MHC restriction of L33-specific T-cell clones. MHC restriction was determined by ELISPOT: L33-specific T-cell clones were assayed for IFNγ secretion in response to stimulation with a panel of matched, MHC class II homozygous APCs. Only STEINLIN cells pulsed with peptide L33, but not Lol P1 or P33 peptides, induced IFNγ secretion. On the basis of the combined MHC class II expression pattern of the 3 APCs, it could be deduced that L33 peptide recognition by the T-cell clones is restricted by HLA-DRB3*0101. Similar results were obtained with both D7T1 and D7T4 T-cell clones. Duplicate assays with D7T4 cells are shown. The MHC alleles expressed by the 3 APCs are shown in Table 3.
MHC restriction of L33-specific T-cell clones. MHC restriction was determined by ELISPOT: L33-specific T-cell clones were assayed for IFNγ secretion in response to stimulation with a panel of matched, MHC class II homozygous APCs. Only STEINLIN cells pulsed with peptide L33, but not Lol P1 or P33 peptides, induced IFNγ secretion. On the basis of the combined MHC class II expression pattern of the 3 APCs, it could be deduced that L33 peptide recognition by the T-cell clones is restricted by HLA-DRB3*0101. Similar results were obtained with both D7T1 and D7T4 T-cell clones. Duplicate assays with D7T4 cells are shown. The MHC alleles expressed by the 3 APCs are shown in Table 3.
Neither of the other 2 common HLA-DRB3 allelic variants, *0202 (expressed by DUCAF) and *0301 (expressed by EMJ), nor any of the other MHC class II molecules expressed on STEINLIN can present the L33 peptide to the clonal T cells, because these are also present on either DUCAF or EMJ, which did not induce T-cell responses. Together, these results show that the isolated T-cell clones D7T1 and D7T4 recognize the L33 peptide in association with HLA-DRB3*0101, and the reactivity of these T-cell clones are therefore restricted by this molecule.
T-cell clones proliferate on stimulation with native HPA-1a antigen derived from platelets
To assess whether the T-cell clones specific for synthetic peptide also recognize native antigen derived from platelets, HLA-DRB3*0101–positive monocytes were cocultured overnight with platelets (HPA-1aa, HPA-1ab, or HPA-1bb) and were then used to stimulate T cells in proliferation assays. These responses were determined by the extent of reduction in dye intensity in CFSE-labeled T cells. The T-cell clones D7T1 and D7T4 showed specific proliferation on stimulation with monocytes pulsed with L33 peptide, whereas no proliferation was seen with the control peptides P33 and Lol P1 (Figure 4A). Vigorous proliferation of the clonal T cells was also seen when stimulating with HPA-1a–positive platelets whereas HPA-1bb platelets did not stimulate the T cells to proliferate (Figure 4B). The Proliferation Score (PS) for clonal T cells was determined, and representative data (clone D7T4) are given in Figure 4C. T-cell proliferation was only seen with the use of HLA-DRB3*0101–positive monocytes with HPA-1aa platelets (PS = 3.36) and HPA-1ab platelets (PS = 3.34). No proliferation was seen when using HPA-1bb platelets or HLA-DRB3*0101–negative monocytes (PS values < 0.95). Thus, the clonal T cells D7T1 and D7T4 recognize native antigen derived from platelets, processed and presented by HLA-DRB3*0101–positive monocytes.
L33-specific T-cell clones respond specifically to HLA-DRB3*0101–positive monocytes pulsed with either synthetic peptide or HPA-1a–positive platelets. Proliferative responses induced by stimulation with peptide-pulsed or platelet-pulsed monocytes. Both clones proliferate specifically in response to (A) L33, but not Lol P1 and P33 control, peptides, and (B) HPA-1a–positive, but not HPA-1a–negative, platelets. (C) D7T1 and D7T4 T-cell clones proliferate in response to stimulation with HPA-1a–positive platelets (HPA-1aa or HPA-1ab), but not HPA-1a–negative (HPA-1bb) platelets, combined with HLA-DRB3*0101–positive, but not negative, monocytes. Representative data for clone D7T4 is shown.
L33-specific T-cell clones respond specifically to HLA-DRB3*0101–positive monocytes pulsed with either synthetic peptide or HPA-1a–positive platelets. Proliferative responses induced by stimulation with peptide-pulsed or platelet-pulsed monocytes. Both clones proliferate specifically in response to (A) L33, but not Lol P1 and P33 control, peptides, and (B) HPA-1a–positive, but not HPA-1a–negative, platelets. (C) D7T1 and D7T4 T-cell clones proliferate in response to stimulation with HPA-1a–positive platelets (HPA-1aa or HPA-1ab), but not HPA-1a–negative (HPA-1bb) platelets, combined with HLA-DRB3*0101–positive, but not negative, monocytes. Representative data for clone D7T4 is shown.
Discussion
This study formally shows that HPA-1a–specific T cells can be isolated from a woman that has given birth to a child with NAIT. We isolated clonal HLA-DRB3*0101–restricted HPA-1a–specific CD4+ T cells from an HPA-1a–immunized woman and established 2 such clones in long-term cultures. The specificity of the isolated T-cell clones was confirmed both by IFNγ secretion and by proliferation in response to stimulation with synthetic HPA-1a peptide. The HLA-DRB3*0101 restriction was shown with the use of a panel of B-LCLs as antigen-presenting cells that were matched for the expression of MHC class II molecules with the exception of the HLA-DRB3 locus. We also show that the clones recognize native antigen derived from HPA-1a–positive platelets when the antigen is presented by HLA-DRB3*0101–positive monocytes. Together, these findings show that HPA-1a–specific HLA-DRB3*0101–restricted CD4+ T cells are present in alloimmunized women.
The maternal immune response that results in platelet-reactive antibodies and NAIT probably depends on help from antigen-specific T cells. Previous reports have provided considerable support to the notion that HPA-1a–specific T cells are present in mothers of newborns affected by NAIT by demonstration of specific proliferative responses to various peptides containing the HPA-1a determinant.17-19 In the study published by Maslanka et al,18 the investigators used TCR spectratyping and PCR techniques with clonotypic primers to track T cells growing in these cultures stimulated with a peptide containing the HPA-1a epitope. The clonotypic primers were also used to show the presence of these T cells in PBMCs at various time points in relation to pregnancy and in platelet-stimulated cultures. Sukati et al19 showed a linkage between proliferative responses to various peptides containing the HPA-1a epitope with anti–HPA-1a antibody levels and both HLA-DRB3*01 and HLA-DQB1*02 alleles. Jackson et al17 also determined that most affected women tested had proliferative responses to peptides containing the HPA-1a, but not the HPA-1b, epitope. However, in these reports the proliferating cells were not isolated or identified. The demonstration of clonally isolated HPA-1a–specific T cells in the present study provides unambiguous evidence for the association of T-cell responses with NAIT. The established HPA-1a–specific clonal cell lines will permit in vitro studies, both to characterize HPA-1a–specific immune responses and to test the potential to manipulate the T-cell response with the aim to reduce or prevent anti–HPA-1a antibody responses from taking place.
The technique we used for identification and cloning, FACS isolation of CFSE-labeled cells, allows direct isolation of single cells that proliferate in response to stimulation with specific peptide. This technique has also been used by others.20,21
It has been known for some time that immunization of women against fetal HPA-1a in connection with pregnancy is strongly associated with HLA-DRB3*0101.1,2,6 Therefore, the finding that the isolated T-cell clones are restricted by this MHC molecule provides considerable support to the notion that T cells are involved in immune responses that result in NAIT. Moreover, peptides containing the HPA-1a epitope have been shown to bind the HLA-DRB3*0101 MHC class II molecule and have served as a model to define the peptide-binding motif of the HLA-DRB3*0101 molecule11 : peptides that can associate with HLA-DRB3*0101 have hydrophobic anchor residues at positions P1 and P9 and a charged residue at position P4. Recently, the crystal structure of HLA-DRB3*0101 was resolved with a modified HPA-1a peptide in the peptide-binding cleft.10 This structure confirmed the 3 dominating pockets corresponding to P1, P4, and P9 and found that 2 β-chain arginine residues, β-11 and β-74, are largely responsible for the negative charge of the P4 pocket. Interestingly, in the 2 other common HLA-DRB3 variants, HLA-DRB3*0202 and HLA-DRB3*0301, these 2 arginines are substituted with hydrophobic and polar amino acids, respectively. Predictably, HPA-1a peptides will be less likely to associate with these 2 HLA-DRB3 variants because of the lack of a dominant charged P4 pocket. Here, we show that HPA-1a peptide–pulsed APCs that carry the allelic variants HLA-DRB3*0202 or HLA-DRB3*0301 cannot stimulate the HPA-1a–specific T-cell clones (Figure 3). The B-LCLs were not matched for expression of MHC class I molecules. However, it is unlikely that L33 was presented by MHC class I, because binding of the coreceptor CD4 to MHC class II is required for activation of most T cells.
Although the 2 HPA-1a–specific clones we isolated in this study were HLA-DRB3*0101 restricted, it cannot be ruled out that T cells restricted by other MHC molecules may also play a role in NAIT. In fact, in a study that has received relatively little attention, it was shown that a second allele, HLA-DQB1*0201, also was associated with immunization against HPA-1a.2 Although the prevailing view seems to be that NAIT is solely associated with HLA-DRB3*0101, most of the HPA-1a–immunized women examined in the above-mentioned study had both HLA-DRB3*0101 and HLA-DQB1*0201 allelic variants. This opens the possibility that different T-cell populations are involved in the immune response that leads to immunization against HPA-1a: one population restricted by HLA-DRB3*0101, that is, the T cells characterized in the present study, and a second putative T-cell population restricted by HLA-DQB1*0201. If so, it will be of great interest to determine the role of the different T cells in the immune response that result in immunization of HPA-1a–negative women carrying an HPA-1a–positive fetus. Alternatively, the HLA-DRB3*0101 allele or other genetic elements that are important for HPA-1a immunization may be in linkage disequilibrium with HLA-DQB1*0201. In either case, identification of the second element or T-cell population in addition to HLA-DRB3*0101–restricted T cells, seemingly required for HPA-1a immunization, may show novel mechanisms related to the initiation and/or regulation of antibody responses.
The woman that gave rise to the T-cell clones in the present study had relatively low anti–HPA-1a antibody levels. However, the neonate was severely thrombocytopenic. This suggests that maternal anti–HPA-1a antibodies were present during pregnancy although at low levels at delivery: a drop in anti–HPA-1a antibody levels at the later part of pregnancy can be observed in affected women.22 This phenomenon is interesting and the mechanism is unknown.
The HPA-1a–specific clones analyzed in this study secreted predominantly IFNγ in response to stimulation with synthetic peptide. In addition, it has been shown (by intracellular cytokine staining) that stimulation of the D7T4 clone with native antigen presented by monocytes induced production of IFNγ but not IL-4 (M.T.A. and T.B.S., unpublished data, August 7, 2008).
Together, these findings suggest that these clones have a cytokine profile consistent with a type 1 immune response. Normal pregnancies are usually biased toward type 2 immunity, whereas immune-mediated complications during pregnancy are associated with type 1 immune responses.23 Therefore, it will be of great interest to determine whether NAIT is associated with type 1 or type 2 immune responses. Proper evaluation of the cytokine profile of HPA-1a–specific T-cell responses is pending isolation of more clones and analysis of T-cell responses in several different immunized women.
For this and for other detailed studies of the nature of the underlying immune responses leading to anti-HPA-1a antibody production and subsequent NAIT, it is important to develop reagents, such as MHC class II tetramers, for direct evaluation of in vivo responses and for isolation and characterization of the responding cells. The HPA-1a–specific T-cell clones we have established in long-term cultures will be invaluable in confirming the functionality and specificity of such reagents.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the immunized women participating in this study for donating blood samples and the University Hospital of North Norway (Tromsø, Norway) Blood Bank for providing blood products.
This work was supported by a grant from the North Norway Regional Health Authority (Bodø, Norway).
Authorship
Contribution: M.T.A. and T.B.S. designed the study, performed the experiments, and wrote the manuscript; and A.H., M.K.K., and B.S. provided donor blood samples from HPA-1a–immunized women, discussed the data, and critically reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tor B. Stuge, Department of Immunology, University of Tromsø, N-9037 Tromsø, Norway; e-mail: tor.brynjar.stuge@uit.no.