Abstract
Although cytogenetic abnormalities are important prognostic factors in myeloid malignancies, they are not included in current prognostic scores for primary myelofibrosis (PMF). To determine their relevance in PMF, we retrospectively examined the impact of cytogenetic abnormalities and karyotypic evolution on the outcome of 256 patients. Baseline cytogenetic status impacted significantly on survival: patients with favorable abnormalities (sole deletions in 13q or 20q, or trisomy 9 ± one other abnormality) had survivals similar to those with normal diploid karyotypes (median, 63 and 46 months, respectively), whereas patients with unfavorable abnormalities (rearrangement of chromosome 5 or 7, or ≥ 3 abnormalities) had a poor median survival of 15 months. Patients with abnormalities of chromosome 17 had a median survival of only 5 months. A model containing karyotypic abnormalities, hemoglobin, platelet count, and performance status effectively risk-stratified patients at initial evaluation. Among 73 patients assessable for clonal evolution during stable chronic phase, those who developed unfavorable or chromosome 17 abnormalities had median survivals of 18 and 9 months, respectively, suggesting the potential role of cytogenetics as a risk factor applicable at any time in the disease course. Dynamic prognostic significance of cytogenetic abnormalities in PMF should be further prospectively evaluated.
Introduction
Primary myelofibrosis (PMF) is a myeloid malignancy characterized by progressive marrow fibrosis and extramedullary hematopoiesis. The median survival is 5 years from the time of diagnosis, but there is marked interpatient variability, with some patients dying within 12 months of presentation and others surviving for more than 10 years.1,2 Therefore, there exists a significant need for effective prognostic systems to guide life expectancy and therapy selection. The existing prognostic systems are uncomplicated scores based mainly on peripheral blood parameters, with the Lille score (based on anemia and abnormal white cell count) being the most commonly used.1,2 In younger patients, the presence of thrombocytopenia, monocytosis, circulating blasts, and constitutional symptoms may also be prognostic.2,3 Pitfalls with the current systems include the inability to identify the patients with extremely poor prognosis (median survival, ≤ 12 months), the exclusion of key biologic characteristics, such as karyotypic and molecular aberrations, and the applicability of these scores only to patients at the time of initial diagnosis.1-3
Cytogenetic abnormalities are an integral part of prognostic systems in other myeloid malignancies, including acute myeloid leukemia and myelodysplasia.4-7 In these malignancies, studies of recurrent karyotypic aberrations led to key discoveries in molecular pathways, including the recognition of the AML1-ETO and PML-RARA translocations in acute myeloid leukemia, and the deletion of RPS14 in 5q- myelodysplasia.8-10 In contrast, the study of cytogenetic abnormalities in PMF is in its infancy. The 3 most important studies of cytogenetic aberrations in PMF were composed of 47, 106, and 165 patients, reported by investigators from Lille, Sheffield, and the Mayo clinic, respectively.11-14 These studies showed that karyotypic abnormalities occur in 32% to 48% of patients at diagnosis and that their presence is generally associated with reduced survival, with the possible exception of patients with deletions in 13q or 20q, who appear to have similar survival as normal diploid patients.11-14 Beyond this, little is understood about the prognostic implications of other common aberrations, including those traditionally regarded as being adverse in acute myeloid leukemia and myelodysplasia. In addition, it is not known about whether the acquisition of adverse karyotypic abnormalities is associated with reduced subsequent survival, analogous to the situation with chronic myeloid leukemia, where karyotypic evolution is an important mechanism of progression to accelerated and blast phases.15
To address these questions and to describe recurrent karyotypic aberrations for correlative scientific studies, we analyzed our institutional experience with the results of conventional karyotypic analysis in a well-characterized cohort of PMF patients.
Methods
Institutional review board approval was obtained for this study from the University of Texas M. D. Anderson Cancer Center, and patient informed consent was obtained in accordance with the Declaration of Helsinki. Between January 1984 and April 2007, 268 patients with primary myelofibrosis (agnogenic myeloid metaplasia, chronic idiopathic myelofibrosis), as defined by the World Health Organization,6,16 presented to the University of Texas M. D. Anderson Cancer Center for evaluation and treatment. All patients underwent physical examination, laboratory studies (including full blood count, biochemical panel, liver function tests, and lactate dehydrogenase), and bone marrow examination, including full karyotypic analysis. Conventional cytogenetic analysis was performed on metaphases obtained from unstimulated bone marrow aspirate cultures using standard techniques. Results were reported using the International System for Human Cytogenetic Nomenclature. Eighty-two patients were also evaluated for the presence of the JAK-2 V617F mutation using previously reported assays.17
Cytogenetic testing was considered to be “at initial diagnosis” if the patient was referred and assessed at our center within 4 months of confirmation of primary myelofibrosis; all other patients were considered “beyond initial diagnosis.” This operational definition was chosen based on the practice in the current literature to facilitate comparison between studies.13 Several patients had cytogenetic analysis repeated during their disease course and were evaluable for karyotypic evolution. To exclude those patients who had repeat testing because of progressive or uncontrolled disease, patient charts were manually reviewed and the reason for repeat cytogenetic study documented. Only those patients with stable disease who had a repeat study as part of routine clinical care (as recorded by the treating physician) or as part of scheduled trial reassessments were included in the analysis of the prognostic significance of karyotypic evolution.
The primary endpoint was overall survival. This was calculated from the date of cytogenetic testing rather than from the date of initial diagnosis as a preliminary examination of the data showed that karyotypic changes evolve during the disease course, and cytogenetics was best considered, therefore, as a dynamic risk factor. The Lille score was calculated according to published criteria (adverse prognostic risk factors are: hemoglobin < 10 g/dL, white blood cells < 4 or > 30 × 109/L; risk group: 0 factor indicates low; 1 factor, intermediate; 2 factors, high).1 Categorical data were compared using the Fisher exact or χ2 tests, and continuous data using the Mann-Whitney test, as appropriate. Survival was evaluated using the Kaplan-Meier method, and the impact of baseline factors on survival was evaluated using log-rank analyses. The impact of multiple factors on survival was evaluated with Cox proportional hazards analysis, using a backward selection strategy. All P values were 2-sided.
Results
Patient population
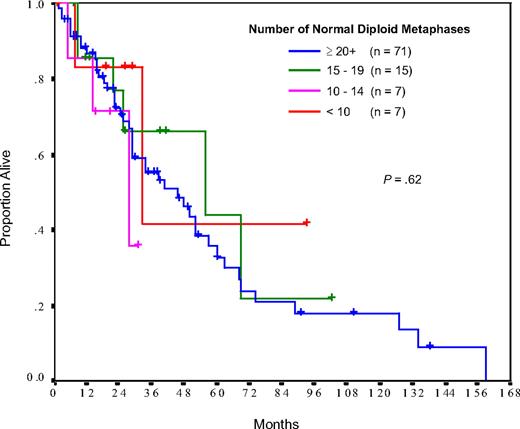
Of the 268 patients for whom karyotypic analysis was performed, specimens obtained from 217 (81%) yielded either a clonal abnormality or more than or equal to 20 normal diploid metaphases. The median number of evaluable metaphases for all patients was 20 (range, 4-35), and 73% of patients had more than or equal to 20 evaluable metaphases. A low cytogenetic yield (diploid karyotype in < 20 metaphases, no mitotic cells, or nonclonal abnormalities) was obtained in 51 samples (19%). Low cytogenetic yields were more common in patients with low white cell counts, occurring in 35% and 17% of patients with a white cell count of less than 4.0 and more than or equal to 4.0 × 109/L, respectively (P = .02). The majority (67%) of patients with low cytogenetic yields had a normal diploid karyotype in 10 to 19 metaphases, and their survival was identical to that of patients with more than or equal to 20 normal diploid metaphases (Figure 1). For this reason, patients with 10 to 19 normal diploid metaphases were grouped together with those having more than or equal to 20 normal diploid metaphases for all subsequent analyses. This grouping was consistent with the practice of other investigators.12
Comparison of survival between patients with various numbers of normal diploid metaphases, when evaluated at diagnosis.
Comparison of survival between patients with various numbers of normal diploid metaphases, when evaluated at diagnosis.
The final study population was thus 256 patients: 91 (36%) patients with a clonal abnormality and 165 (64%) with a normal diploid karyotype. Of these patients, 131 (51%) were evaluated at diagnosis, and 125 (49%) were evaluated beyond initial diagnosis. Compared with patients evaluated at diagnosis, patients evaluated beyond initial diagnosis had features of more advanced disease, including abnormal performance status, exposure to hydroxyurea or erythropoietin, anemia or transfusion dependency, and elevated monocyte counts (Table 1; P < .05 for comparisons with patients evaluated at initial diagnosis). In addition, patients evaluated beyond initial diagnosis were more likely to have an abnormal karyotype (42% vs 29%, P = .03; Table 1). The median survival from the time of cytogenetic assessment was shorter for patients tested beyond initial diagnosis (26 months) than for patients tested at diagnosis (46 months, P = .02). However, this difference was accounted for by the difference in the median time to cytogenetic testing in these 2 groups (28 months vs 1 month, respectively).
The impact of karyotype on survival
For the analysis of the impact of karyotypic abnormalities on survival, patients were stratified according to whether they were tested at diagnosis or tested beyond initial diagnosis. Interestingly, the same hierarchy of cytogenetic abnormalities emerged in both groups:
Favorable abnormalities.
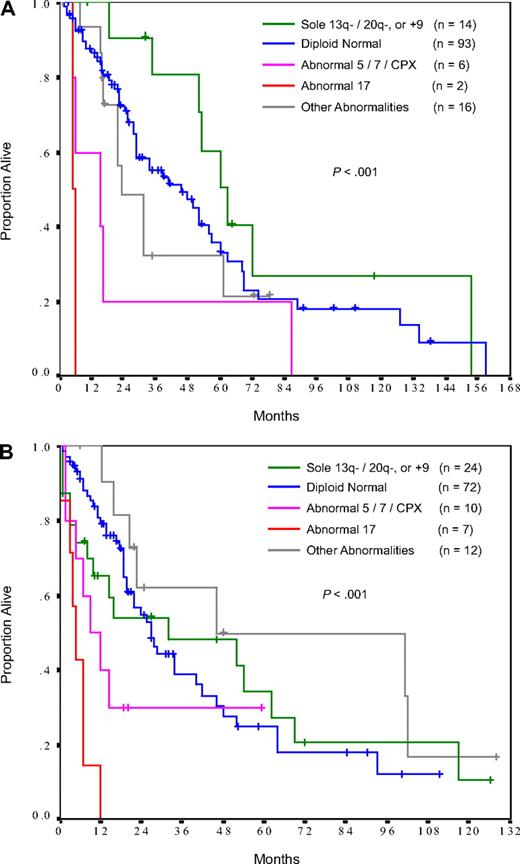
Three categories of cytogenetic abnormalities emerged as being associated with survival comparable with that of patients with a normal diploid karyotype: sole deletion in 20q (n = 19), sole deletion in 13q (n = 14), and trisomy 9 plus or minus 1 other abnormality (n = 5). The median survival was 53 months, 63 months, and not reached, for patients evaluated at diagnosis, and 37 months, 25 months, and not reached, respectively, for patients evaluated beyond initial diagnosis (Figure 2; Table 2).
Survival by cytogenetic category. (A) Patients evaluated at diagnosis. (B) Patients evaluated beyond initial diagnosis. CPX indicates complex (≥ 3) abnormalities.
Survival by cytogenetic category. (A) Patients evaluated at diagnosis. (B) Patients evaluated beyond initial diagnosis. CPX indicates complex (≥ 3) abnormalities.
Unfavorable abnormalities.
Patients with chromosome 5 or 7, or complex (≥ 3) aberrations had a survival that was inferior to that of patients with normal diploid karyotypes (P = .05 and .06 for patients evaluated at diagnosis and beyond initial diagnosis, respectively). The median survival was 15 months for those evaluated at diagnosis and 9 months for those evaluated beyond initial diagnosis (Figure 2; Table 2). Many of these abnormalities overlapped in the same person (eg, monosomy 7 and complex karyotype).
Very unfavorable abnormalities.
The poorest survival was in patients with abnormalities of chromosome 17, which was present in 2 patients tested at diagnosis and 7 patients tested beyond initial diagnosis. Four patients had monosomy 17, 3 patients had isochromosome 17q, 1 patient had add(17)(q25), and 1 patient had add(17)(p13). The karyotype was complex in 6 of 9 patients. Regardless of whether the chromosome 17 aberration was discovered at diagnosis or beyond, survival was similarly dismal with all patients projected to have died by 12 months (Figure 2; Table 2).
Relationship with JAK-2 status.
Eighty-two patients had known JAK-2 V617F mutation status. Of the 35 patients who were JAK-2 wild-type, 2 patients (6%) had unfavorable or very unfavorable cytogenetic abnormalities. Of the 47 patients who were JAK-2 V617F mutated, 7 patients (15%) had unfavorable or very unfavorable cytogenetic abnormalities (P = .29).
Cytogenetics has prognostic significance independent of traditional factors
Baseline characteristics prognostic for inferior survival on univariate analysis were age 65 years or older, performance status more than or equal to 1, hemoglobin less than 10 g/dL or transfusion dependency, white blood cell count less than 4 or more than 30 × 109/L, platelet count less than 100 × 109/L, peripheral blood blasts more than or equal to 2%, and unfavorable or very unfavorable cytogenetics (Table 3). Monocyte count more than or equal to 109/L was also prognostic in patients tested at diagnosis (Table 3). On the multivariate models considering relevant baseline factors, adverse cytogenetics findings were confirmed to be independently associated with survival both in patients tested at diagnosis and in patients tested beyond initial diagnosis (hazard ratios, 3.5 and 2.6, respectively; Table 3).
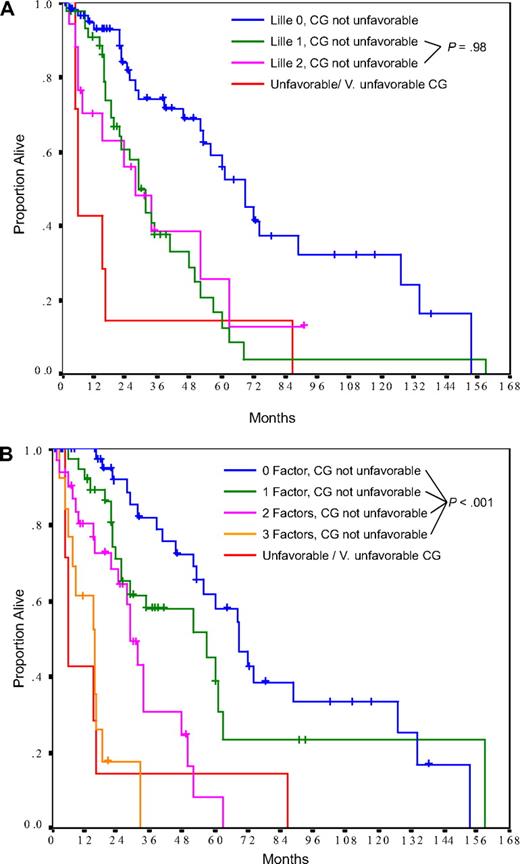
To explore the relationship between karyotypic abnormalities and traditional risk models, the performance of unfavorable or very unfavorable cytogenetics in predicting inferior survival was compared against the Lille score in patients tested at diagnosis. In this group of patients, the Lille score was 0 in 47%, 1 in 36%, and 2 in 18% (Table 1). Patients with Lille scores of 0 or 1 had a significantly higher probability of having a normal diploid karyotype or a favorable abnormality than patients with Lille score 2 (98% vs 74%, respectively, P = .001). When patients were stratified according to their cytogenetic risk and Lille score, patients with unfavorable or very unfavorable cytogenetics were identified as the highest risk group (median survival, 6 months); whereas in the absence of unfavorable cytogenetics, Lille score 1 and 2 patients displayed virtually identical survival curves (Figure 3A, P = .98). We therefore explored the use of 3 prognostic factors identified in the multivariate model (Table 3) in place of the Lille score: hemoglobin less than 10 g/dL or transfusion dependency, platelet count less than 100 × 109/L, and Zubrod performance status more than or equal to 1. In this model, patients without unfavorable cytogenetics were effectively divided into risk categories with median survivals of 69, 57, 29, and 16 months, respectively (for patients with 0, 1, 2, or 3 of these factors, P < .001; Figure 3B). In contrast, patients with unfavorable or very unfavorable cytogenetics had a dismal outcome irrespective of the number of other risk factors present (Figure 3B). Therefore, a combination of karyotypic findings and clinical features appeared to be highly effective in determining survival in patients evaluated at initial diagnosis.
Survival by cytogenetic (CG) category and other prognostic factors in patients tested at diagnosis. (A) In patients without unfavorable cytogenetics, the Lille score failed to differentiate between intermediate-risk (Lille 1) and high-risk (Lille 2) patients. (B) Using the factors identified as being independently significant on the multivariate model (hemoglobin < 10 g/dL or transfusion dependency, platelets < 100 × 109/L, performance status ≥ 1), patients without unfavorable cytogenetics were effectively divided into risk categories with median survivals of 69, 57, 29, and 16 months, respectively (for 0, 1, 2, and 3 factors, P < .001). Patients with unfavorable or very unfavorable karyotypes had a dismal survival (median, 6 months) irrespective of the number of factors present.
Survival by cytogenetic (CG) category and other prognostic factors in patients tested at diagnosis. (A) In patients without unfavorable cytogenetics, the Lille score failed to differentiate between intermediate-risk (Lille 1) and high-risk (Lille 2) patients. (B) Using the factors identified as being independently significant on the multivariate model (hemoglobin < 10 g/dL or transfusion dependency, platelets < 100 × 109/L, performance status ≥ 1), patients without unfavorable cytogenetics were effectively divided into risk categories with median survivals of 69, 57, 29, and 16 months, respectively (for 0, 1, 2, and 3 factors, P < .001). Patients with unfavorable or very unfavorable karyotypes had a dismal survival (median, 6 months) irrespective of the number of factors present.
The prognostic significance of karyotypic evolution
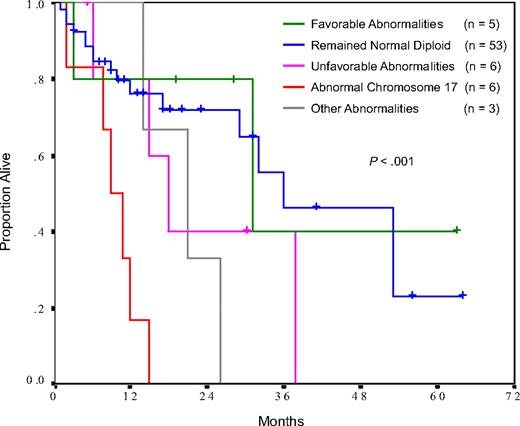
Eighty-eight patients underwent repeat cytogenetic testing for routine clinical care or scheduled clinical trial reasons, of which 73 were in a stable disease phase and were, therefore, evaluable for the impact of karyotypic evolution. Fifty-three patients (73%) showed normal diploid karyotypes at baseline that remained unchanged at reassessment, and their median survival was 36 months from the time of repeat testing (Figure 4; Table 4). Five patients (7%) evolved into the favorable category, with a median survival similar to that of patients whose karyotypes remained normal diploid (31 months, P = .78). Six patients (8%) evolved into the unfavorable category, with a median survival (18 months) that was numerically shorter, but not statistical significantly inferior, to that of patients with normal diploid karyotypes (P = .25). Similar to our observations at baseline, patients who developed a chromosome 17 abnormality had a dismal median survival of only 9 months (Figure 4; Table 4).
Survival subsequent to karyotypic evolution in patients undergoing repeat cytogenetic assessment. Patients with progressive disease or development of blast phase at the time of re-evaluation were excluded from this analysis.
Survival subsequent to karyotypic evolution in patients undergoing repeat cytogenetic assessment. Patients with progressive disease or development of blast phase at the time of re-evaluation were excluded from this analysis.
Discussion
This retrospective study demonstrates the importance of karyotypic abnormalities in the largest group of PMF patients with detailed cytogenetic information published to date. At the time of diagnosis, patients were divided into those with a normal diploid karyotype, favorable abnormalities (sole 13q-, 20q-, or trisomy 9 ± 1 other abnormality), unfavorable abnormalities (rearrangements of chromosome 5 or 7, or ≥ 3 aberrations), and very unfavorable abnormalities (abnormalities of chromosome 17), with median survivals of 63, 46, 15, and 5 months, respectively. In addition, we presented evidence to show that the prognostic risk of cytogenetic abnormalities may not be static but may rather evolve with the disease course. This observation is not surprising as the presence of karyotypic abnormalities is an intrinsic property of the malignant clone, and there are no compelling reasons why a factor predictive of poor survival at diagnosis should not also be important later in the disease course. In contrast, traditional risk scores may not be reliable at time points other than at initial diagnosis as they rely on peripheral blood parameters that may be modified by external factors, such as transfusions, growth factor therapy, and treatment side effects.
The ability to select a priori those patients with the worst outcome is particularly important in PMF as the only therapy capable of modifying its natural history is allogeneic stem cell transplantation, a procedure that carries a substantial treatment-related mortality of up to 34%.18-21 Therefore, patients selected to proceed to transplantation or other high-risk therapies should be those predicted to be at the highest risk of death from the disease. Based on our analysis of the interaction between cytogenetics and other prognostic markers, we identified 2 groups of patients with the highest risks of death: those with unfavorable or very unfavorable cytogenetics and those with the combination of low hemoglobin, low platelet count, and impaired performance status. However, before these findings can be generalized for application in the clinic setting, several limitations of the current study must be addressed. First, the study is retrospective in nature, and the numbers of patients within each cytogenetic category remain relatively small. Although the karyotypic evolution analysis supported the baseline karyotypic abnormality hierarchy, it was not possible in a retrospective study to fully control for all potential sources of selection bias, and the karyotypic evolution information should be regarded as a hypothesis-generating observation deserving prospective exploration. Second, although we were able to examine the prognostic impact of cytogenetics in the context of routine clinical and laboratory studies, the relationship between cytogenetics and novel molecular markers (such as JAK-2 mutation) was not able to be fully elucidated. Lastly, the interpretation of apparently normal diploid karyotypes in patients with fewer than 20 evaluable metaphases remains incompletely resolved. Because of the fibrotic nature of the marrow, aspirate specimens from patients with PMF may be scant, and in the case of patients with fewer than 20 metaphases it is not known whether the malignant clone was truly normal diploid or insufficient tumor cells were present. In this regard, however, our demonstration that patients with various numbers of apparently normal diploid metaphases had similar survival suggests that it is unlikely that many patients with unfavorable karyotypes were missed because of inadequate specimen. For all of these reasons, the findings of the current study should be the subject of further prospective evaluation by a cooperative working group.
The identification of recurrent chromosomal abnormalities in this study, in combination with reports published by other authors,11-14 provides researchers with an opportunity to gain insights into the pathogenesis of PMF. The deletion on 20q in PMF spans 20q11 to 20q13,11-14 but this abnormality is not unique to PMF and has also been described in myelodysplasia7 and polycythemia vera.22 The common deleted region in myeloid diseases has been progressively narrowed to a 1.7-Mb interval, with the causative gene remaining elusive.23,24 The deletion in 13q in PMF span 13q12 to 13q22,11-14 and has also been described in chronic lymphocytic leukemia (where it is a favorable finding)25 and multiple myeloma (where it is an unfavorable finding).26 Although the RB1 tumor suppressor gene locus at 13q14 is commonly deleted in multiple myeloma,26 a previous study had shown that this locus was not consistently involved in myeloproliferative diseases.27 Recently, a systemic dissection of the 13q locus in chronic lymphocytic leukemia led to the discovery of deletions of the microRNAs mir-15 and mir-16 as being the probable pathogenic culprits in this disease.28 Trisomy 9 was described in the Mayo clinic series where it was present in 12 patients, 11 of whom had one or more additional lesions.13 Although only 5 favorable patients had trisomy 9 in the current study, it is nevertheless striking that none of them had died at a follow-up of up to 126 months. Interestingly, the JAK-2 V617F mutation was reported in a recent study to be associated with amplifications of 9p24 region (the JAK-2 locus).29 Other recurrent PMF cytogenetic findings that had no prognostic impact in the current study but nevertheless present interesting targets for genetic evaluation include abnormalities of chromosome 1 (especially 1q partial duplications). Finally, the coincidence of the same poor-risk cytogenetic abnormalities in PMF, acute myeloid leukemia, myelodysplasia, and chronic lymphocytic leukemia point to common pathogenic pathways in hematologic malignancies that deserve further investigation.
Authors' note.
During the preparation of the second revision of this manuscript, results of the International Working Group for Myelofibrosis Research and Treatment (IWG-MRT) Prognostic Scoring System (PSS) were prepublished.30 This landmark study evaluated baseline (at diagnosis) characteristics in 1054 patients from 7 centers, of whom 409 had available cytogenetic information. The presence of one or more karyotypic abnormalities was associated with reduced survival on multivariate analysis, although this was independent of the new PSS only in the intermediate-risk groups. In the PSS study, the detailed examination of specific cytogenetic subgroups was not possible because of incomplete data. Therefore, the authors regard the current study as being an important complement to the new PSS.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
The authors thank Dr Ayalew Tefferi for his advice during statistical analysis.
Authorship
Contribution: C.S.T., S.P., H.K., and S.V. designed the research and analyzed the data; C.S.T. and S.V. wrote the paper; L.V.A. was responsible for cytogenetic analysis and coauthored the paper; A.L. assisted in data collection and analysis; and S.V., K.I.L., J.C., M.J.K., and D.A.T. assisted in the preparation of data, analysis of results, initial revisions to the paper, and final proofreading.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Srdan Verstovsek, Leukemia Department, University of Texas M. D. Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: sverstov@mdanderson.org.