Abstract
Methotrexate polyglutamates (MTXPGs) determine in vivo efficacy in acute lymphoblastic leukemia (ALL). MTXPG accumulation differs by leukemic subtypes, but genomic determinants of MTXPG variation in ALL remain unclear. We analyzed 3 types of whole genome variation: leukemia cell gene expression and somatic copy number variation, and inherited single nucleotide polymorphism (SNP) genotypes and determined their association with MTXPGs in leukemia cells. Seven genes (FHOD3, IMPA2, ME2, RASSF4, SLC39A6, SMAD2, and SMAD4) displayed all 3 types of genomic variation associated with MTXPGs (P < .05 for gene expression, P < .01 for copy number variation and SNPs): 6 on chromosome 18 and 1 on chromosome 10. Increased chromosome 18 (P = .002) or 10 (P = .036) copy number was associated with MTXPGs even after adjusting for ALL subtype. The expression of the top 7 genes in leukemia cells accounted for more variation in MTXPGs (46%) than did the expression of the top 7 genes in normal HapMap cell lines (20%). The top 7 inherited SNPs in patients accounted for approximately the same degree of variation (17%) in MTXPGs as did the top 7 SNP genotypes in HapMap cell lines (20%). We conclude that acquired genetic variation in leukemia cells has a stronger influence on MTXPG accumulation than inherited genetic variation.
Introduction
Acute lymphoblastic leukemia (ALL) is the most common form of childhood cancer. Subtypes of ALL have been characterized based on the presence of gene deletions, amplifications, and translocations of regions of DNA.1,2 Treatment outcome of ALL has steadily increased over the past 4 decades, with current studies reporting 5-year event-free survival rates of more than 80%.3-8 These increased cure rates are partly the result of improved risk-directed treatment stratification and optimization of delivery of standard medications.9-11
Methotrexate (MTX) is a component of all treatment protocols for ALL, and interindividual variability in MTX disposition has resulted in variation in clinical response to the drug.12,13 The ability of cells to polyglutamylate MTX to form active MTX polyglutamates (MTXPGs) and therefore retain the drug for a longer period of time is a major determinant of both in vivo efficacy and toxicity.14-16 The concentrations of MTXPGs have been related to its pharmacodynamic effects, but genomic determinants of MTXPG accumulation have yet to be fully elucidated.
Levels of MTXPGs vary widely between different subtypes of ALL, with highest levels seen in B-hyperdiploid and lowest levels seen in E2A-PBX1(TCF3-PBX1), T-ALL, and TEL-AML1(ETV6-RUNX1) subtypes.17,18 Using candidate gene and pathway-directed approaches, differences in intracellular polyglutamate accumulation have also been attributed to the proportion of leukemia cells in S-phase, inhibition of de novo purine synthesis, and the expression levels of SLC19A1, FPGS, ABCG2, and DHFR, which function in the transport or metabolism of MTX.14,17,19-23 However, none of these factors can completely account for the extent of variation in MTXPG accumulation observed in patient leukemia cell samples within and between ALL subtypes.
Herein we used multiple genome-wide approaches to characterize the inherited and somatically acquired genomic determinants of MTXPG accumulation in childhood ALL.
Methods
Patients
Children were treated at St Jude Children's Research Hospital for ALL on previously published protocols Total XIIIA, XIIIB and XV.24-26 The Institutional Review Board at St Jude Children's Research Hospital approved the investigation, and signed informed consent was obtained from patients or parents before enrollment, as appropriate, in accordance with the Declaration of Helsinki.
MTXPG accumulation was measured in bone marrow samples obtained from 248 patients at 42 to 44 hours after a single treatment of MTX, at 1 g/m2 intravenously infused over 4 hours or 24 hours as preinduction window therapy (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). This time point is the latest practical time for sampling that still precedes administration of leucovorin, is consistent with our prior studies,14,27-31 and is likely to reflect steady-state intracellular accumulation (based on serial sampling in peripheral blood ALL blasts).29 The median percentage leukemia cells in the post-MTX bone marrow samples was 96%.
The patients were diagnosed with ALL based on immunophenotyping as well as molecular and cytogenetic analysis3 and were grouped into T-lineage ALL or B-lineage ALL with a hyperdiploid karyotype (> 50 chromosomes; B-hyperdiploid), t(1;19) (E2A-PBX1 or TCF3-PBX1), t(12;21) (TEL-AML1 or ETV6-RUNX1), or those patients who had none of these chromosomal aberrations (B-other). We did not include patients with MLL-AF4 and BCR-ABL1 subtypes resulting from small patient numbers (Table 1). The median percentage of leukemic blasts in the diagnostic bone marrow samples was 97%; 17 of 248 (6.9%) patients with evaluable MTXPG had fewer than 80% blasts in bone marrow.
Plasma MTX concentrations were measured at 1, 6, 23, and 42 hours from the start of MTX; pharmacokinetic parameters, including MTX clearance (mL/min per m2), were estimated using a 2-compartment first-order model as described29 using the maximum a posteriori probability estimator in ADAPT II32 with priors based on previous St Jude Total protocols.
Cell lines
Ninety Epstein-Barr virus (EBV)–transformed B lymphoblastoid cell lines derived from 30 trios (mother, father, and child) of Utah residents with ancestry from Northern and Western Europe (CEU) and 90 EBV-transformed B lymphoblastoid cell lines derived from 30 trios from a Yoruba community in Ibadan, Nigeria (YRI) were purchased from Coriell Institute for Medical Research (Camden, NJ). These constitute the core cell lines genotyped by the International HapMap project (www.hapmap.org). The cell lines were maintained in RPMI 1640 media with 2 mM l-glutamine (Invitrogen, Carlsbad, CA) and 15% fetal bovine serum (HyClone Laboratories, Logan, UT) in a humidified atmosphere at 37°C with 5% CO2. Four cell lines (3 CEU and 1 YRI) were excluded from further analysis because of low viability.
Drug
MTX in the form of MTX sodium used to treat the patients was obtained from Bedford Laboratories (Bedford, OH). MTX (NSC-740) used to treat the cell lines was obtained from Sigma-Aldrich (St Louis, MO).
MTXPG accumulation
The bone marrow cells from patients were prepared by centrifugation in a Ficoll-hypaque gradient and washed 3 times using a solution of N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, Hank buffered salt solution, and heparin as described earlier.15 Supernatants were resuspended in water, boiled in a water bath for 5 minutes, and frozen at −80°C until analysis.
Two aliquots of 5 million cells from each of the 176 CEU and YRI cell lines were plated at a density of 500 000 cells/mL, incubated at 37°C with 1 μM MTX and 670 μM glycine, 37 μM adenosine, and 41 μM thymidine for 24 hours, centrifuged, washed twice with ice-cold phosphate-buffered saline, resuspended in H2O, and boiled in a water bath for 5 minutes.33 Supernatants were then frozen at −80°C until analysis by high-pressure liquid chromatography.
After high-pressure liquid chromatography separation, MTXPGs were quantified in patient bone marrow sample supernatants by a radiolabeled enzymatic assay (REA)–based method or by a fluorescent method of detection19,34-36 ; all results were normalized to the REA scale of values (details in “Supplemental Methods” in Document S1, Figure S2).
Gene expression
High-quality total RNA was extracted using TriReagent (MRC, Cincinnati, OH) from cryopreserved mononuclear cell suspensions from bone marrow at diagnosis of ALL from 145 patients (Figure S1). Because there was no significant difference (P = .082, t test) in MTXPG accumulation between the 145 patients with available gene expression data versus 103 patients without these data, it appears there is minimal selection bias in the group for whom gene expression data were evaluable. The Affymetrix HG-U133A 2.0 Array (Affymetrix, Santa Clara, CA) comprising more than 22 000 probe sets representing approximately 14 500 characterized genes was used to interrogate the expression of RNA as described.37 The data were analyzed with Bioconductor 2.1 using the Affymetrix MAS5.0 algorithm (www.bioconductor.org). Genes with an average intensity of less than 6 were considered not expressed and therefore excluded from further analysis. The expression signals were log transformed. Quality control of the gene expression data was carried out as described previously, leaving 15 119 probe sets for further analysis.38 Gene expression analysis of the CEU and YRI cell lines using the Affymetrix GeneChip Human Exon 1.0 ST Array comprising approximately 1.4 million probe sets was downloaded and processed as previously described (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE7761).39
Somatic leukemia copy number variation
DNA was extracted from diagnostic bone marrow of 82 patients for genomic analysis of copy number variation (CNV; Figure S1). Because there was no significant difference (P = .314, t test) in MTXPG accumulation between the 82 patients with versus the 166 patients without leukemia cell DNA CNV data, there appears to be little selection bias in the patients evaluated for cell CNV. A total of 500 ng of each DNA sample was digested with HindIII and XbaI or NspI and StyI restriction enzymes, amplified, labeled, and hybridized to the Affymetrix GeneChip Human Mapping 100K set and 500K set, respectively. DNA copy number analysis was performed using dChipSNP40 as described previously.41,42 Two CNV datasets were used: normalized single inherited single nucleotide polymorphism (SNP) intensities, including 615 922 SNPs and large chromosome genomic segments inferred from DNACopy.41 If segmental analysis indicated gain or loss of more than 80% of the chromosome, it was considered as a gain or loss of the whole chromosome. We considered CNV only if it was observed in at least 3 patients.
Somatic leukemia cell cytogenetic analysis
Inherited (constitutional) DNA genotyping
DNA was extracted using the QIAGEN Blood and Cell Culture DNA kit (Valencia, CA) from whole blood obtained from 144 patients (Figure S1) after they achieved complete remission. Because there was no significant difference (P = .66, t test) in MTXPG accumulation between the144 patients with versus the 104 patients without inherited DNA genotyping data available, the possibility of selection bias is minimized. Germline DNA samples were genotyped using the Affymetrix GeneChip Human Mapping 100K set and 500K set as described in “Somatic leukemia copy number variation.” The chips were scanned, and genotype calls were made using the BRLMM algorithm as implemented in the GTYPE software (http://www.affymetrix.com/products/software/specific/gtype.affx). SNPs with call rates of less than 95% in all patients and minor allele frequencies of less than 1% were filtered out, leaving 447 287 SNPs for further analysis. Publicly available genotyping data for the 87 CEU cell lines and 89 YRI cell lines were downloaded from the International HapMap Project website, release 22 (www.hapmap.org).
Inherited CNV
Methods used for assessing inherited CNV are provided in “Supplemental Methods” in Document S1.
Statistical analyses
Multiple linear regression was used to evaluate the association between MTXPG accumulation (normalized for detection method as detailed in “Supplemental Methods” in Document S1) in patient samples and window treatment arm, ALL subtype, MTX plasma clearance during window treatment, and method of MTXPG detection (REA or fluorescence) using all patients with available data (n = 248; Table S1). The residuals from the multiple regression model were used to estimate adjusted MTXPG levels (adjusted for all significant covariates), and adjusted MTXPG levels were used as the dependent variable in subsequent association analyses.
Linear regression analysis was performed to assess the association between MTXPGs and gene expression, percentage blasts in the bone marrow, leukemia cell gene or chromosome CNV, and genotype of the inherited SNPs.
In the gene expression analysis, we performed a linear regression analysis between MTXPGs and the gene expression levels for each probe set represented on the Affymetrix U133A expression array. Probe sets were rank ordered by their P values. Pearson χ2 test was used to test whether the number of genes associated with MTXPGs on each chromosome were overrepresented.
In the CNV analysis, we used the normalized signal intensity for each SNP as the independent variable to perform a linear regression analysis between MTXPGs and the signal intensities for each of the approximately 600 000 SNPs on the SNP Chip. Pearson χ2 test was used to test whether the number of SNPs within any chromosome regions were overrepresented. In addition, we also performed a regression analysis for each of the 22 autosomes using the status of whole chromosome gain/loss as the independent variable.
In the genotype analysis, we treated the genotypes of all approximately 600 000 SNPs on the SNP arrays as the independent variables (genotypes were coded based on an additive model: 0, 1, and 2 for AA, AB, and BB, respectively). Race was included as a covariate in the model. A SNP was annotated to a gene if it was within 10 kb of the transcription start or end site. If there were fewer than 3 patients in a homozygous genotype group, these patients were combined with the heterozygous genotype group for that particular SNP. The deviation from Hardy-Weinberg equilibrium was calculated for each SNP in the largest patient group (whites), and those with P less than 10−7 were excluded from further analysis.
Similar analyses were performed in the CEU and YRI cell lines between MTXPGs and gene expression or SNP genotypes. We applied a linear mixed effects model to account for the relatedness of the family trios included in the analysis. Race was included as a covariate in the linear mixed effect model.
All statistical analyses were performed using the statistical environment R2.6.1 (R Development Core Team, http://www.r-project.org).
Results
MTXPG accumulation in patient leukemia cells and cell lines
As expected, there was wide interindividual variation in MTXPG accumulation in the leukemia cells of the 248 patients, and it displayed a normal distribution (Figure S3A). We found that MTXPG accumulation was significantly associated with ALL subtype (B-hyperdiploid patients had the highest MTXPG accumulation), MTX treatment arm (MTX infused over 24 hours had higher MTXPGs than after a 4-hour infusion), MTX plasma clearance (higher MTXPGs with lower plasma MTX clearance), and method of detection (higher MTXPGs for fluorescence than for the REA method of detection; Table S1). Thus, the subsequent genome-wide association analyses used MTXPG measures that were adjusted for these covariates. We did not observe a significant correlation between total MTXPG1-7 accumulation and the percentage of blasts in bone marrow (r2 = 0.01, P = .09). There was a very high correlation (r2 = 0.93) between total MTXPG1-7 accumulation and long-chain MTXPG5-7 in the leukemia cells of the 248 patients. Therefore, all analyses were conducted using total MTXPG accumulation.
There was wide interindividual variation in MTXPG accumulation of the CEU and YRI HapMap cell lines used in this study, and it also showed normal distribution (Figure S3B).
Whole genome approach
Leukemia cell gene expression associated with leukemia cell MTXPG accumulation in patients.
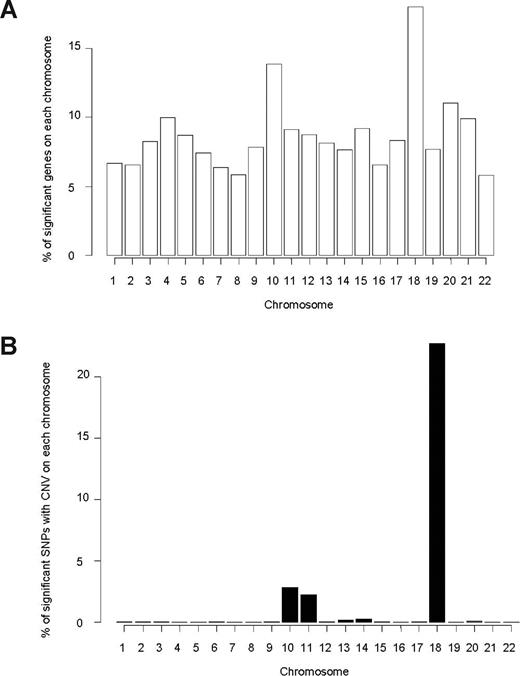
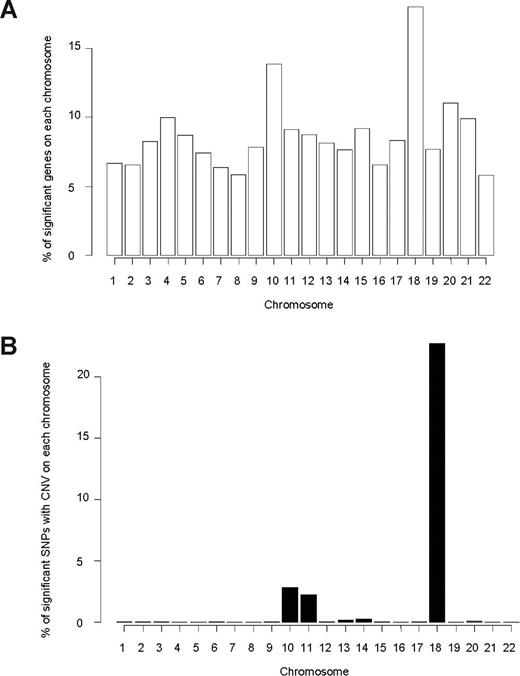
Among 11 783 evaluable genes (15 119 probe sets) in 145 patients, we found that expression of 1075 genes (1232 probe sets) was significantly associated with MTXPG accumulation (P < .05; Figure S4A). Genes whose expression was associated with MTXPG accumulation were overrepresented on chromosome 18 (P = 1.3 × 10−6, χ2 test) and on chromosome 10 (P = 1.1 × 10−7, χ2 test; Figure 1A; Table S2).
Chromosome distribution of genes and SNPs associated with MTXPG. (A) Genes whose expression is significantly associated (P < .05) with MTXPG in 145 patients are overrepresented on chromosome 18 (P = 1.3 × 10−6, χ2 test) and chromosome 10 (P = 1.1 × 10−7, χ2 test). (B) Single SNP CNV in 82 patients significantly associated (P < .01) with MTXPG is overrepresented on chromosome 18 (P < 1.0 × 10−7, χ2 test) and on chromosome 10 (P < 1.0 × 10−7, χ2 test).
Chromosome distribution of genes and SNPs associated with MTXPG. (A) Genes whose expression is significantly associated (P < .05) with MTXPG in 145 patients are overrepresented on chromosome 18 (P = 1.3 × 10−6, χ2 test) and chromosome 10 (P = 1.1 × 10−7, χ2 test). (B) Single SNP CNV in 82 patients significantly associated (P < .01) with MTXPG is overrepresented on chromosome 18 (P < 1.0 × 10−7, χ2 test) and on chromosome 10 (P < 1.0 × 10−7, χ2 test).
Somatic leukemia cell single SNP CNV associated with leukemia cell MTXPG accumulation in patients.
We assessed the association of single SNP CNV with MTXPG accumulation in diagnostic leukemia cells of all 82 patients with CNV data. Of those SNPs with CNV (n = 534 630 SNPs; 26 726 regions), 6065 were associated with MTXPG accumulation (P < .01), implicating 156 genes (Table S3). The chromosomal locations of these SNP CNVs associated with MTXPGs were overrepresented on chromosome 18 (P < 1.0 × 10−7, χ2 test), on chromosome 10 (P < 1.0 × 10−7, χ2 test), and on chromosome 11 (P < 1.0 × 10−7, χ2 test; Figure 1B).
Inherited genetic variation and leukemia cell MTXPG accumulation in patients.
Of 447 287 inherited SNP genotypes in 144 patients, 4632 SNPs had allele frequencies that were significantly associated with MTXPG accumulation (P < .01), and these SNPs represented 1005 unique genes and all autosomes (Figures S4B, S5).
Inherited CNV was also assessed in these 144 patients (based on 600K SNP-chip data) but was not strongly associated with MTXPG accumulation (“Supplemental Methods” in Document S1; Table S4).
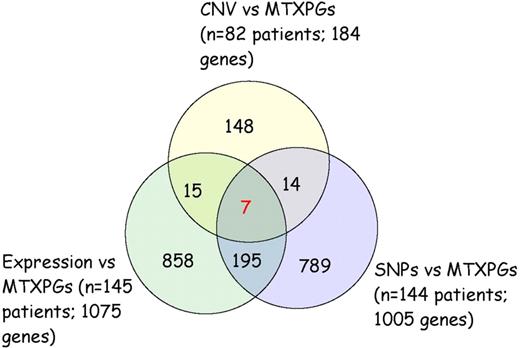
Overlap between leukemia cell gene expression, somatic leukemia cell CNV, and inherited SNP genotype versus MTXPG accumulation association analyses in patients.
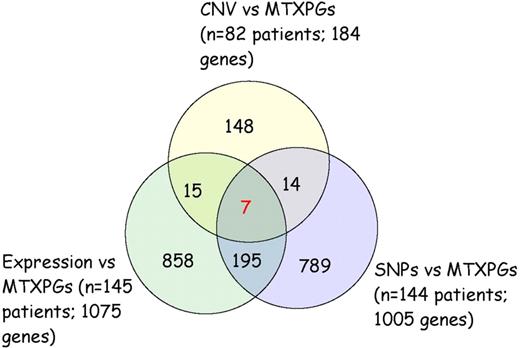
Combining the genes associated with MTXPGs based on leukemia cell gene expression (n = 145; 1075 genes), leukemia cell CNV (n = 82; 184 genes), and inherited SNP genotypes (n = 144; 1005 genes), there were 7 genes that overlapped for all 3 levels of evidence: FHOD3, IMPA2, ME2, RASSF4, SLC39A6, SMAD2, and SMAD4 (Figure 2; Table S5). Of these 7 genes, 6 were located on chromosome 18; RASSF4 is located on chromosome 10. The expression of these 7 genes along with ALL subtype, MTX clearance, and method of measurement of MTXPGs accounted for 59% of the variation in MTXPGs observed among the 145 patients with gene expression data.
Seven genes overlap from the association analyses of MTXPG accumulation versus leukemia cell gene expression, leukemia cell single SNP CNV, and inherited SNP genotypes.
Seven genes overlap from the association analyses of MTXPG accumulation versus leukemia cell gene expression, leukemia cell single SNP CNV, and inherited SNP genotypes.
Analysis of chromosomes 18 and 10
Somatic leukemia cell whole chromosome CNV associated with leukemia cell MTXPG accumulation.
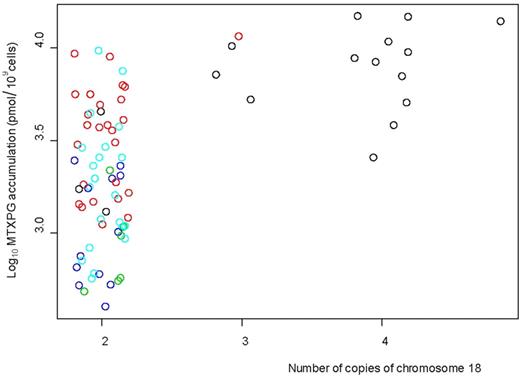
We assessed the presence of whole chromosome (> 80% of a chromosome affected by either a loss or gain of copy number) CNV (based on the 600K SNP-chip data) associated with MTXPG accumulation in the leukemia cells of 82 evaluable patients. Increased CNV of chromosome 18 had the strongest association with MTXPG accumulation (P = 3.13 × 10−8, analysis of variance [ANOVA]; Table S6; Figure 3) with 15 of the 82 patients having a gain of chromosome 18. Of the 15 patients with a gain of chromosome 18, 14 had B-hyperdiploid ALL subtype; when we adjusted the association analysis for ALL subtype, chromosome 18 CNV was still significantly associated with MTXPG accumulation (P = .002, ANOVA).
Increased copies of chromosome 18 in the patient leukemia cells (n = 82, evaluated by 600 000 SNP-chip data) were associated with higher MTXPG accumulation (P = 3.13 × 10−8, ANOVA). Fourteen of the 15 patients with a gain of copy number of chromosome 18 had B-hyperdiploid subtype. Black circles represent B-hyperdiploid; red circles, B-other; light blue circles, T-lineage; dark blue circles, TEL-AML1; green circles, E2A-PBX1.
Increased copies of chromosome 18 in the patient leukemia cells (n = 82, evaluated by 600 000 SNP-chip data) were associated with higher MTXPG accumulation (P = 3.13 × 10−8, ANOVA). Fourteen of the 15 patients with a gain of copy number of chromosome 18 had B-hyperdiploid subtype. Black circles represent B-hyperdiploid; red circles, B-other; light blue circles, T-lineage; dark blue circles, TEL-AML1; green circles, E2A-PBX1.
Increased CNV of chromosome 10 was also associated with higher MTXPG accumulation (P = 1.1 × 10−5, ANOVA; Figure 4; Table S6), with 15 of 82 patients having a gain of chromosome 10. Twelve of 15 patients had B-hyperdiploid ALL subtype; when we adjusted the association analysis for ALL subtype, the chromosome 10 CNV was still significantly associated with MTXPG accumulation (P = .036, ANOVA).
Increased copies of chromosome 10 in the patient leukemia cells (n = 82, evaluated by 600 000 SNP-chip data) were associated with higher MTXPG accumulation (P = 1.08 × 10−5, ANOVA). Twelve of the 15 patients with a gain of copy number of chromosome 10 had B-hyperdiploid subtype. Black circles represent B-hyperdiploid; red circles, B-other; light blue circles, T-lineage; dark blue circles, TEL-AML1; green circles, E2A-PBX1.
Increased copies of chromosome 10 in the patient leukemia cells (n = 82, evaluated by 600 000 SNP-chip data) were associated with higher MTXPG accumulation (P = 1.08 × 10−5, ANOVA). Twelve of the 15 patients with a gain of copy number of chromosome 10 had B-hyperdiploid subtype. Black circles represent B-hyperdiploid; red circles, B-other; light blue circles, T-lineage; dark blue circles, TEL-AML1; green circles, E2A-PBX1.
Cytogenetic analysis of somatic whole chromosome copy number was available for a larger number of patients (n = 213; 82 of whom also had SNP-chip-defined CNV, with 95% concordance between the 2 methods for chromosome 10 and 94% for chromosome 18). There was a significant positive correlation between cytogenetically defined chromosome 18 copy number and MTXPG accumulation (P = 1.0 × 10−9, Figure S6A), which remained when the analysis was adjusted for ALL subtype (P = .002), and also when only patients with B-hyperdiploid ALL were analyzed (P = .012; n = 56 total; n = 38 with chromosome 18 gain). There was also a significant positive correlation between cytogenetically defined chromosome 10 copy number and MTXPG accumulation (P = 1.9 × 10−8, Figure S6B) that remained after adjusting the analysis for ALL subtype (P = .030) and within B-hyperdiploid patients only (P = .013; n = 56 total; n = 40 with chromosome 10 gain). Cytogenetically defined chromosome 17 copy number was significantly positively correlated with MTXPG accumulation (P = 1.8 × 10−6, Figure S6C), but this association did not remain after adjusting the analysis for ALL subtype (P = .455).
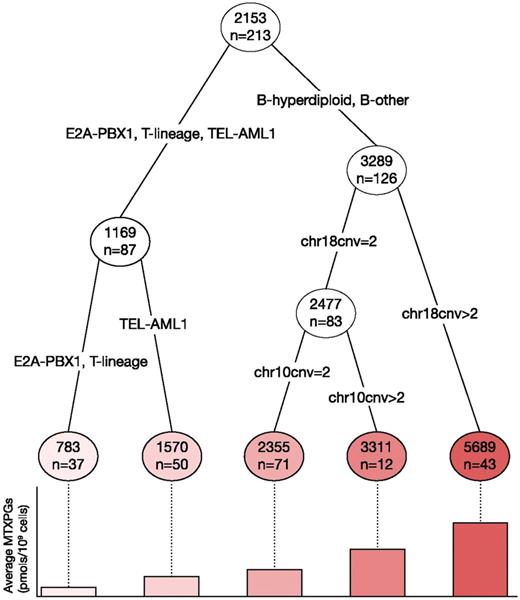
In a classification and regression tree analysis carried out using the 213 patients with cytogenetic analysis data available (Figure 5), ALL subtype was the strongest predictor of MTXPG accumulation (P = 8.7 × 10−5), followed by chromosome 18 copy number in B-hyperdiploid and B-other patients (P = 1.6 × 10−5). In the patients with 2 or fewer copies of chromosome 18, chromosome 10 copy number was marginally associated with MTXPG accumulation (P = .19).
Classification and regression tree based on all 213 patients with cytogenetically determined chromosome copy number for predictors of MTXPG accumulation. ALL subtype was the strongest predictor of MTXPG accumulation (P = 8.71 × 10−5), followed by chromosome 18 copy number (chr18cnv) (P = 1.6 × 10−5; adjusted for subtype), and chromosome 10 copy number (chr10cnv) in patients with no chromosome 18 gain (P = .192; adjusted for subtype). Numbers in circles indicate the mean MTXPG concentration in patient leukemia cells (pmol/109 cells), also depicted in the bar chart below.
Classification and regression tree based on all 213 patients with cytogenetically determined chromosome copy number for predictors of MTXPG accumulation. ALL subtype was the strongest predictor of MTXPG accumulation (P = 8.71 × 10−5), followed by chromosome 18 copy number (chr18cnv) (P = 1.6 × 10−5; adjusted for subtype), and chromosome 10 copy number (chr10cnv) in patients with no chromosome 18 gain (P = .192; adjusted for subtype). Numbers in circles indicate the mean MTXPG concentration in patient leukemia cells (pmol/109 cells), also depicted in the bar chart below.
Leukemia cell gene expression association with somatic leukemia cell CNV and MTXPG accumulation.
Because extra copies of chromosomes 18 and 10 were associated with increased MTXPGs, we performed a directed analysis to test whether expression of genes on these chromosomes was associated with MTXPGs (n = 82 patients). Of the 189 genes interrogated on the Affymetrix HG-U133A array that were on chromosome 18, 44 were significantly positively correlated with MTXPG accumulation (P < .05). After excluding the 15 patients with chromosome 18 gains identified in the aforementioned analysis, the expression of 3 of the 44 genes (IMPA2, PHLPP, and TYMS) on chromosome 18 remained significantly associated (P < .05) with MTXPG accumulation; these associations remained when the analysis was limited to patients with more than 80% blasts in bone marrow (Table 2). The correlation between the expression of these 3 genes and MTXPG accumulation also differed by ALL subtype, with the strongest effect observed within the B-hyperdiploid and B-other subtype groups (Table S7). A substantial proportion (∼ 43%) of the variation in MTXPG accumulation in B-lineage ALL (n = 42; B-hyperdiploid and B-other subtypes pooled) was explained by the expression of these 3 genes on chromosome 18.
Of the 447 genes interrogated on chromosome 10 by the U133A array, 131 were significantly positively correlated with MTXPG accumulation (P < .05). After excluding the 15 patients with a gain of chromosome 10, the expression of 10 of the 131 genes remained significantly associated (P < .05) with MTXPG accumulation (Table S8). The correlation between the expression of these 10 genes and MTXPG accumulation differed by ALL subtype, with the strongest effect observed within the B-other subtype (Table S9). A substantial proportion of the variation in MTXPG accumulation in B-hyperdiploid (∼ 45%), B-other (∼ 32%), and TEL-AML1 (∼ 52%) ALL subtypes can be accounted for by the expression of these 10 genes.
By combining the 3 genes (IMAP2, PHLPP, and TYMS) on chromosome 18 and the 10 genes (BUB3, CDC2, GHITM, GLRX3, GOT1, KAZALD1, KIF11, PRDX3, VDAC2, and ZWINT) on chromosome 10 whose expression was most significantly associated with MTXPG accumulation, we found that the expression of these 13 genes accounted for 66% of the variation in MTXPG accumulation among patients. Stepwise variable selection from among these 21 genes indicated that the expression of 3 genes, IMPA2, PHLPP, and VDAC2, together accounted for 62% of the variation in MTXPGs in patient samples.
Additional chromosomes with somatic leukemia cell CNV associated with leukemia cell MTXPG accumulation
MTXPG accumulation in normal lymphoblastoid cell lines
Gene expression versus MTXPG accumulation analysis.
We interrogated the association of the expression of approximately 1.4 million probe sets (from the Affymetrix GeneChip Human Exon 1.0 ST Array) with MTXPG accumulation in the CEU and YRI cell lines. We found that the expression of 612 genes was significantly associated (P < .05) with MTXPG accumulation. Of these 612 genes, only 35 were found to be significantly and concordantly associated (P < .05) with MTXPG accumulation in the patient leukemia cells (Table S10). Nonetheless, 7 of the 35 (20%) overlapping genes are on chromosome 18, an overrepresentation compared with what would be expected by chance (1.5%, P = 8.26 × 10−7). Identical to the patient analysis, we found a significantly positive correlation between IMPA2 expression (P = .033, Pearson correlation) or PHLPP expression (P = .041, Pearson correlation) and MTXPG accumulation (Figure S9). To explore how well gene expression variation accounted for MTXPG variation in patient leukemia cells versus normal lymphoblastoid cell lines, we estimated the percentage of variation in MTXPGs accounted for by the top 7 genes in each dataset. The expression of the top-ranked 7 genes (based on P value) explains approximately 46% of the variation in MTXPGs seen in 145 patients, whereas the expression of the top-ranked 7 genes explains only approximately 20% of the variation in MTXPGs observed in the 176 cell lines. There was no overlap in the top 7 genes for patient leukemia cells versus normal lymphoblastoid cell lines.
SNP genotype versus MTXPG accumulation analysis.
Using the publicly available genotyping data from the International HapMap project for the CEU and YRI cell lines, we found that 8310 SNP genotypes were significantly associated (P < .01) with MTXPG accumulation. Of these, 35 SNP genotypes were also found to be significantly and concordantly associated with MTXPG accumulation in the patient leukemia cells (Table S11). The 35 SNPs were not overrepresented on chromosomes 18 or 10. To explore how well inherited SNP genotypes accounted for variation in MTXPG accumulation in patient leukemia cells versus normal lymphoblastoid cell lines, we estimated the percentage of variation in MTXPGs accounted for by the top 7 inherited SNPs (based on P value) in each dataset. The top 7 inherited SNP genotypes explain approximately 17% of the variation in MTXPGs observed in the 144 patients, whereas the top 7 SNP genotypes explain approximately 20% of the variation in MTXPGs observed in the 176 cell lines. There was no overlap in the top-ranked 7 SNPs for inherited DNA from patients versus normal lymphoblastoid cell lines.
Discussion
The variation in MTXPG accumulation among ALL subtypes has been previously described,17,18 but the inherited and acquired genomic determinants of this variation remain unclear. Using a multimodality genome-wide approach, the most important genomic variations associated with variation in MTXPG accumulation were related to somatically acquired genomic variation, with a lesser component resulting from inherited genetic variation. Specifically, gains of chromosome 10 or 18 in leukemia cells were associated with higher MTXPG levels. This effect can be in large part attributed to the expression of 3 genes found on chromosome 18 (ie, IMPA2, PHLPP, and TYMS; accounting for ∼ 43% of the variation) and 10 genes found on chromosome 10 (ie, BUB3, CDC2, GHITM, GLRX3, GOT1, KAZALD1, KIF11, PRDX3, VDAC2, and ZWINT; accounting for ∼ 45% of the variation). The combination of these 13 genes explained approximately 66% of the variation in MTXPG accumulation, with 62% of this variation accounted for by the expression of only 3 genes: IMPA2, PHLPP, and VDAC2. Further confirmatory and functional experiments are needed to determine whether these genes play a causal role and the mechanisms by which they might affect accumulation of MTX in leukemia.
Genetically distinct subtypes of ALL such as B-lineage ALL with hypodiploid or hyperdiploid karyotypes, or translocations such as BCR-ABL, E2A-PBX1, and TEL-AML1, MLL rearrangements and T-lineage ALL, can be readily distinguished by their unique gene expression profiles.44-46 ALL subtypes are also distinct in their accumulation of MTXPGs, with B-hyperdiploid subtype having the highest accumulation and E2A-PBX1 subtype having the lowest accumulation. Herein, we expanded our approach to the entire genome, which has revealed additional important genomic determinants of MTXPGs, few of which directly overlap with the folate pathway.
There are additional mechanisms that could account for the remaining approximately 34% of variation in MTXPG accumulation that were not interrogated in these analyses. Rare SNP and insertion/deletion polymorphisms not well surveyed on the Affymetrix chips, alternatively spliced variant transcripts, and posttranscriptional mechanisms of variation could all result in variable expression or activity of proteins that were not captured in our surveys but that could impact on MTXPG concentrations.
In children with B-hyperdiploid ALL, gains of chromosome 18 are frequent,47-50 and our results replicate these findings (14 of 17 B-hyperdiploid patients with chromosome 18 gain determined by SNP-chip; 29 of 42 B-hyperdiploid patients with chromosome 18 gain determined by cytogenetics). In addition, increased copy number of chromosome 18 has been associated with better event-free survival47,48 and indeed overall survival47 in patients with B-hyperdiploid ALL using both univariate and multivariate models, consistent with a favorable prognosis associated with hyperdiploidy and with increased intracellular exposure to MTX.
The expression of 3 genes on chromosome 18 (IMPA2, PHLPP, and TYMS) was significantly associated with CNV of this chromosome and was also associated with MTXPG accumulation; indeed, expression levels of these genes combined explained approximately 43% of the variation in MTXPG accumulation observed in our B-hyperdiploid and B-other ALL subtypes combined. Both IMPA2 and TYMS are plausible candidate genes. IMPA2 (inositol monophosphatase 2) is directly linked to glucuronate homeostasis51 and could affect availability of substrate for polyglutamylation of MTX. TYMS (thymidylate synthetase) is directly linked to folate homeostasis as it catalyzes the methylation of dUMP to dTMP using 5,10-methylenetetrahydrofolate as a cofactor. The relationship between TYMS and MTX effects is complicated: because it is a primary target of the drug, higher expression can translate into clinical and preclinical drug resistance. However, increased TYMS expression is also associated with an increase in the percentage of tumor cells in cycle and therefore with sensitivity to MTX52 ; prior work has indeed demonstrated that, across heterogeneous cell types, increased TYMS expression is associated with increased MTX polyglutamylation52,53 and efficacy.23,52 We should acknowledge the fact that all our measures of MTXPGs were made in “bulk” tumor at diagnosis; it is possible that residual ALL cells present later in therapy might differ from diagnostic bulk tumor in their MTXPG or in the relative contribution of TYMS to drug sensitivity (via increased MTXPG) vs resistance (via increased pharmacologic target). Interestingly, the expression of both IMPA2 and PHLPP (but not TYMS) was associated with MTXPG accumulation in the HapMap cell lines (Figure S9).
Gains of chromosome 10 have been associated with increased in vitro MTXPG accumulation in leukemic cells from children with B-hyperdiploid ALL and, indeed, improved response to chemotherapy.54 Herein, we found that gains of chromosome 10 were associated with MTXPG accumulation in vivo after MTX administration to patients and that this association remained significant when we adjusted for ALL subtype. Of the 10 genes on chromosome 10 whose expression was associated with CNV and with MTXPG accumulation, one (GOT1) plays a role in glutamate metabolism, amino acid metabolism, and the urea and tricarboxylic acid cycles, which could be plausibly linked with the MTXPG accumulation we observed. Others, such as PRDX3 (a target of the c-myc oncogene)55,56 and GLRX3, have been linked to cancer response.57
As in other studies of pediatric ALL,47,48,50,58 we found that gains in copy number of chromosomes 4, 17, and 21 were common. Although gains of chromosomes 4 and 21 were significantly associated with MTXPG accumulation, these associations were not significant when the analyses were adjusted for ALL subtype, and the genes on these chromosomes were not overrepresented based on gene expression and SNP analyses. In a previous study,14 gain of chromosome 21 was associated with an increase in MTXPG accumulation, but only in patients treated with low-dose MTX (30 mg/m2 orally), not those treated with high-dose MTX as was true for the patients reported herein. Trisomies of chromosomes 4, 10, and 17 have been shown to be associated with increased event-free survival by some groups,59,60 although the mechanism underlying this finding is not clear.
Whether the inherited genetic variation of the host or the somatically acquired genetic variation of the tumor is more important for drug-responsiveness is a fundamental question of cancer pharmacogenetics.3,58,61 Although we found inherited genetic variation in the form of SNP genotypes associated with MTXPG accumulation in the patient's leukemia cells, we found that they explain less of the variation in MTXPGs observed in patients than the expression of genes in the leukemia cells (17% vs 43% when we take the top 7 SNP genotypes or gene expression values, respectively). In addition, the amount of variation in MTXPGs in the normal lymphoblastoid HapMap cell lines that is accounted for by SNP genotypes or by gene expression is approximately the same (20% vs 20%, respectively, when we take the top 7 SNPs/genes from each analysis) as is true for inherited SNPs in patient samples. We also found little overlap between the results of the association analyses from the patient leukemia cells and the normal HapMap cell lines. Together, these data indicate that acquired somatic lesions in the leukemia cells (evident in CNV or leukemic cell gene expression) have a stronger influence on the variable accumulation of MTXPGs than does inherited genetic variation (evident in inherited SNP genotypes or in gene expression), although it is worthwhile to note that our findings are considering the entire population of leukemia cells and that there may be somatic lesions that distinguish subpopulations of these cells, such as those that persist as minimal residual disease. This is the first human cancer for which this has been shown; and given the frequency of DNA and chromosomal aberrations in cancer cells, it merits investigation in other human malignancies.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the patients and their families, and our research faculty and staff for participating; Dr J. Carl Panetta for the pharmacokinetic analysis; Nancy Kornegay, Diana Chan, and Mark Wilkinson for database and computer expertise; and Yaqin Chu, May Chung, Natalya Lenchik, Margaret Needham, Emily Melton, and Siamac Salehy for outstanding technical assistance.
This work was supported by National Cancer Institute (CA51001, CA78224, CA21765), the National Institutes of Health/National Institute of General Medical Sciences Pharmacogenetics Research Network and Database (U01GM61393, UO1GM61374; www.pharmgkb.org PS207998 and PS207999), the Phelan Foundation, a Center of Excellence grant from the State of Tennessee, and the American Lebanese Syrian Associated Charities.
National Institutes of Health
Authorship
Contribution: M.V.R. designed research, collected and interpreted data, and drafted the manuscript; D.F. analyzed and interpreted data and drafted the manuscript; W.Y. and C.C. performed statistical analysis; S.C.R. performed cytogenetics and ALL classification; C.G.M. and J.R.D. determined acquired abnormalities based on genomics; and W.E.E. and C.-H.P. were principal investigators of the laboratory and clinical protocols, respectively.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mary V. Relling, Department of Pharmaceutical Sciences, St Jude Children's Research Hospital, 262 Danny Thomas Pl, Memphis, TN 38105-2794; e-mail: mary.relling@stjude.org.