Abstract
Leukocyte adhesion deficiency-1/variant (LAD1v) syndrome presents early in life and manifests by infections without pus formation in the presence of a leukocytosis combined with a Glanzmann-type bleeding disorder, resulting from a hematopoietic defect in integrin activation. In 7 consanguineous families, we previously established that this defect was not the result of defective Rap1 activation, as proposed by other investigators. In search of the genetic defect, we carried out homozygosity mapping in 3 of these patients, and a 13-Mb region on chromosome 11 was identified. All 7 LAD1v families share the same haplotype, in which 3 disease-associated sequence variants were identified: a putative splice site mutation in CALDAGGEF1 (encoding an exchange factor for Rap1), an intronic 1.8-kb deletion in NRXN2, and a premature stop codon (p.Arg509X) in FERMT3. Two other LAD1v patients were found to carry different stop codons in FERMT3 (p.Arg573X and p.Trp229X) and lacked the CALDAGGEF1 and NRXN2 mutations, providing convincing evidence that FERMT3 is the gene responsible for LAD1v. FERMT3 encodes kindlin-3 in hematopoietic cells, a protein present together with integrins in focal adhesions. Kindlin-3 protein expression was undetectable in the leukocytes and platelets of all patients tested. These results indicate that the LAD1v syndrome is caused by truncating mutations in FERMT3.
Introduction
The leukocyte adhesion deficiency-1/variant (LAD1v) syndrome was first described by us in 1997; it consists of 2 major hallmarks: a moderate LAD-1–like syndrome and a severe Glanzmann-like bleeding tendency.1 To date, the natural history of the patients is remarkable for clinical features related to defects in granulocyte function that result in leukocytosis, infections, and/or inflammation without pus formation in all patients and poor wound healing or late detachment of the umbilical cord in some patients.2 A platelet disorder manifests by early hemorrhagic diathesis in a number of patients, resulting in severe intracranial bleedings.2 The need for regular erythrocyte transfusions can be understood by the repeated skin and mucosal bleedings from oropharynx and, to a lesser extent, from the gastrointestinal tract.
For adhesion, various molecules are important, among which are a unique family of heterodimeric structures known as the integrins. In particular, the members of the subfamilies of β1 and β2 integrins on leukocytes and β3 integrins on platelets are required for the induction of strong adhesion, which is regulated by cell activation. Many molecules involved in signal transduction have been identified in regulating integrin activity, but the insight in the signaling pathways that control the inside-out signaling is still limited.3,4
In LAD1v patients, both the leukocytes and platelets have a normal level of integrin expression, but these integrins do not function properly. In vitro, the absence of chemotaxis and the lack of adhesion of neutrophils were noted, without any obvious signaling defect. The same is true for the platelets in the patients because effector functions induced in the platelets are intact as long as the β3 integrin activation is not a determinant step in the process.1,2
Alon et al have suggested that the syndrome, which they named LAD-III, is caused by an aberrant activation of Rap1.5,6 Recently, this group has found in 2 patients mutations in a guanine nucleotide exchange factor (GEF) for Rap1.7 These patients were described to be homozygous for a splice junction mutation in the CALDAGGEF1 (alias RASGRP2) gene encoding this exchange factor. Both mRNA and protein levels of this GEF were reported to be diminished in LAD-III lymphocytes, neutrophils, and platelets. Moreover, the caldaggef1−/− mouse model clearly underpinned the phenotype of the Glanzmann-type of platelet disorder, combined with the defective leukocyte extravasation and inflammatory response in a peritonitis model.8
Rap1 functions in spatial and temporal control of cell polarity.9-12 Similar to all other small GTPases, Rap1 binds either guanosine diphosphate (GDP) or guanosine triphosphate (GTP), and the conformational change between these 2 states represents a molecular switch: ie, the GTP-bound form is “on” (able to react with downstream effector molecules) and the GDP-bound form is “off.”9,13 The GDP/GTP exchange is regulated by tissue- and cell-specific GEFs and GTPase-activating proteins. In contrast to the results described in LAD-III,5 we did not find any effect of the putative splice site mutation on CalDAG-GEF1 mRNA levels, splicing, or CalDAG-GEF1 protein levels in our patients.14 Moreover, Rap1 activation was normal, suggesting a normal activation and deactivation cycle and hence a normal CalDAG-GEF1 activity.2
Over the years, we had identified a series of 9 LAD1v patients from 7 families.2 We have now used homozygosity mapping with single nucleotide polymorphism (SNP) array methodology to identify homology regions in the genome. Such an approach has been successfully used to identify the gene locus and subsequent gene defect in a series of genetic disorders. A large homozygous region of 13 Mb is identified in the LAD1v families. These families all shared that same haplotype, strongly suggesting that the LAD1v locus is located in this region. We have now identified mutations in FERMT3, encoding kindlin-3, as the cause of LAD1v syndrome, and we have excluded mutations in CALDAGGEF1 or a combination of the mutations in both CALDAGGEF1 and FERMT3, as was most recently suggested.15
Methods
Purification of different blood cells
Heparinized venous blood was collected from healthy donors and from LAD1v patients and their family members, after informed consent was obtained. The study was approved by the Academic Medical Center institutional medical ethical committee in accordance with the standards laid down in the 1964 Declaration of Helsinki.
The whole-blood samples were first centrifuged for 15 minutes at 300g to obtain platelet-rich plasma. After removal of the platelet-rich plasma fraction, the interface was diluted with an equal volume of phosphate-buffered saline (PBS) containing 0.5% (wt/vol) human serum albumin, and peripheral mononuclear cells (PBMCs) and granulocytes were separated by Percoll density gradient separation as described.16,17 The interface on top of the Percoll containing the PBMCs was washed twice in PBS and resuspended to a concentration of 10 × 106 cells/mL before further analysis. After lysis of the erythrocytes in the pellet fraction in isotonic NH4Cl,17 the granulocytes were washed twice in PBS containing 0.5% (wt/vol) human serum albumin and resuspended to a concentration of 10 × 106 cells/mL before further analysis. Purity of neutrophils obtained was always more than 95%.
Platelets were washed free of plasma by dilution in a 10-fold excess of N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid buffer (132 mM NaCl, 6 mM KCl, 1 mM MgSO4, 1.2 mM K2HPO4, 20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.4) with acid citric dextrose (85 mM trisodium citrate, 71 mM citric acid, and 111 mM dextrose) added at one-tenth of the total volume resulting in a pH of 6.7 to 6.8. After centrifugation (500g for 10 minutes), platelets were washed again in the same buffer and finally resuspended at 50 × 106 cells/mL in N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid buffer containing 5 mM glucose and 1 mM CaCl2.
Platelet activation and PAC-1 antigen detection
Washed platelets (50 × 106 cells/mL) were incubated for 30 minutes at 37°C and then stimulated for 5 minutes with 1 U/mL thrombin. Subsequently, samples were incubated for 20 minutes at room temperature with fluorescein isothiocyanate (FITC)–labeled PAC-1 (10 μg/mL), a monoclonal antibody selectively recognizing the high-affinity conformation of αIIbβ3.18 Parallel incubations were performed with 10 μg/mL CD61-FITC (clone C17) to measure total αIIbβ3 expression and control mouse IgG-FITC to measure background binding. After staining, the platelet samples were fixed by 10-fold dilution in PBS containing 0.5% (wt/vol) formaldehyde, and PAC-1 binding was subsequently measured with a FACScan flow cytometer (BD Biosciences, San Jose, CA).
Western blotting
Cell lysates of neutrophils, PBMCs, or platelets were prepared by addition of an equal volume of Laemmli sample buffer containing 50 mM dithiothreitol and 1% β-mercaptoethanol and heated for 30 minutes at 95°C. Samples were put on ice and clarified by centrifugation at 14 000 rpm in an Eppendorf centrifuge for 10 minutes at 4°C. The samples were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (Schleicher & Schuell, Dassel, Germany). CalDAG-GEF1 was detected with 2 different mouse monoclonal antibodies and 2 polyclonal rabbit antisera (a kind gift of Dr J. R. Crittenden, Department of Brain and Cognitive Sciences, Massachusetts Institute of Technology, Cambridge, MA), followed by horseradish peroxidase–coupled goat anti–mouse-Ig (GE Healthcare, Little Chalfont, United Kingdom), and detection by enhanced chemiluminescence (GE Healthcare).
For kindlin-3 protein detection, rabbits were immunized with a specific peptide from human kindlin-3 (EPEEELYDLSKVVLA; amino acids 156-170), which was coupled to Imject Maleimide Activated mcKLH (Pierce Chemical, Rockford, IL). Antisera were affinity-purified with a commercial kit (SulfoLink Kit; Pierce Chemical). High-affinity antibodies were eluted from the column with 100 mM glycine buffer at pH 2.7 and then dialyzed against PBS.
Western blots of total cell lysates were incubated with one of these affinity-purified rabbit antisera, encoded ahk1837, followed by incubation with goat anti–mouse-IgG IRDye 800CW. Quantification of bound antibodies was performed on an Odyssey Infrared Imaging system (LI-COR Biosciences, Lincoln, NE). Immunostaining for glyceraldehyde-3-phosphate dehydrogenase served as a control for equal loading and electrophoretic transfer of each lane in the gels.
Molecular studies
Homozygosity mapping.
Genomic DNA from PBMCs and fibroblasts from the patients and their parents and healthy siblings were extracted by standard methods. A genome-wide search was performed with the Affymetrix GeneChip Human Mapping 10k, version 2.0 array (Affymetrix, Santa Clara, CA). This array contains 10 204 SNPs distributed across the genome. The GeneChips were processed by standard Affymetrix protocols (ServiceXS, Leiden, The Netherlands). The SNP genotype call rate was more than 98%.
Sequence analysis.
Direct sequencing of polymerase chain reaction (PCR) products from either genomic DNA or cDNA was used. The coding region of the human FERMT3 and CALDAGGEF1 was amplified in separate PCRs (primer sequences are available on request). After isolation of mRNA from fresh blood samples or cell lines generated in the past of these patients, cDNA was made for mutation analysis. Sequencing was performed with the Applied Biosystems Bigdye terminator kit, version 1.1, and products were analyzed on a Genetic Analyzer (ABI3130XL; Applied Biosystems, Foster City, CA).
Results
Homozygosity mapping
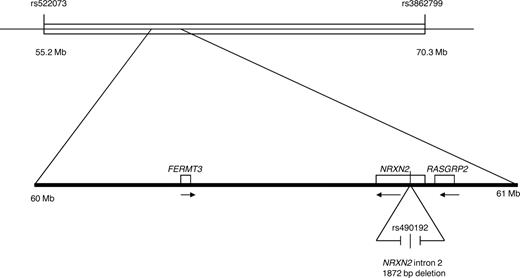
We previously identified 7 families with 9 members with the LAD1v syndrome, including the index case originally described in 1997.1 We have now analyzed 3 patients from 3 of these 7 apparently unrelated families by Affymetrix 10k SNP array. All patients shared the same haplotype in a region of 13 Mb, flanked by rs522073 and rs3862799 on chromosome 11q13, which showed complete homozygosity in all patients (Figure 1).
Chromosome 11q13 homozygosity mapping. The top line indicates the region of homozygosity for a shared haplotype of a 13-Mb region in the families reported before.2 Several SNPs and an intronic deletion were identified. The private SNP in CALDAGGEF1 (c.1592-3C>A; reference sequence NM_001098670.1) and the small intronic deletion in NRXN2 gene (1872 bp between position 64227796 and 64229668, human reference sequence build 36.3). The position of the FERMT3 gene, the locus for LAD1v syndrome, is also indicated.
Chromosome 11q13 homozygosity mapping. The top line indicates the region of homozygosity for a shared haplotype of a 13-Mb region in the families reported before.2 Several SNPs and an intronic deletion were identified. The private SNP in CALDAGGEF1 (c.1592-3C>A; reference sequence NM_001098670.1) and the small intronic deletion in NRXN2 gene (1872 bp between position 64227796 and 64229668, human reference sequence build 36.3). The position of the FERMT3 gene, the locus for LAD1v syndrome, is also indicated.
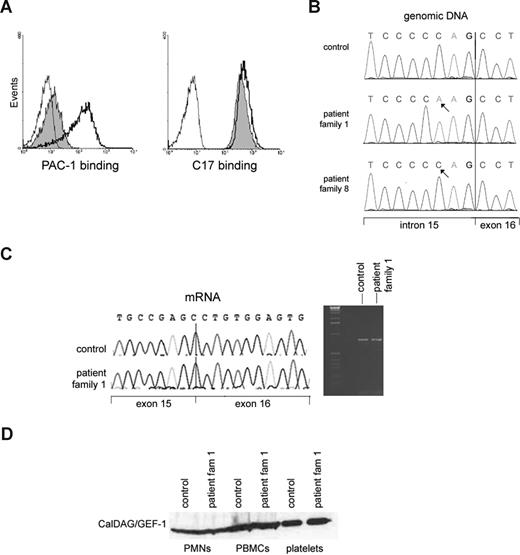
The region contains 404 known genes and annotated open reading frames according to the NCBI map build 36.3. Although multiple genes had been considered, we initially focused on CALDAGGEF1. This gene encodes a Rap-1 GEF, also known as RasGRP2, which has been implicated as a key regulator of agonist-induced αIIbβ3 activation in platelets.19 The platelets of the patients show indeed a strongly disturbed αIIbβ3 activation, as indicated by the lack of PAC-1 epitope expression after platelet activation18 (Figure 2A), which is further proof of the Glanzmann-like bleeding tendency in the presence of normal αIIbβ3 expression. The LAD1v syndrome, as first described in 1997,1 is thus characterized by the inability to activate different types of integrins, which are themselves present at normal or near-normal levels.
Lack of GPIIbIIIa (αIIbβ3) activation on platelets in the presence of normal CalDAG-GEF1 levels in the LAD1v syndrome. (A) PAC-1 binding to activated αIIbβ3 integrins as detected by flow cytometry (left panel) shows strongly decreased binding to patient platelets after thrombin stimulation (shaded histogram) compared with control platelets (open histogram, solid black line). The binding of CD61 antibody to measure total αIIbβ3 expression, as measured in parallel samples, is shown in the right panel. In both panels, the binding of negative mouse Ig-FITC to the stimulated patient platelets (thin black line) is shown for comparison. The slight shift observed with PAC-1 in the patient platelets was not observed without thrombin stimulation (not shown). (B) A homozygous sequence variant of CALDAGGEF1 in intron 15 is present in the patient. (C) The intronic gene variation results in normal splicing of mRNA. Both reverse-transcribed PCR and sequence analysis of control and patient cDNA that contains exons 12 to 18 gave the expected wild-type products of 802 bp, as shown here. The arrow in the sequence traces indicates the exon 15–exon 16 boundary. (D) Normal CalDAG-GEF1 protein expression on Western blotting at the expected relative molecular mass of approximately 75 kDa is found. Results are representative for 5 of the patients from families 1 through 7 tested in total.
Lack of GPIIbIIIa (αIIbβ3) activation on platelets in the presence of normal CalDAG-GEF1 levels in the LAD1v syndrome. (A) PAC-1 binding to activated αIIbβ3 integrins as detected by flow cytometry (left panel) shows strongly decreased binding to patient platelets after thrombin stimulation (shaded histogram) compared with control platelets (open histogram, solid black line). The binding of CD61 antibody to measure total αIIbβ3 expression, as measured in parallel samples, is shown in the right panel. In both panels, the binding of negative mouse Ig-FITC to the stimulated patient platelets (thin black line) is shown for comparison. The slight shift observed with PAC-1 in the patient platelets was not observed without thrombin stimulation (not shown). (B) A homozygous sequence variant of CALDAGGEF1 in intron 15 is present in the patient. (C) The intronic gene variation results in normal splicing of mRNA. Both reverse-transcribed PCR and sequence analysis of control and patient cDNA that contains exons 12 to 18 gave the expected wild-type products of 802 bp, as shown here. The arrow in the sequence traces indicates the exon 15–exon 16 boundary. (D) Normal CalDAG-GEF1 protein expression on Western blotting at the expected relative molecular mass of approximately 75 kDa is found. Results are representative for 5 of the patients from families 1 through 7 tested in total.
In support of a suspected defect in cellular signaling, we recently observed that the β2 and β3 integrins on the neutrophils and platelets of the patients can be activated by exposure to ultraviolet-C light, which induces a conformational change in these integrins independent of signal transduction.20
Similar to our patients, the leukocyte extravasation was also defective in caldaggef1 knockout mice.8 We therefore performed complete genomic sequencing of all coding exons from the CALDAGGEF1 gene. We did not find any missense or nonsense mutations, as we reported previously.14 We did detect an intronic sequence variant c.1592-3C>A (reference sequence NM_001098670.1; Figure 2B). This variant was also described in 2 LAD-III patients.5 Neither this intronic variation nor the NRXN2 intronic deletion (discussed below in this section) was found in 80 Turkish controls.
To test whether this variant affects mRNA splicing, we analyzed CalDAG-GEF1 at the mRNA and protein level. No abnormal transcripts were observed by reverse-transcribed PCR, thus excluding any splicing defect that would possibly lead to lack of expression or mislocalization of the protein within the cell (Figure 2C). To confirm the expected expression of the CalDAG-GEF1 protein, 2 monoclonal and 2 polyclonal antibodies were used for immunoblotting lysates from various blood cells of 3 different LAD1v patients from the original 7 families tested before. No decrease in the level of the protein was observed in neutrophils, PBMCs, or platelets of these patients (Figure 2D). Rap1 activation was also normal, as we had previously published.2
After having excluded CALDAGGEF1 as the causative gene defect, we further analyzed the homozygous region in detail. Surprisingly, all 3 patients analyzed on the Affymetrix Gene Chip used in this study showed no signal of the probe for the detection of rs490192 (“no call”), which is located in the neurexin 2 gene (NRXN2) at position 64227945 (human reference sequence NCBI build 36.3). Because this SNP usually gives a good signal, we considered this a true negative signal. The presence of a deletion in this region was confirmed by PCR. All our patients showed a homozygous deletion in this region. By long PCR and sequence analysis, we identified the borders of this intronic 1872-bp deletion. However, NRXN2 is not expressed in neutrophils, PBMCs, or platelets; therefore, it is not probable that this deletion is causing the disease.
Further evidence that neither the CALDAGGEF1 mutation nor the NRXN2 deletion was causing the LAD1v syndrome came from a novel patient we recently identified, who had a different chromosome 11q13 haplotype (Table 1, patient 8). In the absence of invasive infections, impaired wound healing, or anemia, this 2-year-old girl showed a similar leukocytosis and clinical bleeding tendency as the other LAD1v patients. On testing of the blood cells, similar neutrophil and platelet defects were observed as previously described for the patients from the 7 consanguineous families.2 This patient, the fourth child of consanguineous parents of a family that originated from a more southeastern region of Turkey than the other families, carried neither the CALDAGGEF1 mutation nor the NRXN2 intron 2 deletion.
Within the genomic region encompassing the q12-13 band, several genes of potential importance for integrin activation were considered, encoding proteins such as Rel-A, PLC-β3, PPP1R14B, or MAP4K2 (alias GC for “germinal center” kinase), which may be linked to signaling via nuclear factor-κB activity, phosphoinositides, phosphatase activity, or diacylglycerol, or shows homology to serine-threonine protein kinases, respectively. Most of these genes could be excluded by the information available from databases on expression arrays, functional datasets, tissue expression patterns, or direct sequencing of genomic DNA.21-23
FERMT3 mutated in the LAD1v syndrome
Candidate actin-binding integrin partners, such as talin, filamin, and calreticulin, associate with all leukocyte and platelet integrins.24-26 Although these key integrin adaptors are not restricted to cells of the hematopoietic lineage, it was suggested before2 that homologous hematopoietic cell–specific adaptors may be defective in our LAD-like syndrome. The presence of FERMT3 (encoding kindlin-3) is located at a distance of approximately 500 kb centromeric of the CALDAGGEF1 and NRXN2 genes. A 30-kDa so-called FERM domain is found in a family of proteins that mediate linkage of the cytoskeleton to transmembrane proteins. This family includes erythroid protein 4.1, ezrin, radixin, moesin (FERM).27 These FERM domain–containing proteins are thought to bind either directly or indirectly to the plasma membrane through the FERM domain and can anchor actin microfilaments. The FERM domain of talin is implicated as the binding domain for β-integrin cytoplasmic tails, consisting of the N-terminal 433 amino acids.28
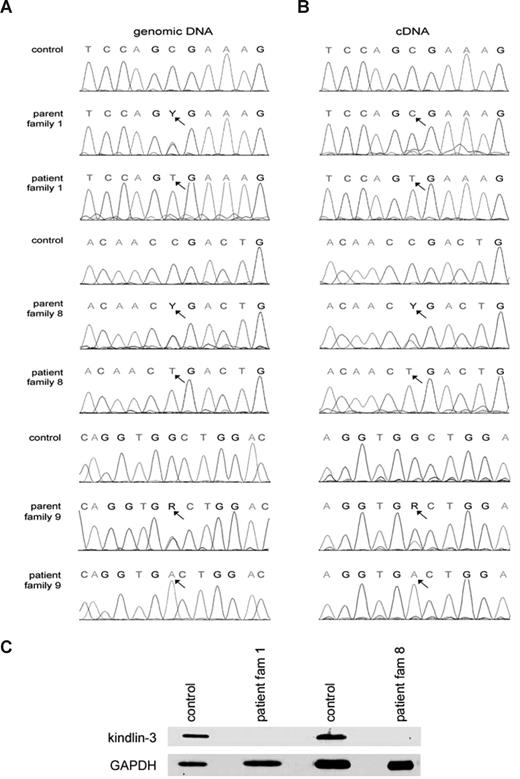
We sequenced the human FERMT3 gene, starting with the mRNA from LAD1v patients, and identified a homozygous mutation in exon 12 (c.1525C>T) in all LAD-1v patients reported previously (reference sequence NM 031471.4).2 This mutation results in a premature stop at codon 509 (p.Arg509X). The parents were all heterozygotes for this mutation and were healthy. Because this premature stop codon is expected to give nonsense-mediated decay, we analyzed the transcript levels in heterozygous carriers. There was preferential breakdown of the transcript derived from the mutated allele, as demonstrated by the reduction of the affected transcript to 10% to 50% compared with the normal transcripts in the heterozygous parents (Figure 3A). The presence of this mutation was confirmed by direct sequencing of exon 12 (Figure 3B). In the new 2-year-old LAD1v patient 8, we observed a different homozygous nonsense mutation (c.1717C>T, p.Arg573X) in exon 14 of FERMT3. The parents of this patient were both heterozygotes of this mutation. In yet another patient (Table 1, patient 9), the first child from consanguineous Arabic parents,29 DNA sequencing showed no abnormalities in CALDAGGEF1 or NRXN2. In the DNA from this patient, we found a homozygous nonsense mutation (c.687G>A, p.Trp229X) in exon 6 of FERMT3. Again, both the parents of this patient were heterozygotes of this mutation.
Nonsense mutations FERMT3, encoding kindlin-3, result in the LAD1v syndrome. (A) A homozygous c.1525 C>T mutation in exon 12 results in p.Arg509X in patients from the previously described families 1 to 7, and a homozygous c.1717 C>T mutation in exon 14 yields p.Arg573X in family 8. Y and R refer to base position mixtures of C/T and G/A, respectively.(B) Heterozygotes show preferential presence of normal transcripts over mutated FERMT3 transcripts, as shown for families 1 to 7. (C) Irrespective of the mutation, the protein kindlin-3 is undetectable at the expected relative molecular mass of 76 kDa in the platelets of LAD1v patients (and leukocytes, data not shown); the 25-kDa control protein glyceraldehyde-3-phosphate dehydrogenase was observed in all lanes serving as loading control.
Nonsense mutations FERMT3, encoding kindlin-3, result in the LAD1v syndrome. (A) A homozygous c.1525 C>T mutation in exon 12 results in p.Arg509X in patients from the previously described families 1 to 7, and a homozygous c.1717 C>T mutation in exon 14 yields p.Arg573X in family 8. Y and R refer to base position mixtures of C/T and G/A, respectively.(B) Heterozygotes show preferential presence of normal transcripts over mutated FERMT3 transcripts, as shown for families 1 to 7. (C) Irrespective of the mutation, the protein kindlin-3 is undetectable at the expected relative molecular mass of 76 kDa in the platelets of LAD1v patients (and leukocytes, data not shown); the 25-kDa control protein glyceraldehyde-3-phosphate dehydrogenase was observed in all lanes serving as loading control.
The mutations in the Turkish LAD1v patients all result in a stop codon and are located in the C-terminal, second so-called FERM domain.26 Expression of a (truncated) kindlin-3 protein was undetectable in the patients' platelets with polyclonal antisera raised against human kindlin-3, whereas positive staining was present in control platelets (Figure 3). Specificity of the antisera was indicated by competition with the kindlin-3 peptide used as the immunogen in control lysates from blood cells and platelets, and not by an irrelevant peptide. Moreover, these antisera did not show any binding to nonhematopoietic cell lines, such as CHO and HeLa cells, which are expected to be negative for kindlin-3 (data not shown).
Discussion
The data from 9 patients in 7 unrelated families indicate that the LAD1v syndrome can be recognized early in life by necrotic lesions without pus formation, poor wound healing, and a severe life-threatening bleeding tendency. Infections range from bacterial pneumonia and early septicemia to fungal disease. Severe intracranial hemorrhages may affect neonates, followed by a period of persistent mucosal bleeding and fragile gums because of chronic periodontitis during childhood. Two patients were thus far transplanted successfully, demonstrating that bone marrow transplantation may be curative. There may be phenocopies of the syndrome with a different genetic background. After our first case,1 a number of similar patients have been reported with defective LAD-Glanzmann-like syndrome.4-6,30,31 All these patients have the typical complex of clinical features that were highly similar to the original patient we have described.1 Typically, our patients (families 1-7) are of Turkish descent, and all originate from a central region of Turkey, until a new patient was recently diagnosed from a family from the southeastern region in Turkey (family 8). This last patient as well as an Arabic patient (family 9) had nonsense mutations in FERMT3 different from the other patients tested thus far. Moreover, these last 2 patients had no CALDAGGEF1 mutations and lacked the common ancestral allele identified in the other families.
Alon et al suggested in the past that the syndrome (identified by that group as LAD-III5 ) is caused by an aberrant activity of Rap1, as found in a single EBV-transformed B-cell line from one patient.7 Rap1 activity is involved in integrin-mediated adhesion.6 Although there was no evidence for defective Rap1 activation in any of the patients described with the LAD1v syndrome,2 Pasvolsky et al reported mutations in the gene encoding CalDAG-GEF1,7 which seemed to corroborate the caldaggef1−/− knockout mouse model.8,19 The 2 LAD-III patients have been described to carry a mutation in CALDAGGEF1, reported to cause aberrant splicing, 30-fold lower levels of mRNA, and the complete lack of CalDAG-GEF1 protein.7 This site would cause a shifted reading frame and premature stop codon, resulting in nonsense-mediated decay of the mRNA and lack of protein expression. In our original LAD1v patients from 7 unrelated Turkish families, which all share this homozygous sequence variant at intron 15 of the CALDAGGEF1 gene, no missplicing of the mRNA and no loss of mRNA or protein were, however, detected in neutrophils, lymphocytes, or platelets. The normal CalDAG-GEF1 protein expression in the patients is further supported by the normal Rap1 activation reported previously.2
We conclude that this private SNP is part of an ancestral haplotype carrying several genes in which a disease-associated mutation could be located. The silent CALDAGGEF1 mutation and the NRXN2 deletion are not disease causing but in linkage disequilibrium with the nonsense mutation in exon 12 of the FERMT3 gene in the patients from the 7 previously described families. The FERMT3 mutations result in undetectable kindlin-3 protein expression, whereas CalDAG-GEF1 is normally expressed. Most recently, after the genetic analysis of our LAD1v patients had been completed, Mory et al15 have reported the same mutation in the FERMT3 gene in 3 LAD-III patients as we detected in all but 2 of our LAD1v patients. They suggested that the syndrome is caused by a combined defect in both CalDAG-GEF1 and kindlin-3. Based on our results and those obtained in kind3−/− mice, it is not necessary to invoke such an unprecedented coincidence of events. The absence of kindlin-3 protein in the absence of any mutation in the CALDAGGEF1 gene, as observed in the other, newly diagnosed patients, shows that the phenotype observed in the patients' neutrophils and platelets can be caused by mutations in FERMT3 alone. Indeed, kind3−/− mice show, besides a platelet aggregation defect, defective leukocyte extravasation (Richard Fässler, Department of Molecular Medicine, Max Planck Institute of Biochemistry, Martinsried, Germany, personal communication, September 2008), confirming this notion.
The leukocytic kindlin-3 (or URP2) variant encodes a 663-amino acid protein with a calculated molecular mass of approximately 76 kDa. URP2 (kindlin-3) shares 55.4% identity with MIG2 (kindlin-2) and 58.7% identity with URP1 (alias kindlin-1).27 All 3 proteins share weak but significant homology with talin-1 and talin-2, in particular, in their FERM domain. Whereas kindlin-1 and -2 are largely confined to epithelial cells and muscle cells, respectively, kindlin-3 is confined to hematopoietic cells, with the highest expression in megakaryocytes.32 Most recently, Moser et al reported that kind3−/− mice die within a week of birth with pronounced osteopetrosis and severe hemorrhages in the gastrointestinal tract, skin, brain, and bladder, phenomena that were already apparent during development.33 Chimeric animals with kind3−/− livers also had a pronounced hemostatic defect, suggesting platelet dysfunction of hematopoietic origin, which is in line with our data on successful bone marrow transplantation in patients.
Platelet counts were normal in kind3−/− chimeric mice, but the platelets were unable to activate their β3 integrins. Activation of these integrins normally occurs after platelets contact a wounded vessel and is mediated by binding of talin to β-integrin cytoplasmic tails. Kindlin-3 binds to regions of the β-integrin tails distinct from those bound by talin33 and constitutes an essential element for platelet integrin activation. Interestingly, a structural defect of the red cell membrane skeleton has also been suggested to contribute to the severe anemia in kind3−/− mice.34 In our patients, we did not observe such a severe anemia or major red cell abnormalities, but defective kindlin-3 in human red cells may contribute to transfusion dependency in some patients, which may well relate to the coinciding and often persistent mucosal diathesis (Table 1). Clinical osteopetrosis has a wide clinical spectrum as a consequence of a variety of gene defects.35,36 The clinical and radiographic features of moderate to severe osteopetrosis were not a prominent feature in the patients described previously,2 although we had the impression of a firm and dense bone structure on performance of bone marrow punctures.
The clinical variation in symptoms among the first 7 families2 (carrying the same FERMT3 mutation) and families 8 and 9 suggests that there is no strict genotype-phenotype relationship (Table 1). Although phenocopies of the LAD1v syndrome may exist as a consequence of true mutations in CALDAGGEF1, such mutations must be different from the suggested splice site mutation that was reported in LAD-III.5 To date, all patients have a mutation in FERMT3, but only some of them in combination with a private SNP in CALDAGGEF1, providing evidence that the FERMT3 mutation is responsible for the disease in LAD1v patients.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Drs A. Metin, S. Berrak, and E. Ozek for their help and collaboration with patient data and material, Mr R. Verhaar and Mrs I. de Cuyper for performing the PAC-1 binding studies, Dr R. Fässler for his kind cooperation and discussions on the mouse model, and Prof R. van Lier for critical reading of the paper and helpful suggestions.
This work was supported by the Landsteiner Foundation for Blood Transfusion Research (LSBR-0619).
Authorship
Contribution: T.W.K., M.d.B., T.K.v.d.B., D.R., A.J.V., and F.B. were responsible for various parts of the research design as well as for the data analysis; E.v.d.V., M.d.B., M.A.J.W., A.T.J.T., and M.E.J. performed most of the experiments; M.M. generated new anti–human kindlin-3–specific antisera; T.W.K., K.S., Ö.S., S.Ü., and M.Ç. took care of the patients and collected patient materials and clinical data; and T.W.K., D.R., A.J.V., and F.B. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Taco W. Kuijpers, Emma Children's Hospital, Academic Medical Center, Room G8-205, Meibergdreef 9, 1105 AZ Amsterdam, The Netherlands; e-mail: t.w.kuijpers@amc.uva.nl.