Abstract
In patients with severe congenital neutropenia (SCN) and mice with growth factor independent-1 (Gfi1) loss of function, arrested myeloid progenitors accumulate, whereas terminal granulopoiesis is blocked. One might assume that Gfi-null progenitors accumulate because they lack the ability to differentiate. Instead, our data indicate that Gfi1 loss of function deregulates 2 separable transcriptional programs, one of which controls the accumulation and lineage specification of myeloid progenitors, but not terminal granulopoiesis. We demonstrate that Gfi1 directly represses HoxA9, Pbx1, and Meis1 during normal myelopoiesis. Gfi1−/− progenitors exhibit elevated levels of HoxA9, Pbx1 and Meis1, exaggerated HoxA9-Pbx1-Meis1 activity, and progenitor transformation in collaboration with oncogenic K-Ras. Limiting HoxA9 alleles corrects, in a dose-dependent manner, in vivo and in vitro phenotypes observed with loss of Gfi1 in myeloid progenitor cells but did not rescue Gfi1−/− blocked granulopoiesis. Thus, Gfi1 integrates 2 events during normal myeloid differentiation; the suppression of a HoxA9-Pbx1-Meis1 progenitor program and the induction of a granulopoietic transcription program.
Introduction
A hierarchical network of lineage-specifying transcription factors regulates hematopoietic self-renewal, differentiation, survival, and proliferative cell fate decisions; however, the underlying molecular integration of these decisions remains enigmatic. Here, we show that a single transcription factor integrates separable myeloid transcriptional programs governing progenitor biology and terminal differentiation.
The Hox genes encode homeodomain proteins that are essential regulators of embryogenesis, hematopoietic development, and transformation.1 HoxA9 is highly expressed in the hematopoietic stem cell (HSC) fraction of bone marrow,2 and forced expression of HoxA9 in mouse bone marrow results in enhanced proliferation of pluripotent hematopoietic stem and progenitor cells.3 In contrast, HoxA9−/− mice display a reduction in total leukocytes and lymphocytes and diminished granulocytosis in response to recombinant G-CSF.4
HoxA9 forms heterodimeric DNA binding complexes with members of the Pbx and/or Meis family of homeodomain proteins.5 HoxA9 interactions with Pbx1 increase DNA binding affinity,6 whereas interactions between Pbx1 and Meis1 may serve to improve nuclear localization.7 Meis proteins interact with and block the Pbx nuclear export signal, leaving the Pbx nuclear localization signal (NLS) to mediate Pbx nuclear translocation and abundance.7–9 Similarly, HoxA9 and Meis1 interact without6 and on DNA.5 Meis1 interaction mediates HoxA9 nuclear localization, but HoxA9 nuclear retention is regulated by an NLS on either side of the homeodomain.10 The HoxA9-Pbx1-Meis1 transcription factor complex targets genes involved in HSC and progenitor expansion, including Pim1, c-Myb, Cd34, and Flt3.11–13
The transcriptional repressor growth factor independent-1 (Gfi1) is a zinc-finger DNA binding protein that is the target of common Moloney murine leukemia virus proviral insertion sites.14–16 Gfi1 is composed of an N-terminal SNAG transcriptional repressor domain,17 and a C-terminal Zn-finger domain that is necessary for DNA binding.18 A single point mutation in the SNAG domain (Gfi1P2A) destroys Gfi1-mediated biology.17,19–21 Gfi1 is critically required for the development of mature neutrophils,22,23 and Gfi1−/− myeloid progenitors proliferate abnormally.24 In humans, a GFI1N382S mutation is associated with severe congenital neutropenia (SCN).25 SCN-associated Gfi1N382S mutations generate a dominant negative-acting protein that is causal in blocking granulopoiesis.20 Both Gfi1−/− mice and patients with GFI1N382S SCN show the abnormal accumulation of arrested myeloid progenitors.22,23,25
We recently illustrated direct antagonism between the Drosophila orthologs of Gfi1 versus HoxA9, Pbx1, and Meis1.26 Here, we demonstrate that this relationship extends to mammals. Specifically, Gfi1 directly regulates the expression of the HoxA9-Pbx1-Meis1 transcription factor complex. Moreover, deregulation of Gfi1 increases myeloid progenitor in vitro persistence and in vivo accumulation. Notably, these Gfi1−/− phenotypes critically require HoxA9. However, unlike the Gfi1 target gene Csf1,20 modulation of HoxA9 alleles did not rescue Gfi1 loss-of-function–blocked granulopoiesis. Thus, myeloid progenitor accumulation in both Gfi1−/− mice and patients with GFI1N382S SCN correlates with deregulation of HoxA9-directed progenitor transcriptional programs that can be uncoupled from Gfi1-controlled granulopoiesis.
Methods
Mice
Gfi1Δex4-5 and Gfi1fex4-5 mice27 were backcrossed onto a C57Bl/6 background (The Jackson Laboratory, Bar Harbor, ME). Polymerase chain reaction (PCR) typing of the Gfi1fex4-5 allele was performed with primers 5′-CAGTCCGTGACCCTCCAGCAT-3′ and 5′-CTGGGAGTGCACTGCCTTGTGTT-3′, whereas detection of the Gfi1Δex4-5 allele was performed with primers 5′-CAGTCCGTGACCCTCCAGCAT-3′ and 5′-CCATCTCTCCTTGTGCTTAAGAT-3′. Data from germline Gfi1Δex4-5/Δex4-5 mice (termed Gfi1−/− in the remaining text) are shown throughout the rest of the article; however, essential points were checked with germline Gfi1Δex2-3/Δex2-3 mice23 and Gfi1Δex4-5/Δex2-3 mice and found to be similar. Mx-1Cre transgenic mice were obtained from The Jackson Laboratory. K-RasLSLG12D knock-in mice were obtained from the Mouse Models of Human Cancer repository (National Cancer Institute [NCI], Fredrick, MD). HoxA7−/−28 and HoxA9−/−29 were maintained on C57Bl/6 background. Rosa-Cre-ERT2 mice30 were kindly provided by Anton Berns (Netherlands Cancer Institute, Amsterdam, The Netherlands). Mice were bred and housed by Cincinnati Children's Hospital Medical Center (CCHMC) Veterinary Services, and mouse manipulations were reviewed and approved by the Children's Hospital Research Foundation Institutional Animal Care and Use Committee.
Leukemia
The Mx1Cre+LSL-K-RasG12D and Mx1Cre+LSL-K-RasG12D Gfi1fex4-5/fex4-5 mice were injected intraperitoneally with 100 μg polyinosinic-polycytidylic acid (pIpC) on alternate days for 3 doses. All mice were kept in microisolator cages, were monitored for disease progression every other day initially and every day after the first appearance of symptoms, and were killed when moribund. Statistical analysis was performed using Prism 4 software (GraphPad Software, San Diego, CA). Cumulative survival was plotted against days after treatment with pIpC (log-rank test). Secondary transplantations were performed by injecting 106 splenic cells from killed mice into the lateral tail vein of sublethally irradiated (6 Gy) CD45.1 congenic (BoyJ) recipients. Disease progression was monitored as mentioned earlier.
Retroviral constructs
The murine Gfi1 cDNA was cloned into the EcoRI site of the MSCV-IRES-eGFP, SF91-IRES-Venus, and MSCV-Puro retroviral vectors. Gfi1P2A and Gfi1N382S mutants20 were cloned into the SF91-IRES-Venus vector. Virus stocks were produced by transfecting the Phoenix-Eco packaging cell line.
Bone marrow cell transduction
Bone marrow from 6- to 8-week-old mice was flushed from femurs, tibias, and iliac crests using DMEM (Invitrogen, Carlsbad, CA) with 15% heat-inactivated fetal bovine serum (HyClone Laboratories, Logan, UT) and labeled with mouse Lineage Cell Depletion kit (Miltenyi Biotec, Auburn, CA) followed by separation on an AutoMacs magnetic sorter according to the manufacturer's instructions (Miltenyi Biotec). Cells were maintained in DMEM plus 15% heat-inactivated FBS supplemented with IL-3 (10 ng/mL), IL-6 (20 ng/mL), SCF (25 ng/mL), and TPO (25 ng/mL; all from PeproTech, Rocky Hill, NJ). The cells were expanded in culture for 48 hours before being subjected to retroviral transduction. Briefly, 6-well non-tissue culture–treated plates were coated with RetroNectin (Takara Bio, Kyoto, Japan) according to the manufacturer's protocol and preloaded with viral particles (2-3 MOI/cell). Cells (2 × 106) were plated per well, and spinfected at 1000g for 1.5 hours at room temperature, then incubated overnight with the viral supernatants.
In vitro colony assays
Lin− bone marrow (BM) cells or cells sorted by fluorescence-activated cell sorting (FACS) were plated in methylcellulose medium (M3534; StemCell Technologies, Vancouver, BC) containing IL-3, IL-6, and SCF. Colonies were enumerated and scored after 8 days of culture. For serial replating assays, colonies were pooled, and the number of cells used for the initial plating was replated in fresh methylcellulose medium. Rosa-Cre-ERT2Gfi1fex4-5 BM cells were plated in methylcellulose media with or without 1 μM tamoxifen.
Real-time quantitative PCR analysis of gene expression
RNA from Lin− BM cells or sorted cell populations was purified using either Trizol reagent (Sigma-Aldrich, St Louis, MO) or the RNeasy kit (QIAGEN, Valencia, CA). Purified RNA (1-10 μg) was reverse transcribed using the cDNA High Capacity Archive Kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. The reaction products were diluted to 20 ng/μL, and 2 μL was subjected to real-time PCR, which was performed in triplicate using Taqman primer/probe sets on either an ABI Prism 7500 or 7900 amplification system (Applied Biosystems). Taqman primer/probe sets for c-Myb (Mm00501741_m1), CD34 (Mm00519283_m1), Cxcr4 (Mm01292123_m1), Ebf1 (Mm00395519_m1), Flt3 (Mm00439016_m1), Gfi1 (Mm00515853_m1), Gfi1b (Mm00492319_m1),Hoxa4 (Mm01335255_g1), Hoxa5 (Mm00439362_m1), Hoxa6(Mm00550244_m1), Hoxa7 (Mm00657963_m1), Hoxa9 (Mm00439364_m1),Hoxa10 (Mm00433966_m1), Hoxa11 (Mm00439360_m1), Meis1 (Mm00487664_m1), Meis3 (Mm00485209_m1), Pbx1 (Mm00435507_m1), Pbx2 (Mm01134981_g1), Pbx3 (Mm01333270_m1), Pim-1 (Mm00435712_m1), Pu.1 (Mm00488140_m1), and GAPDH (Mm99999915_g1; as the endogenous internal control) were purchased from Applied Biosystems.
Morphologic analysis of bone marrow cells
Bone marrow cells were harvested in Medium 199 (Invitrogen) and then briefly lysed in ACK Lysing Buffer (Invitrogen). Cells (n = 50 000) were resuspended in PBS with 0.5% FBS, and cytospins were made with a Shandon EZ Single Cytofunnel (Thermo Electron, Pittsburgh, PA). Slides were disassembled from the funnel and allowed to dry. A HEMA3 stain set (Fisher Scientific, Kalamazoo, MI) was used to perform a hematoxylin-eosin staining on the cells by exposing the slides for 1 minute to each solution. A cover slide was applied with a low-viscosity medium Cytoseal 60 (Richard-Allan Scientific, Kalamazoo, MI). Cells were analyzed on a Zeiss Axiovert 200M microscope (Carl Zeiss, Thornwood, NY) with a 100× objective (Zeiss MicroImaging, Thornwood, NY). Photomicrographs were made with an AxioCam MRc color camera and AxioVision 3.1 software (Carl Zeiss).
FACS analysis of bone marrow cells
Tibias and femurs of mice were harvested and flushed with Medium 199 (Invitrogen) to collect bone marrow cells. Red blood cells were lysed briefly in ACK Lysing Buffer. Bone marrow cells were then incubated in a cocktail of predetermined optimal concentrations of lineage (Lin) antibodies: PE-Cy5–conjugated anti-CD3 (clone CT-CD3), PE-Cy5–conjugated anti-CD4 (clone RM4-5), PE-Cy5–conjugated anti-CD8 (clone 5H10), PE-Cy5–conjugated anti-CD19 (clone 6D5), PE-Cy5–conjugated anti–CD45R-B220 (clone RA3-6B2), PE-Cy5–conjugated anti–Gr-1 (clone RB6-8C5; all from Invitrogen) and PE-Cy5–conjugated anti-TER119 (clone TER119; eBioscience, San Diego, CA). Lin+ cells were then depleted using sheep anti–Rat-IgG (Fc)–conjugated immunomagnetic beads (Invitrogen). Isolated Lin− cells were stained with PE-conjugated anti-FcRγII/III (clone A7-R34), Pacific Blue–conjugated anti–IL-7Rα (eBioscience), APC-conjugated anti–c-Kit (clone 2B8), FITC-conjugated anti-CD34 (clone RAM34), biotinylated anti–Sca-1 (clone E13-161.7; BD PharMingen, San Diego, CA) and subsequently stained with APC-Cy7–conjugated streptavidin (Invitrogen).
For the analysis of mature myeloid cell populations from whole bone marrow, cells were stained with FITC-conjugated F4/80 (clone CI: A3-1), Alexa-647–conjugated anti-7/4 (clone 7/4; both from AbD SEROTEC, Raleigh, NC), PE-Cy7–conjugated anti–Gr-1 (clone RB6-8C5) and PE-conjugated anti-CD11bα chain (clone M1/70; BD PharMingen). Flow cytometric analyses were conducted on a FACS LSRII (BD Biosciences, San Jose, CA). Data were exported and analyzed with FlowJo software (TreeStar, Ashland, OR). Statistical analysis from at least 3 independent experiments was performed.
ChIP assays
For chromatin immunoprecipitation (ChIP) analysis, HL60 cells (from Patrick Zweidler-McKay, M. D. Anderson, Houston, TX) were cross-linked with 1% formaldehyde for 10 minutes on ice. Soluble chromatin was prepared using a Bioruptor (Cosmo Bio USA, Carlsbad, CA) to generate DNA fragment size of 200 to 800 bp. Using anti-Gfi1 (2.5D1.7) and control mouse IgG (GE Healthcare, Little Chalfont, United Kingdom) the chromatin-protein complex was immunoprecipitated, and the cross-links were reversed by incubating with NaCl at 65°C for 12 hours. Chromatin was used as input for PCR amplification. The following oligonucleotides were used to amplify the promoter regions to which Gfi1 (and/or HoxA9) binds: HoxA9 promoter (forward, 5′-CTGTTGGTCGTTTCCGACTT-3′; reverse, 5′-CAAATCGCATTGTCGCTCTA-3′), Meis1 promoter (forward, 5′-CACTGGCTGGTTGGAGACTT-3′; reverse, 5′-CCCAGACCTCCATCTCTCAA-3′), Pbx1 promoter (forward, 5′-GCCGGGAGCCCATTTCTG-3′; reverse, 5′-CCACTTGGCGAAAAGAAATCAG-3′), HoxA9 3′UTR (forward, 5′-TTAAGTGTTCTCGGGGATGC-3′; reverse, 5′-CCGCATTTTTAAGGTGGAGA-3′), Meis1 3′UTR (forward, 5′-GGGCTTGAATTTGCATGTCT-3′; reverse, 5′-TGCAGTTTTTCCCATCCTTC-3′); Pbx1 3′UTR (forward, 5′-TCGAAGCAATCAGCAAACAC-3′; reverse, 5′-GGCTGAAATAGCATCCCAAA-3′), b-actin (forward, 5′-AGCGCGGCTACAGCTTCA-3′; reverse, 5′-CGTAGCACAGCTTCTCCTTAATGTC-3′).
Immunoblot analyses
Protein extracts were obtained from Lin− bone marrow cells by lysing cells directly in Complete-M lysis buffer (Roche, Basel, Switzerland). Samples were resolved by 10% SDS–polyacrylamide gel electrophoresis and electrophoretically transferred to PVDF membrane (Immobilon-P; Millipore, Billerica, MA). Immunoblot analysis was performed using the anti-HoxA9 (Upstate Biotechnology, Charlottesville, VA), anti-Pbx1 (Novus Biologicals, Littleton, CO), anti-Meis1 (Abcam, Cambridge, MA) and anti–β-actin (Sigma-Aldrich), and HRP-conjugated goat anti–mouse or anti–rabbit secondary antibody (GE Healthcare) with the ECL-Plus detection kit (Pierce, Rockford, IL).
Results
Gfi1 coordinately regulates the HoxA9-Pbx1-Meis1 complex
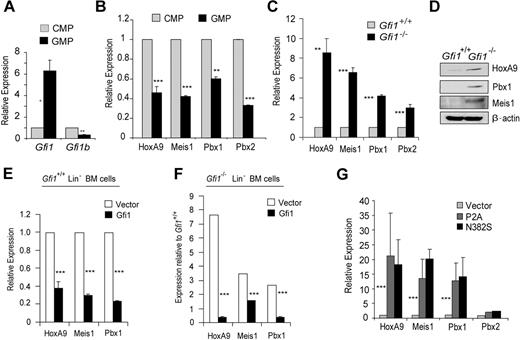
Recently, we showed that the Drosophila orthologs of Gfi1 (Sensless) and HoxA9 (Abd-B) form a molecular switch antagonizing target gene regulation in embryonic anterior-posterior patterning.26 We hypothesized that this mechanism of Gfi1-HoxA9 antagonism would be important in mammalian hematopoiesis. Because Gfi1 is known to be critical for granulopoiesis, we examined gene expression in sorted common myeloid progenitors (CMPs) to granulocyte-monocyte progenitors (GMPs). Gfi1b (a known Gfi1 target gene that mediates erythroid development) is expressed more in CMPs than in GMPs (Figure 1A). Erythroid potential is much greater in CMPs than in GMPs.31 Thus, the pattern of Gfi1b expression validates the sorted isolation of these populations. Gfi1 expression is induced during the transition from CMPs to GMPs (Figure 1A). Interestingly, the expression of HoxA9, Meis1, Pbx1, and Pbx2 is inverse to Gfi1 expression (compare Figure 1A with Figure 1B). Moreover, we found that HoxA9, Meis1, and Pbx1 mRNA and protein were deregulated in Gfi1−/− Lin− bone marrow cells (Figure 1C,D). Furthermore, this relationship appears to be direct because the forced expression of Gfi1 in Gfi1+/+ Lin− bone marrow cells lowered HoxA9, Meis1, and Pbx1 transcript levels (Figure 1E), and Gfi1 expression in Gfi1−/− Lin− bone marrow cells rescued HoxA9, Meis1, and Pbx1 expression (Figure 1F). In contrast, forced expression of Gfi1 dominant-negative mutants (Gfi1P2A or Gfi1N382S), which vary from wild type by the mutation of a single amino acid, increased HoxA9, Meis1, and Pbx1 transcript levels in wild-type Lin− bone marrow cells (Figure 1G). We conclude that Gfi1 coordinately regulates the level of HoxA9-Pbx1-Meis1 complex proteins.
Gfi1 coordinately regulates the expression of the HoxA9-Pbx1-Meis1 complex during myeloid progenitor differentiation. (A) Quantitative real-time gene expression analysis of Gfi1 and Gfi1b in sorted CMPs and GMPs from wild-type Lin− bone marrow cells. (B) Quantitative real-time gene expression analysis of HoxA9, Meis1, Pbx1, and Pbx2 in RNA from panel A. (C) Quantitative real-time gene expression analysis of HoxA9, Meis1, Pbx1, and Pbx2 in RNA from wild-type or Gfi1−/− littermate Lin− bone marrow cells. (D) Immunoblot analysis of HoxA9, Pbx1, and Meis1 in protein extracts from wild-type or Gfi1−/− littermate Lin− bone marrow cells. (E) Quantitative real-time gene expression analysis of HoxA9, Meis1, and Pbx1 in RNA from wild-type Lin− bone marrow cells transduced with an empty retroviral vector (Vector) or 1 expressing Gfi1 (Gfi1). (F) Quantitative real-time gene expression analysis of HoxA9, Meis1, and Pbx1 in RNA from Gfi1−/− Lin− bone marrow cells transduced with an empty retroviral vector (Vector) or 1 expressing Gfi1 (Gfi1). (G) Quantitative real-time gene expression analysis of HoxA9, Meis1, Pbx1, and Pbx2 in wild-type Lin− bone marrow cells transduced with retrovirus expressing Gfi1 dominant-negative mutants (P2A or N382S) or an empty vector control (Vector). Error bars indicate SD. *P ≤ .05, **P ≤ .01, ***P ≤ .001.
Gfi1 coordinately regulates the expression of the HoxA9-Pbx1-Meis1 complex during myeloid progenitor differentiation. (A) Quantitative real-time gene expression analysis of Gfi1 and Gfi1b in sorted CMPs and GMPs from wild-type Lin− bone marrow cells. (B) Quantitative real-time gene expression analysis of HoxA9, Meis1, Pbx1, and Pbx2 in RNA from panel A. (C) Quantitative real-time gene expression analysis of HoxA9, Meis1, Pbx1, and Pbx2 in RNA from wild-type or Gfi1−/− littermate Lin− bone marrow cells. (D) Immunoblot analysis of HoxA9, Pbx1, and Meis1 in protein extracts from wild-type or Gfi1−/− littermate Lin− bone marrow cells. (E) Quantitative real-time gene expression analysis of HoxA9, Meis1, and Pbx1 in RNA from wild-type Lin− bone marrow cells transduced with an empty retroviral vector (Vector) or 1 expressing Gfi1 (Gfi1). (F) Quantitative real-time gene expression analysis of HoxA9, Meis1, and Pbx1 in RNA from Gfi1−/− Lin− bone marrow cells transduced with an empty retroviral vector (Vector) or 1 expressing Gfi1 (Gfi1). (G) Quantitative real-time gene expression analysis of HoxA9, Meis1, Pbx1, and Pbx2 in wild-type Lin− bone marrow cells transduced with retrovirus expressing Gfi1 dominant-negative mutants (P2A or N382S) or an empty vector control (Vector). Error bars indicate SD. *P ≤ .05, **P ≤ .01, ***P ≤ .001.
Gfi1 loss of function predisposes to leukemia
In mammals, forced expression of HoxA9 in mouse bone marrow results in enhanced proliferation of pluripotent hematopoietic stem and progenitor cells,3 eventually leading to a fatal, transplantable acute myeloid leukemia (AML).32,33 Our data indicate that HoxA9 is deregulated onGfi1 loss of function (Figure 1). To determine the biologic sequelae of HoxA9 deregulation in Gfi1−/− cells, we performed in vitro and in vivo assays for myeloid progenitor immortalization and transformation.
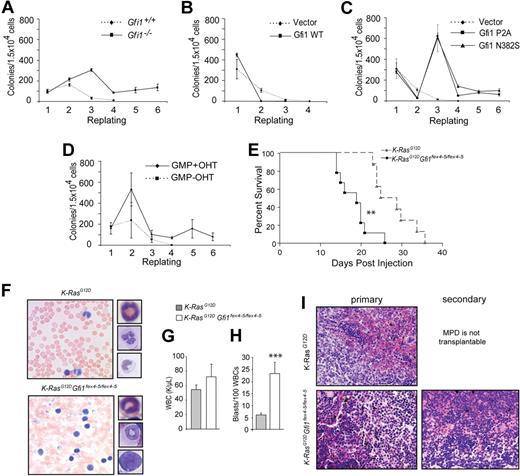
In vitro, colonies were serially plated past the time when Gfi1+/+ colony-forming potential was exhausted. Specifically, Gfi1+/+ and Gfi1−/− littermate bone marrow cells were plated in methylcellulose containing stem cell factor, IL-3, and IL-6, and resulting colonies were enumerated. Wild-type cells typically failed to form colonies after the second or third replating. However, cells from Gfi1−/− mice consistently formed monocytic-type colonies during 6 serial replatings (Figure 2A). In fact, Gfi1−/− CFUs were robust past the 10th replating (data not shown). Next, we forced expression of Gfi1 or Gfi1 dominant-negative mutants20 and determined the effect on CFU replating capacity. Wild-type Lin− bone marrow cells were transduced with retrovirus vectors expressing Gfi1 and enhanced yellow fluorescent protein (eYFP). Transduced cells were further expanded for 36 hours, and eYFP+ cells were sorted and plated to assess CFUs. Forced expression of wild-type Gfi1 extinguished all replating capacity (Figure 2B). In sharp contrast, forced expression of Gfi1 dominant-negative mutants gave consistent patterns of CFU during serial platings (Figure 2C). A burst of CFUs was induced in the third plating by both Gfi1P2A and Gfi1N382S, but this was followed by a contraction to a lower number of CFUs that could be replated past that of empty vector transduced cells. This pattern resembles that of CFUs formed by germline Gfi1−/− marrow cells during serial plating.
Gfi1 loss of function predisposes to leukemia. (A) Methylcellulose colony-forming assay with serial replating of wild-type and Gfi1−/− (Gfi1Δex4-5/Δex4-5) Lin− bone marrow cells. (B) Methylcellulose colony-forming assay with serial replating of sorted Lin− wild-type bone marrow cells transduced with retrovirus vectors expressing Gfi1 (Gfi1 WT) or an empty vector control (Vector). (C) Methylcellulose colony-forming assay with serial replating of sorted wild-type bone marrow cells transduced with retrovirus vectors expressing Gfi1 dominant-negative mutants (P2A or N382S) or an empty vector control (Vector). (D) Methylcellulose colony-forming assay with serial replating of Rosa-CreERT2+Gfi1fex4-5 fex4-5 sorted GMPs with or without tamoxifen (OHT) added in vitro to activate the CreERT2 protein and delete floxed Gfi1 alleles. (E) Survival curve of Mx1Cre+K-ras lslG12D (n = 7) or Mx1Cre+K-ras lslG12D Gfi1fex4-5/fex4-5 (n = 7) mice beginning at time of pIpC injections. (F) Photomicrographs (40 ×) of peripheral blood smears from animals upon humane killing from panel E with insets showing myeloid forms and blasts. (G) Peripheral white blood cell (WBC) counts from mice in panel E. (H) Peripheral blood myeloblast counts from mice in panel E. (I) Photomicrographs (5 ×) of H&E-stained splenic tissue from mice in panel E (primary) or recipients of 105 spleen cells from moribund Mx1Cre+K-ras lslG12D Gfi1fex4-5/fex4-5 animals in panel E (secondary). Error bars indicate SEM. **P ≤ .01, ***P ≤ .001.
Gfi1 loss of function predisposes to leukemia. (A) Methylcellulose colony-forming assay with serial replating of wild-type and Gfi1−/− (Gfi1Δex4-5/Δex4-5) Lin− bone marrow cells. (B) Methylcellulose colony-forming assay with serial replating of sorted Lin− wild-type bone marrow cells transduced with retrovirus vectors expressing Gfi1 (Gfi1 WT) or an empty vector control (Vector). (C) Methylcellulose colony-forming assay with serial replating of sorted wild-type bone marrow cells transduced with retrovirus vectors expressing Gfi1 dominant-negative mutants (P2A or N382S) or an empty vector control (Vector). (D) Methylcellulose colony-forming assay with serial replating of Rosa-CreERT2+Gfi1fex4-5 fex4-5 sorted GMPs with or without tamoxifen (OHT) added in vitro to activate the CreERT2 protein and delete floxed Gfi1 alleles. (E) Survival curve of Mx1Cre+K-ras lslG12D (n = 7) or Mx1Cre+K-ras lslG12D Gfi1fex4-5/fex4-5 (n = 7) mice beginning at time of pIpC injections. (F) Photomicrographs (40 ×) of peripheral blood smears from animals upon humane killing from panel E with insets showing myeloid forms and blasts. (G) Peripheral white blood cell (WBC) counts from mice in panel E. (H) Peripheral blood myeloblast counts from mice in panel E. (I) Photomicrographs (5 ×) of H&E-stained splenic tissue from mice in panel E (primary) or recipients of 105 spleen cells from moribund Mx1Cre+K-ras lslG12D Gfi1fex4-5/fex4-5 animals in panel E (secondary). Error bars indicate SEM. **P ≤ .01, ***P ≤ .001.
Gfi1 expression is induced during the normal transition between CMPs and GMPs (Figure 1A). We noted that Gfi1 expression inversely correlated with replating ability, because isolated CMPs consistently replated more times than GMPs (data not shown). To provide a molecular link between the level of Gfi1 and GMP colony replating capacity, we used a model in which tamoxifen (OHT)–inducible Cre (ROSA-Cre-ERT2) mediates deletion of Gfi1fex4-5 alleles in isolated progenitors. On OHT-induced Cre-ERT2 activity and Gfi1 deletion, GMPs consistently generated colonies beyond the sixth plating (Figure 2D), whereas Gfi1-sufficient GMP colony formation was exhausted after the third replating. In fact, Gfi1 deletion allowed for GMP colony formation past the 10th replating (data not shown). To address whether this increase was due to a subset of GMPs with increased replating capacity or whether a gain of function was observed in all cells, we sorted individual GMPs into 96-well plates and monitored their replating ability. A similar percentage of individual GMPs was capable of serial replating compared with the bulk GMP population (data not shown).
Taken together, the high levels of HoxA9, Pbx1, and Meis1, in addition to the abnormally long lifespan of GMPs, suggest that Gfi1-deficient animals may be prone to development of myeloid leukemias. Indeed, forced expression of HoxA9 and Meis1 mediate leukemogenesis in vivo.32 That leukemias have not been reported in Gfi1−/− mice may be related to the often lethal complications of Gfi1−/− phenotypes versus the kinetics of HoxA9-Meis1 leukemogenesis (which extends beyond the lifespan of Gfi1−/− mice). To determine whether the Gfi1 null environment is sensitized for leukemia development, we used an established model of myeloproliferative disorder (MPD) and tested the ability of Gfi1 loss of function to progress the disease to leukemia. Oncogenic RAS mutations are among the most frequently detected genetic alterations in patients with AML.34 However, activation of oncogenic K-RasG12D in murine bone marrow yields a lethal MPD with short latency and complete penetrance.35,36 To directly test the utility of Gfi1 loss of function in leukemogenesis, Mx1Cre+ mice with a K-raslslG12D gene and either wild-type or floxed Gfi1 alleles (Mx1Cre+K-raslslG12DGfi1+/+ versus Mx1Cre+K-raslslG12DGfi1fex4-5/fex4-5) were injected with pIpC and monitored daily. Consistent with published reports,35,36 induction of K-RasG12D expression resulted in a rapid, lethal MPD with a mean latency of 25 days (Figure 2E dotted line). However, although both genotypes lead to increased white blood cells in the peripheral blood (Figure 2F), Mx1Cre+K-raslslG12DGfi1fex4-5/fex4-5 mice displayed significantly decreased latency (P < .01) to approximately 16 days (Figure 2E solid line). Moreover, although K-RasG12D expression induced the expansion of differentiated myeloid lineage cells in the peripheral blood, Mx1Cre+K-RaslslG12DGfi1fex4-5/fex4-5 phenotypes included abnormal blast cells in the peripheral blood (P < .01; Figure 2F). To validate the leukemogenic capacity of such blasts, splenic cells from killed mice of both experimental groups were transplanted into sublethally irradiated CD45.1 congenic recipients. In agreement with published reports,35,36 cells from the spleen of Mx1Cre+K-raslslG12DGfi1+/+ mice did not produce a phenotype in transplant recipients. In contrast, transplantation of cells from the spleen of Mx1Cre+K-raslslG12DGfi1fex4-5/fex4-5 mice induced an acutely lethal disease characterized by increased white blood cell (WBC) counts and the presence of circulating undifferentiated blasts within 2 weeks (Figure 2I). We conclude that Gfi1 loss of function induces the progression of the K-RasG12D–associated MPD to AML.
Gfi1 loss of function transcriptionally reprograms GMPs through HoxA9
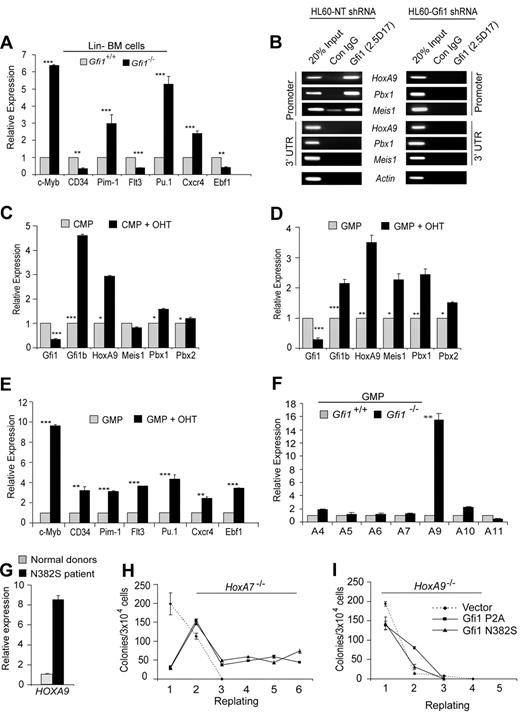
We next wanted to determine whether the elevated levels of HoxA9, Pbx1, and Meis1 in Gfi1−/− Lin− bone marrow cells resulted in functional transcriptional programming. Therefore, we examined confirmed HoxA9-target genes c-Myb, CD34, and Pim1,11–13 as well as putative HoxA9 target genes Flt3, Pu.1, Cxcr4, and Ebf1.37 We found that the steady-state levels of c-Myb, Pim1, Pu.1, and Cxcr4 transcripts were significantly increased in Gfi1−/− compared with Gfi1+/+ Lin− bone marrow cells (Figure 3A). However, the level of CD34, Flt3, and Ebf1 were not increased (Figure 3A). Given the disparity that some but not all HoxA9 target genes were up-regulated in the absence of Gfi1, we questioned whether Gfi1 directly regulates HoxA9, Pbx1, and Meis1. First, we performed ChIP on human myeloid leukemia cells to find binding to the promoter, but not the 3′ untranslated region of HoxA9, Pbx1, and Meis1 (Figure 3B). Next, we knocked down Gfi1 expression in the same cell line, which ablated Gfi1 binding to all 3 genes (Figure 3B) and up-regulated endogenous HoxA9 expression. We conclude that Gfi1 directly and specifically regulates HoxA9, Pbx1, and Meis1.
Gfi1 loss of function directly deregulates HoxA9, Pbx1, and Meis1 in GMPs. (A) Quantitative real-time gene expression analysis of putative HoxA9-target genes c-Myb, CD34, Pim1, Flt3, Pu.1, Cxcr4, and Ebf1 in RNA from wild-type or Gfi1−/− littermate Lin− bone marrow cells. (B) Chromatin immunoprecipitation (ChIP) analysis with a Gfi1-specific monoclonal antibody (2.5D.17) or isotype control IgG (Con IgG) showing Gfi1 physically bound to conserved promoter elements in HoxA9, Pbx1, and Meis1 (Promoter), but not 3′-untranslated regions of these genes (3′UTR). Gfi1 binding was detected in control nontargeting shRNA-treated HL60 cells (HL60-NT shRNA) but not in Gfi1-specific shRNA-knock-down HL60 cells (HL60-Gfi1 shRNA). (C,D) Quantitative real-time gene expression analysis of Gfi1, Gfi1b, HoxA9, Meis1, Pbx1, and Pbx2 in sorted CMPs (C) and GMPs (D) from RosaCreERT2+Gfi1fex4-5/fex4-5 Lin− bone marrow cells with or without tamoxifen (OHT). (E) Quantitative real-time gene expression analysis of putative HoxA9-target genes c-Myb, CD34, Pim1, Flt3, Pu.1, Cxcr4, and Ebf1 in RNA from panel D. (F) Quantitative real-time gene expression analysis of HoxA locus gene expression in RNA from wild-type or Gfi1−/− littermate isolated GMPs. (G) Quantitative real-time gene expression analysis of HoxA9 in CD34+ bone marrow cells from 3 healthy donors or a GFI1N382S patient. Error bars indicate SD. (H,I) Methylcellulose colony-forming assay with serial replating of HoxA7−/− (H) or HoxA9−/− (I) Lin− bone marrow cells transduced with retrovirus expressing Gfi1 dominant-negative mutants (P2A or N382S) or an empty vector control. Error bars indicate SEM.
Gfi1 loss of function directly deregulates HoxA9, Pbx1, and Meis1 in GMPs. (A) Quantitative real-time gene expression analysis of putative HoxA9-target genes c-Myb, CD34, Pim1, Flt3, Pu.1, Cxcr4, and Ebf1 in RNA from wild-type or Gfi1−/− littermate Lin− bone marrow cells. (B) Chromatin immunoprecipitation (ChIP) analysis with a Gfi1-specific monoclonal antibody (2.5D.17) or isotype control IgG (Con IgG) showing Gfi1 physically bound to conserved promoter elements in HoxA9, Pbx1, and Meis1 (Promoter), but not 3′-untranslated regions of these genes (3′UTR). Gfi1 binding was detected in control nontargeting shRNA-treated HL60 cells (HL60-NT shRNA) but not in Gfi1-specific shRNA-knock-down HL60 cells (HL60-Gfi1 shRNA). (C,D) Quantitative real-time gene expression analysis of Gfi1, Gfi1b, HoxA9, Meis1, Pbx1, and Pbx2 in sorted CMPs (C) and GMPs (D) from RosaCreERT2+Gfi1fex4-5/fex4-5 Lin− bone marrow cells with or without tamoxifen (OHT). (E) Quantitative real-time gene expression analysis of putative HoxA9-target genes c-Myb, CD34, Pim1, Flt3, Pu.1, Cxcr4, and Ebf1 in RNA from panel D. (F) Quantitative real-time gene expression analysis of HoxA locus gene expression in RNA from wild-type or Gfi1−/− littermate isolated GMPs. (G) Quantitative real-time gene expression analysis of HoxA9 in CD34+ bone marrow cells from 3 healthy donors or a GFI1N382S patient. Error bars indicate SD. (H,I) Methylcellulose colony-forming assay with serial replating of HoxA7−/− (H) or HoxA9−/− (I) Lin− bone marrow cells transduced with retrovirus expressing Gfi1 dominant-negative mutants (P2A or N382S) or an empty vector control. Error bars indicate SEM.
To reconcile the failure of Gfi1 loss of function to deregulate all HoxA9 target genes, we next examined CMPs and GMPs isolated from ROSA-Cre-ERT2+Gfi1fex4-5/fex4-5 bone marrow cells cultured with or without OHT for the expression of HoxA9, Pbx1, and Meis1. Interestingly, although tamoxifen-activated Cre-mediated deletion of floxed Gfi1 alleles induced the expression of HoxA9 and Pbx1 in both CMPs and GMPs, we noted that Meis1 was induced only in GMPs (compare Figure 3C with 3D). The reason for Meis1 induction in GMPs but not CMPs is not known, but it corresponds to the dramatic expansion of GMP on conditional Gfi1 deletion (Figure 2D). We therefore reasoned that the disparity in transcriptional programming (Figure 3A) might be resolved in GMPs, where Meis1 is significantly induced onGfi1 deletion (Figure 3D). Indeed, the expression of c-Myb, CD34, Pim1, Flt3, Pu.1, Cxcr4, and Ebf1 were significantly increased in GMPs after conditional deletion of floxed Gfi1 alleles (Figure 3E). We conclude that loss of Gfi1 transcriptionally reprograms GMPs.
Germline deletion of Hox genes can affect the regulation of surrounding genes.38 Thus, it is formally possible that Gfi1 regulation of HoxA9 could affect multiple nearby HoxA locus genes. Other than HoxA9, which was increased approximately 8-fold in Gfi1−/− Lin− bone marrow cells (Figure 1C), we did not detect significant deregulation of other HoxA genes in Lin− bone marrow cells (data not shown). Because Gfi1 loss of function affects progenitor numbers (which could skew gene expression changes) we refined our analyses to purified Gfi1+/+ and Gfi1−/− GMPs. The steady-state levels of HoxA9 transcripts were increased approximately 16-fold in purified GMPs (Figure 3F). Notably, other HoxA locus genes were not as dramatically deregulated (Figure 3F). Mouse Gfi1−/− and human GFI1N382S progenitors accumulate in vivo.22,23,25 Thus, we next examined the level of HOXA9 in GFI1N382S patient CD34+ bone marrow cells. Compared with 3 independent samples of normal CD34+ bone marrow cells, the level of HoxA9 was induced 8-fold in the GFI1N382S sample (Figure 3G). Together, these data indicate that regulation of HoxA9 by Gfi1 is specific and conserved.
We next analyzed the consequence of HoxA9 loss to the in vitro phenotype mediated by Gfi1 dominant-negative mutants. We included HoxA7−/− mice in our analyses as a control for genetic manipulation of this locus. Wild-type, HoxA7−/−, and HoxA9−/− Lin− bone marrow cells were transduced with retroviral vectors expressing Gfi1 dominant-negative mutants (P2A or N382S) and were plated to analyze CFU capacity. Colonies were enumerated, and cells were serially plated until CFUs were exhausted. Although the expression of Gfi1 dominant-negative mutants in wild-type or HoxA7−/− bone marrow cells induced increased colony replating (Figure 3H), HoxA9−/− bone marrow cells were not responsive, with HoxA9−/− cells lacking CFUs after the third plating with or without Gfi1 dominant-negatives (Figure 3I). These data indicate that HoxA9 is critically required to induce the in vitro persistence of myeloid progenitors induced by Gfi1 loss of function.
Gfi1 and HoxA9 epistasis
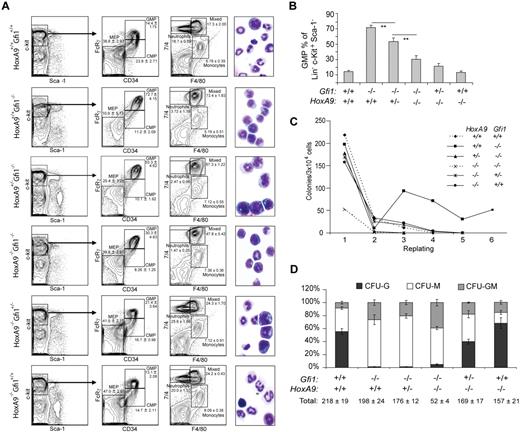
Given these strong in vitro phenotypes, we next examined Gfi1-HoxA9 epistasis in vivo by mating Gfi1+/− and HoxA9+/− mice to obtain Gfi1+/−HoxA9+/− progeny, which were then intercrossed to obtain mice with variable numbers of Gfi1 and HoxA9 alleles. Similar experiments with HoxA7−/− mice were performed as a control and show that altering HoxA7 alleles does not alter Gfi1−/− phenotypes. Gfi1−/− mice display increased GMPs and decreased megakaryocyte erythroid progenitors (MEPs). Limiting HoxA9 alleles significantly decreased the number of Gfi1−/− GMPs (Figure 4A,B) and increased the number of Gfi1−/− MEPs in a dose-dependent manner (Figure 4A). Moreover, the in vitro replating capacity of Gfi1−/− bone marrow cells was normalized by limiting even a single allele of HoxA9 (Figure 4C). In vitro, we found that the number of HoxA9 alleles correlated with the number of CFU-Ms, but that limiting Gfi1 alleles increased HoxA9−/− CFU-Ms (Figure 4D). In contrast, limiting HoxA9 alleles did not overcome the Gfi1−/− block to terminal differentiation of granulocytes in vitro (Figure 4D) or in vivo (Figure 4A). Similarly, we did not detect a statistically significant effect of limiting HoxA9 alleles on the number of phenotypic short-term or long-term Gfi1−/− HSCs. We conclude that the in vivo and in vitro phenotypes of Gfi1−/− progenitors critically depend on HoxA9.
HoxA9 controls Gfi1-induced myeloid progenitor differentiation, in vivo accumulation, and in vitro life span. (A) Flow cytometric analyses of bone marrow cells from mice (WT, n = 4; HoxA9−/−Gfi1+/+, n = 4; HoxA9−/−Gfi1+/−, n = 4; HoxA9−/−Gfi1−/−, n = 4; HoxA9+/−Gfi1−/−, n = 2; HoxA9+/+Gfi1−/−, n = 4). Populations ± SD (insets) are shown. Photomicrographs (×100) of cytospins from whole bone marrow preparations of representative animals (right). (B) Graphic representation of GMP in panel A. (C) Methylcellulose serial replating assay of Lin− bone marrow cells from mice in panel A. (D) Methylcellulose colony formation assays of bone marrow cells with percentages displayed are the average of independent analyses from individual mice in panel A. All assays represented (A-D) were initiated from 1 cohort of mice on the same day. Results shown are representative of at least 3 separate analyses on different cohorts. Error bars indicate SEM. **P ≤ .01
HoxA9 controls Gfi1-induced myeloid progenitor differentiation, in vivo accumulation, and in vitro life span. (A) Flow cytometric analyses of bone marrow cells from mice (WT, n = 4; HoxA9−/−Gfi1+/+, n = 4; HoxA9−/−Gfi1+/−, n = 4; HoxA9−/−Gfi1−/−, n = 4; HoxA9+/−Gfi1−/−, n = 2; HoxA9+/+Gfi1−/−, n = 4). Populations ± SD (insets) are shown. Photomicrographs (×100) of cytospins from whole bone marrow preparations of representative animals (right). (B) Graphic representation of GMP in panel A. (C) Methylcellulose serial replating assay of Lin− bone marrow cells from mice in panel A. (D) Methylcellulose colony formation assays of bone marrow cells with percentages displayed are the average of independent analyses from individual mice in panel A. All assays represented (A-D) were initiated from 1 cohort of mice on the same day. Results shown are representative of at least 3 separate analyses on different cohorts. Error bars indicate SEM. **P ≤ .01
Interestingly, the Gfi1−/−HoxA9−/− bone marrow displayed a striking differentiation defect indicated by the abundance of bone marrow cells with abnormal morphology (Figure 4A). Overall, the Gfi1−/−HoxA9−/− bone marrow displayed poor colony formation with total CFU numbers typically 67% to 75% lower than either Gfi1−/− or HoxA9−/− single knockouts (Figure 4D). With CFU-Gs restricted by the lack of Gfi1, the constriction centered mainly on the loss of CFU-Ms (Figure 4D). Phenotypic Gfi1−/−HoxA9−/− CMPs and GMPs were clearly present (Figure 4A), but they failed to efficiently form colonies (Figure 4C,D). Moreover, the colonies that did form were completely devoid of secondary replating capacity (Figure 4C). Thus, the Gfi1−/−HoxA9−/− mice died within 1 month of birth and displayed compounded defects similar to a bone marrow failure syndrome.
Discussion
Gfi1 integrates myeloid progenitor quiescence and differentiation through separable transcriptional programs. Patients with GFI1N382S SCN display the accumulation of differentiation-arrested myeloid progenitors.25 Similarly, Gfi1−/− mice exhibit an accumulation of GMPs and abnormal arrested progenitors,22,23 associated with increased stem and progenitor proliferation.24,39 However, the underlying molecular mechanisms have not been described. Gfi1 loss of function through germline Gfi1 deletion leads to the accumulation of GMPs in vivo.22,23 This phenotype correlates to an extended ability to form colonies in vitro induced by Gfi1 deletion or the expression of SCN-associated Gfi1 dominant-negative mutants, which shows that the phenotype is cell autonomous. The molecular mechanism pivots on Gfi1 regulation of the activity and expression of the HoxA9-Pbx1-Meis1 transcription factor complex, which is normally suppressed by Gfi1 during myeloid progenitor differentiation. Failure to induce the Gfi1 program in GMPs deregulates transcriptional control of normal hematopoietic progenitors and leads to their accumulation, but this can be uncoupled from a failure to form granulocytes.
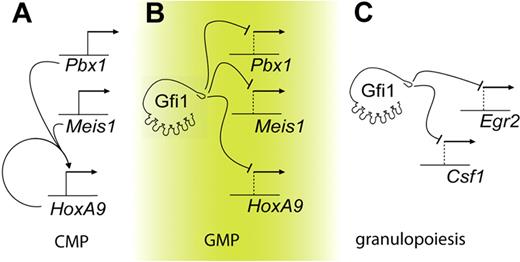
Gfi1 directly regulates the HoxA9-Pbx1-Meis1 transcription factor complex in myeloid progenitors. First, Gfi1 binds to evolutionarily conserved elements in the promoters of the HoxA9, Pbx1, and Meis1 genes in living cells (Figure 5B). The binding results in functional gene regulation because the expression of Gfi1 is inverse to HoxA9, Pbx1, and Meis1 expression during the normal transition from CMPs to GMPs. Moreover, HoxA9, Pbx1, and Meis1 expression is induced on conditional deletion of Gfi1 in GMPs. In contrast, forced expression of Gfi1 lowers HoxA9, Pbx1, and Meis1 expression in wild-type cells and rescues HoxA9, Pbx1, and Meis1 expression in Gfi1−/− cells. In the absence of Gfi1, HoxA9 autoregulation40 may further elevate HoxA9 expression levels. Thus, Gfi1 represses Pbx1 and Meis1 and competes with HoxA9 autoregulation to control HoxA9. The normal physiologic context of Gfi1 versus HoxA9-Pbx1-Meis1 antagonism is the CMP-to-GMP transition in which the induction of Gfi1 directly represses the continued expression of HoxA9-Pbx1-Meis1 factors (Figure 5).
Gfi1 integrates separable transcriptional programs in myeloid progenitors. Graphic representations. (A) In CMPs, HoxA9 is autoregulated. (B) In GMPs, Gfi1 represses Meis1, Pbx1, and HoxA9. Deregulation of Gfi1 leads to the HoxA9-dependent accumulation of myeloid progenitors. (C) Granulopoiesis is critically controlled by Gfi1 repression of target genes such as Csf1 and Egr2.20,47
Gfi1 integrates separable transcriptional programs in myeloid progenitors. Graphic representations. (A) In CMPs, HoxA9 is autoregulated. (B) In GMPs, Gfi1 represses Meis1, Pbx1, and HoxA9. Deregulation of Gfi1 leads to the HoxA9-dependent accumulation of myeloid progenitors. (C) Granulopoiesis is critically controlled by Gfi1 repression of target genes such as Csf1 and Egr2.20,47
SCN phenotypes induced by GFI1 loss of function may derive from deregulation of myelopoiesis at multiple points. Notably, HoxA9-dependent transcriptional programming modulates myeloid differentiation, but lowering HoxA9 alleles in Gfi1−/− progenitors does not permit terminal granulopoiesis. In contrast, we have recently shown that the deregulation of Csf1 by SCN-associated Gfi1 mutants is critical to the Gfi1 loss-of-function impairment of terminal granulopoiesis (Figure 5).20 Therefore, multiple steps in myeloid development may be controlled by Gfi1 and deregulated in patients with GFI1-mutant SCN. First, the ability to induce terminal granulopoietic transcriptional programming is blocked by monopoietic signaling from Csf120 (Figure 5C). Second, elevated expression of HoxA9, Pbx1, and Meis1, which should normally be down-regulated during the transition from CMPs to GMPs (Figure 5A,B), induces the abnormal accumulation of arrested myeloid progenitors. In Gfi1-null mice, such cells express primary granule genes such as MPO and ELA2.23 In ex vivo culture systems, forced expression of HoxA9 and Meis1 in primary murine bone marrow cells transform G-CSF stimulation into a proliferative signal.41 We note that recombinant G-CSF therapy for patients with GFI1-mutant SCN further expands arrested myeloid progenitors to mediate modest protection from infection.25 Thus, our data provide a potential explanation for this clinical observation.
HoxA9 activity is critical to the Gfi1−/− deregulation of progenitor biology but not granulopoiesis. Gfi1 loss of function increases GMP numbers but blocks terminal granulopoiesis in vivo and CFU-Gs in vitro.20,22,23 HoxA9 loss of function lowers CMP numbers (but not GMP) in vivo4 and decreases CFU-Ms in vitro. Gfi1−/− GMPs show an approximate 16-fold induction of HoxA9 expression. In experiments to examine epistasis, limiting alleles of HoxA9 lowered the number of Gfi1−/− GMPs and increased the number of Gfi1−/− MEPs in vivo. These results are underscored by the dose-dependent relationship between HoxA9 alleles and Gfi1−/− GMP accumulation. In vitro, limiting HoxA9 alleles eliminated Gfi1−/− colony replating, showing that the in vitro persistence of Gfi1−/− colony-forming cells critically depends on HoxA9. Thus, these Gfi1−/− phenotypes are hypostatic to HoxA9. Because lack of Gfi1 prevented the formation of CFU-Gs in vitro and granulocytes in vivo (independent of HoxA9 status), HoxA9 and Gfi1 epistasis is not a critical factor in terminal granulopoiesis. One might consider that myeloid progenitors that are unable to differentiate may accumulate as a default; however, we conclude that Gfi1−/− progenitor accumulation (but not terminal granulopoiesis) is controlled by HoxA9.
As evidenced by dramatic expansion of CFUs after inducible Gfi1 deletion in isolated cells in vitro, the expansion of Gfi1−/− GMPs in vivo is most likely due to the abnormal proliferative potential of such cells. We note that the forced expression of Gfi1 dominant negatives and conditional Gfi1 deletion amplify an expansion of CFUs, followed by a contraction to replatable colonies. We speculate that the differentiation state of the progenitors may control the response to loss of Gfi1. Specifically, deregulation of Meis1 appeared to be specific to GMPs, and Meis1 responded differently to Gfi1 expression in both Gfi1+/+ and Gfi1−/− Lin− cells compared with HoxA9 and Pbx1. In vitro, Gfi1−/− marrow has been reported to be hyperresponsive to cytokines,22,23 providing a potential explanation for Gfi1−/− progenitor proliferation. However, these observations were based on total CFUs generated by whole bone marrow and did not accommodate the dramatic changes in progenitor numbers within Gfi1−/− marrow. HoxA9−/− marrow does not display differential cytokine sensitivity.4 Gfi1−/− GMPs display dramatic deregulation of HoxA9 expression. We note that the in vivo accumulation of Gfi1−/− GMPs and the in vitro colony replating effect of Gfi1 loss of function are dependent on HoxA9 dosage. In Gfi1+/+ cells, HoxA9 overexpression induces progenitor proliferation.3 Thus, it is most likely that the proliferative response of isolated progenitors to Gfi1 loss is due to deregulation of HoxA9 and not to cytokine hypersensitivity.
The profound phenotype of Gfi1−/−HoxA9−/− hematopoietic cells highlights the importance of Gfi1 versus HoxA9 transcriptional programming. Loss of both factors in Gfi1−/−HoxA9−/− mice lead to the presence of abnormal bone marrow cells, decreased total CFUs, and survival. Similar to HoxA9−/− phenotypic HSCs,42 the numbers of Gfi1−/−HoxA9−/− phenotypic CMP and GMP progenitors did not correlate to biologic capacity, indicating that such cells are biologically defective. Moreover, Gfi1−/−HoxA9−/− phenotypes are more profound than deletion of either Gfi1 or HoxA9 alone24,42 and are similar to a bone marrow failure syndrome.
Although Gfi1 is one of the most common genes affected by Moloney leukemia virus (MoMLV) integration in lymphoid leukemias,16 it is rarely seen in myeloid leukemias driven by MoMLV-derivative viruses.43 The reason that insertions in Gfi1 are infrequently seen in myeloid tumors is explained by its repression of HoxA9, Pbx1, and Meis1, which are required for progenitor immortalization and expansion. In mice, the activation of oncogenic Ras (and subsequent chronic Map-kinase signaling) leads to both a proliferative and a differentiation stimulus and lethal overproduction of mature myeloid cells.44 Gfi1-Ras leukemogenesis is probably explained by a Gfi1−/− block to Ras-Map-kinase–induced differentiation within self-renewing cells, which also gain Gfi1−/− proliferative potential. Key molecules that antagonize transformation represent attractive candidates for clinical intervention in leukemia and myeloproliferative disorders. Therefore, the transcriptional programs controlled by Gfi1 are attractive targets for manipulation in such diseases.
In sum, the data show that the transcriptional program initiated by a single transcription factor (Gfi1) integrates myeloid lineage differentiation and proliferation by antagonizing HoxA9-Pbx1-Meis1 activity. This integrative function of Gfi1 is similar to the activity of c-Myc that simultaneously stimulates both cell-cycle progression and exposure to apoptosis.45 Although transcription factor pairs such as C/EBPα/Fog1, C/EBPα/Pu.1, Pu.1/Gfi1, and Eklf/Fli are known to antagonize each other to regulate lineage fate decisions,46–49 the transcriptional antagonism mediated by Gfi1 and HoxA9 is unique in that it includes autoregulation and the integration of separable transcriptional programs controlling progenitor life span and differentiation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Christopher Baum and Axel Schambach for the SF91 vector, Jonathan Walsh for the CMMP vectors, Anton Berns for the ROSA-Cre-ERt2 mice, and Michael A. Caligiuri for HoxA9−/− and HoxA7−/− mice. We also thank Michelle Meadows and Avinash Baktula for technical assistance and James Phelan, Brian Gebelein, Anil Jegga, Chris Karp, Jose Cancelas, Jim Mulloy, and Hartmut Geiger for scientific discussions.
This work was supported by the Division of Intramural Research, NIAID, NIH (J.Z. and W.E.P.), by a grant from Cancerfree Kids (Loveland, OH), and by NIH (CA105152 and HL079574; H.L.G.).
National Institutes of Health
Authorship
Contribution: S.R.H. and C.S.V. designed and performed experiments, analyzed results, generated figures, and wrote the paper; T.B. performed flow cytometry; J.Z. and W.E.P. provided mice; A.C. performed leukemogenesis experiments; B.G. provided essential conceptual information and data before publication; and H.L.G. designed experiments and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: H. Leighton Grimes, Division of Immunobiology, Cincinnati Children's Hospital Medical Center, 3333 Burnet Ave MLC7038, Cincinnati, OH 45229; e-mail: lee.grimes@cchmc.org
References
Author notes
*S.R.H. and C.S.V. contributed equally to this study.