Abstract
Children with Down syndrome (DS) have a greatly increased risk of acute megakaryoblastic leukemia (AMKL) and acute lymphoblastic leukemia (ALL). Both DS-AMKL and the related transient myeloproliferative disorder (TMD) have GATA1 mutations as obligatory, early events. To identify mutations contributing to leukemogenesis in DS-ALL, we undertook sequencing of candidate genes, including FLT3, RAS, PTPN11, BRAF, and JAK2. Sequencing of the JAK2 pseudokinase domain identified a specific, acquired mutation, JAK2R683, in 12 (28%) of 42 DS-ALL cases. Functional studies of the common JAK2R683G mutation in murine Ba/F3 cells showed growth factor independence and constitutive activation of the JAK/STAT signaling pathway. High-resolution SNP array analysis of 9 DS-ALL cases identified additional submicroscopic deletions in key genes, including ETV6, CDKN2A, and PAX5. These results infer a complex molecular pathogenesis for DS-ALL leukemogenesis, with trisomy 21 as an initiating or first hit and with chromosome aneuploidy, gene deletions, and activating JAK2 mutations as complementary genetic events.
Introduction
Children with Down syndrome (DS), characterized by constitutional trisomy 21, have a 50-fold increased risk of developing acute leukemia in the first few years of life.1,2 This comprises the normally extremely rare subtype acute megakaryoblastic leukemia (AMKL) as well as the common variety (B-cell precursor) of acute lymphoblastic leukemia (ALL).2-4 Both DS-AMKL and the transient myeloproliferative disorder (TMD) that often precedes it are consistently associated with acquired mutations in the GATA1 gene.5 Concordant GATA1 mutations in the blast cells of identical twins with TMD6 and mutations in the neonatal blood spots of DS newborns7 indicate that this is an early/prenatal event in DS leukemogenesis
Much less is known about the genetic events predisposing to DS-ALL. Candidate genes for activating mutations in DS-ALL include FLT3 and RAS, both mutated in high hyperdiploid ALL (with acquired trisomy 21),8,9 PTPN11 and BRAF, mutated in B-cell precursor ALL.10,11 An additional candidate is the JAK2 pseudokinase domain mutation JAK2ΔIREED, reported in a single case of DS-ALL.12 A different JAK2 pseudokinase domain mutation, JAK2V617F, is frequently found in the myeloproliferative disorders (MPDs) and believed to be an initiating event.13-15 Submicroscopic deletions that involve genes linked functionally to deregulation of cell cycling or B-cell differentiation were also implicated in the molecular pathogenesis of ALL.16-19 In this study we undertook both sequencing of candidate genes and high-resolution single-nucleotide polymorphism (SNP) array analysis to identify genetic events associated with ALL in DS.
Methods
Patients
Patients' samples (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) were collected, informed consent was obtained in accordance with the Declaration of Helsinki, and the study was carried out with approval of the ethical review committee from all participating institutions. DNA was extracted from archival (frozen cell or cytogenetic–fixed pellet) leukemic samples.
Sequencing
Primers were designed to amplify the following candidate genes: FLT3 ITD, FLT3 kinase domain, KIT kinase domain, PTPN11, BRAF, NRAS, and KRAS (Table S2A). Exons 12 to 23 of the JAK2 gene were polymerase chain reaction (PCR)–amplified and sequenced using previously described primers.13 All samples found to be mutated were PCR-amplified and sequenced in a second, independent experiment. Pyrosequencing was carried out in accordance to the manufacturer's instructions (Biotage AB, Uppsala, Sweden) using the primers shown in Table S2B.
Ba/F3 proliferation assay
MSCV-neo-IRES-GFP murine wild-type JAK2 and JAK2R683G constructs were transfected into GP2 packaging cells to produce retroviruses as previously described.13 Ba/F3 cells were cotransduced with MSCV-puro-TpoR retrovirus and selected in media containing 2 mg/mL G418 and 0.5 μg/mL puromycin, and they were further cultured in the presence or absence of interleukin-3 (IL-3) with or without 10 μM tyrosine kinase inhibitor AG490 (Invitrogen, Carlsbad, CA).
SNP arrays
Leukemic DNA from the patient's bone marrow at presentation of ALL and remission DNA (as germ line control) was genotyped with Affymetrix (Santa Clara, CA) Human GeneChip Mapping 250K Nsp1 and Sty1 arrays as previously described.19,20 Array hybridization data were analyzed for loss of heterozygosity and signal intensity using the in-house GOLF software program (for more details, see Figure S1).
Results and discussion
Sequencing of selected exons of the FLT3, KIT, PTPN11, BRAF, NRAS, and KRAS genes in a series of 10 DS-ALL cases did not identify any mutations (Table S1). Sequencing of JAK2 exon 14 in 42 cases of DS-ALL identified a point mutation at the conserved arginine (R683) affected by the IREED deletion12 in 12 (28%) of 42 cases. Ten of these were an identical arginine-to-glycine (JAK2R683G) and 2 an arginine-to-serine (JAK2R683S) substitution. The mutation was acquired in the leukemic blasts (Figure 1A). Pyrosequencing confirmed the mutations and allowed quantitation of the mutant allele. Sequencing of exon 14 in 41 non–DS-ALL cases, including 23 of high hyperdiploidy (with acquired trisomy 21), did not identify any mutations in this group. Similarly, we did not identify any JAK2R683 mutations in 13 DS-AMKL cases (Table 1). Seven DS-ALL cases without JAK2R683 mutations were also screened for mutations in the JAK2 kinase domain (exons 15-23). None were observed.
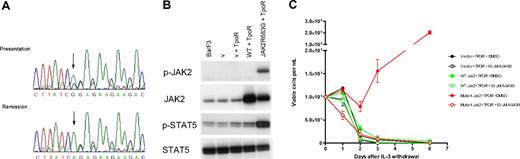
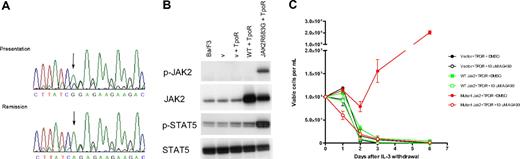
JAK2R683G is an activating mutation in DS-ALL. (A) Sequence analysis shows the JAK2R683G mutation in a DS-ALL patient. (Top) Sequence obtained from bone marrow DNA at presentation and (bottom) complete remission. The A to G substitution is shown by an arrow. During remission the sequence was identical to wild-type, showing that the mutation is acquired. (B) Western blotting analysis of JAK2 and STAT5 protein in IL-3–dependent Ba/F3 cells. Extraction and Western blotting analyses were performed with the use of standard protocols. Antibodies against JAK2 and phospho-JAK2 (p-JAK2), STAT5, and phospho-STAT5 (p-STAT5) were from Cell Signaling Technology (New England Biolabs, Ipswich, MA). Lane 1 indicates untransformed Ba/F3 cells; lane 2, empty vector control (v) (with neo and puro); lane 3, empty vector + TpoR; lane 4, wild-type (wt) JAK2-transformed Ba/F3 cells; lane 5, JAK2R683G mutant–transformed Ba/F3 cells. (C) Ba/F3 proliferation after IL-3 withdrawal. The JAK2R683G mutation was generated in the IMAGE clone 6838318 containing murine JAK2 cDNA using site-directed mutagenesis (Quikchange-Xl; Stratagene; Cambridge, United Kingdom) and confirmed by full-length DNA sequencing. Ba/F3 cells were cotransduced with thrombopoietin receptor (TpoR) and wild-type or mutant (R683G) JAK2. Cells were washed 3 times in PBS and cultured at 105/mL in the absence of IL-3 for 6 days with or without the JAK2 inhibitor AG490. Cell numbers and viability were assessed in duplicate after Trypan Blue exclusion staining.
JAK2R683G is an activating mutation in DS-ALL. (A) Sequence analysis shows the JAK2R683G mutation in a DS-ALL patient. (Top) Sequence obtained from bone marrow DNA at presentation and (bottom) complete remission. The A to G substitution is shown by an arrow. During remission the sequence was identical to wild-type, showing that the mutation is acquired. (B) Western blotting analysis of JAK2 and STAT5 protein in IL-3–dependent Ba/F3 cells. Extraction and Western blotting analyses were performed with the use of standard protocols. Antibodies against JAK2 and phospho-JAK2 (p-JAK2), STAT5, and phospho-STAT5 (p-STAT5) were from Cell Signaling Technology (New England Biolabs, Ipswich, MA). Lane 1 indicates untransformed Ba/F3 cells; lane 2, empty vector control (v) (with neo and puro); lane 3, empty vector + TpoR; lane 4, wild-type (wt) JAK2-transformed Ba/F3 cells; lane 5, JAK2R683G mutant–transformed Ba/F3 cells. (C) Ba/F3 proliferation after IL-3 withdrawal. The JAK2R683G mutation was generated in the IMAGE clone 6838318 containing murine JAK2 cDNA using site-directed mutagenesis (Quikchange-Xl; Stratagene; Cambridge, United Kingdom) and confirmed by full-length DNA sequencing. Ba/F3 cells were cotransduced with thrombopoietin receptor (TpoR) and wild-type or mutant (R683G) JAK2. Cells were washed 3 times in PBS and cultured at 105/mL in the absence of IL-3 for 6 days with or without the JAK2 inhibitor AG490. Cell numbers and viability were assessed in duplicate after Trypan Blue exclusion staining.
Preliminary structural modeling of the JAK2 pseudokinase domain suggests that the JAK2R683G/S mutations probably alter the interdomain interactions with the C-terminal kinase domain (Document S1). Functional studies indicate that the JAK2R683G mutation results in an activated kinase. Western blotting of Ba/F3 cells cotransduced with TpoR and JAK2R683G mutant showed constitutive phosphorylation of JAK2 and STAT5 proteins, whereas the wild-type JAK2 did not (Figure 1B). The R683G mutation allowed IL-3–dependent Ba/F3 cells to survive and proliferate in the absence of IL-3, and this effect was abrogated in the presence of the JAK2 kinase inhibitor AG490 (Figure 1C). These data parallel the effect of the JAK2V617F mutation in MPD.13-15
Cytogenetic analyses of our cases of DS-ALL, with or without the JAK2R683 mutation, indicated the presence of additional chromosomal lesions (Table S1). In common with other reports,21 our series had a lower than usual incidence of common fusion genes (eg, ETV6-RUNX1) and high hyperdiploidy. There do not appear to be any cytogenetic features that distinguish the DS-ALL cases with the JAK2R683 mutation from those without.
High-resolution SNP array analysis of 9 DS-ALL cases identified between 1 and 13 regions of loss or gain per case (Table S3). Small focal deletions, often involving a single gene, were identified in many cases. The most common deletion (4 of 9 cases) involved the ETV6 gene (Figure S1). Focal deletions of other genes implicated in ALL pathogenesis included the CDKN2A (3 of 9 cases), PAX5 (2 of 9 cases) BTLA, EBF1, and RB1 genes (1 case each). The pattern and number of microdeletions in this series, in particular the high incidence of ETV6 deletions, more closely resembled ETV6-RUNX1–positive ALL than other subgroups of ALL.16 There were no microdeletions unique to the JAK2 mutated cases.
Our data indicate that multiple genetic events, including cytogenetically detectable chromosome aneuploidy, submicroscopic deletions of genes, including ETV6, CDKN2A, and PAX5, as well as activating JAK2 mutations, can occur, collectively complementing constitutional trisomy 21 in DS-ALL. The functional studies of the JAK2R683G mutation indicate that this is probably a “driver” mutation22 in a subset of patients with DS-ALL, but the sequence of acquired genetic events, in utero and postnatally, remains to be established. In one case in the present study (patient 9; Table S1), the JAK2R683G mutation was acquired only at relapse. In another (patient 10; Table S1), the mutation appeared to be subclonal, because pyrosequencing found 32% mutant compared with 99% blasts. These data indicate that the mutation can, in some cases, be a late or secondary event.
The absence (or rarity) of JAK2 mutations in non-DS B-cell precursor ALL, including cases with acquired trisomy 21, and the high risk of ALL in DS syndrome indicate that constitutive trisomy 21 provides selective pressure within the early B-cell lineage for the specific JAK2R683 mutation that is different from the JAK2V617F-activating mutation in a stem cell resulting in MPD.14 That this is less consistent than the obligatory GATA1 mutations in myeloid/erythroid progenitors in TMD and AMKL suggests that alternative genetic changes may activate the JAK-STAT pathway in DS-ALL. This would be in accord with the essential role this pathway plays in early B-cell development.23
Children with DS and standard-risk ALL have an inferior clinical outcome to non-DS patients.24 The occurrence of mutational activation of JAK2 in a substantial fraction of DS-ALL therefore may have potential therapeutic implications, as in the MPD with the JAK2V617F mutation.15
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the UK Children's Cancer Group Study and Kathy Pritchard-Jones for patient samples, John Swansbury for helpful advice on cytogenetics, Anthony Renwick for technical help with the pyrosequencing, and David Barford for preliminary structural predictions of the JAK2 pseudokinase domain. The murine JAK2 and TpoR cDNA were both provided by Drs A. Green and LM Scott (Cambridge, United Kingdom) and the pMSCV-neo-IRES-GFP retroviral vector by Dr Louise Van Der Weyden (Cambridge, United Kingdom).
This work was supported by Leukemia Research (London, United Kingdom), the Association for International Cancer Research (AICR; Fife, United Kingdom), and Cancer Research UK (London, United Kingdom).
Authorship
Contribution: L.K. and M.G. designed the research and wrote the paper; A.M.F. supervised the research, analyzed the data, and wrote the paper; D.G.D.C., J.Y., J.P., M.E-I., S.W.H., C.M.B., K.A., and T.C. performed the research; L.K., D.G.D.C., and S.W.H. analyzed the data; and B.D.Y., C.J.H., H.K., and C.W.E.S. provided expertise or vital analytical tools or reagents. All authors critically reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lyndal Kearney, Section of Haemato-Oncology, The Institute of Cancer Research, Brookes Lawley Building, 15 Cotswold Road, Sutton, Surrey SM2 5NG, United Kingdom; e-mail: lyndal.kearney@icr.ac.uk.