Abstract
We conducted comprehensive gene expression profiling (GEP) of primary pulmonary mucosa-associated lymphoid tissue (MALT) lymphoma (n = 33) and compared the results to GEP of other B- and T-cell lymphomas and normal lymphocytes to identify novel markers and deregulated pathways. MALT has a prominent T-cell signature and a marginal zone/memory B-cell profile. Four novel transcripts were specifically overexpressed in MALT, and 2 of these, MMP7 and SIGLEC6, were validated at the protein level. GEP also revealed distinct molecular subsets in MALT. One subset, characterized by MALT1 translocations, showed overexpression of nuclear factor-κB (NF-KB) pathway genes but also was enriched for chemokine signaling pathways. Another subset showed increased plasma cells and a prominent plasma cell gene signature. By analyzing several genes with very high (“spiked”) expression in individual cases, we identified clusters with different biologic characteristics, such as samples with MALT1 translocations having high expression of MALT1 and RARA, samples with plasmacytic differentiation having high FKBP11 expression, and samples with high RGS13 expression tending to have trisomy 3 and reactive follicles. In conclusion, MALT subgroups with distinct pathologic features defined by distinct groups of deregulated genes were identified. These genes could represent novel diagnostic and therapeutic targets.
Introduction
Mucosa-associated lymphoid tissue (MALT) lymphoma, first described by Isaacson and Wright in 1983,1 is a low-grade, extranodal B-cell neoplasm with distinct histologic, molecular, and clinical characteristics. In the most recent World Health Organization classification of lymphoid tumors, MALT lymphoma is classified as a type of marginal zone B-cell lymphoma.2 It is the third most common type of non-Hodgkin lymphoma behind diffuse large B-cell lymphoma and follicular lymphoma, constituting 8% of non-Hodgkin lymphoma in an international survey.3
In recent years, several well-characterized translocations have been described in MALT lymphoma. The prevalence of these translocations differs according to disease sites.4,5 The 2 most common translocations are t(11;18)(q21;q21) and t(14;18)(q32;q21), both of which involve the MALT1 gene locus at 18q21. t(11;18)(q21;q21) results in a functional fusion protein comprising the N terminus of API2 with 3 intact baculovirus inhibitor of apoptosis repeat (BIR) domains and the C-terminal MALT1 region containing an intact caspase-like domain. It is found in 13.5 to 35% of MALT lymphomas and mainly affects gastrointestinal and pulmonary MALT lymphomas. The t(14;18)(q32;q21) occurs in approximately 5% of MALT lymphomas and brings the MALT1 gene under the control of the IGH enhancer on chromosome 14, resulting in the deregulated expression of MALT1. The consequence of both of these translocations is the constitutive activation of the canonical nuclear factor-κB (NF-KB) signaling pathway.6 In normal cells, NF-KB signaling plays a central role in regulating diverse biologic processes involved in immunity, development, growth, and survival.7 In malignant cells, abnormal NF-KB signaling has been implicated in tumor growth and survival and chemoresistance.8
Although progress has been made in understanding the molecular pathogenesis of MALT lymphomas, there are still significant gaps in our knowledge. The availability of high-throughput gene expression profiling (GEP) platform has greatly transformed our ability to investigate changes in the expression of thousands of genes within a cell in a parallel fashion, allowing a snapshot view of global molecular changes. In B-cell lymphomas, the use of GEP has resulted in identification of novel subtypes within known categories of lymphomas,9 normal counterpart of malignant cells,10 prognostic groups,11 and the pathogenic and clinical contributions of infiltrating nonmalignant immune cells.12,13
In this study, we analyze the gene expression profile of a cohort of well-characterized primary pulmonary MALT lymphomas in relation to other lymphomas and normal cells, with the objectives of (1) defining a MALT lymphoma signature or new MALT lymphoma-specific markers, (2) identifying the cell of origin of MALT lymphoma, (3) defining the molecular consequences of translocations involving MALT1, (4) studying the molecular heterogeneity within MALT lymphoma, and (5) identifying genes with “spiked” expression in individual cases, which could represent genes involved in novel translocations with the immunoglobulin heavy chain (IGH) locus14 or novel gene fusions.15 All the MALT lymphomas were primary tumors and were derived from a single anatomic site, to maximize the uniformity of the cohort.
Methods
Tissues
Thirty-five lung MALT lymphoma biopsy specimens from 33 patients were procured for the study after informed consent was obtained in accordance with the Declaration of Helsinki. The study was approved by the Mayo Clinic Institutional Review Board. All tumors represent primary MALT lymphomas of the lung. Two patients had a remote history of undergoing chemotherapy (chlorambucil or cyclophosphamide, doxorubicin, vincristine, and prednisolone) for lymphoma (further details not available). The percentage of malignant B cells, plasma cells, reactive T cells, reactive lymphoid follicles, and uninvolved lung tissue was estimated in each case based on morphology and immunophenotyping performed on frozen sections of the tissue on which the gene expression profiling was performed. All specimens contained at least 15% tumor as estimated by this method (range, 15%-81%; median, 40%). The pathologic features are shown in Table 1. The fluorescence in situ hybridization (FISH) results of these samples have been published previously.4
Pathologic and genetic characteristics of pulmonary MALT lymphoma cases
| ID . | % Tumor . | % Lung . | % T cells . | % Follicles . | FISH . | Sex . |
|---|---|---|---|---|---|---|
| Rem001 | 64 | 20 | 16 | 0 | API2/MALT1 | M |
| Rem002 | 81 | 10 | 9 | 0 | Normal | M |
| Rem003 | 56 | 20 | 24 | 0 | Aneuploid | F |
| Rem004 | 72 | 20 | 8 | 0 | Normal | F |
| Rem005 | 24 | 60 | 16 | 0 | Aneuploid | M |
| Rem006 | 16 | 60 | 24 | 0 | Normal | F |
| Rem007 | 24 | 20 | 56 | 0 | Aneuploid | F |
| Rem008 | 48 | 40 | 12 | 0 | API2/MALT1 | M |
| Rem009 | 35 | 50 | 15 | 0 | Normal | M |
| Rem010 | 40 | 50 | 10 | 0 | Aneuploid | F |
| Rem011 | 24 | 40 | 18 | 18 | Normal | F |
| Rem012 | 32 | 20 | 16 | 32 | Normal | F |
| Rem013 | 24 | 40 | 12 | 24 | Normal | F |
| Rem014 | 24 | 70 | 6 | 0 | IGH/MALT1 | F |
| Rem015 | 28 | 60 | 12 | 0 | Normal | F |
| Rem016 | 64 | 20 | 16 | 0 | Normal | F |
| Rem017 | 40 | 50 | 10 | 0 | Aneuploid | F |
| Rem018 | 28 | 60 | 12 | 0 | Aneuploid | F |
| Rem019 | 56 | 30 | 14 | 0 | API2/MALT1 | F |
| Rem020 | 48 | 40 | 12 | 0 | Aneuploid | M |
| Rem021 | 15 | 50 | 35 | 0 | API2/MALT1 | F |
| Rem022 | 21 | 30 | 14 | 35 | Normal | M |
| Rem023 | 63 | 30 | 7 | 0 | IGH/MALT1 | M |
| Rem024 | 25 | 50 | 25 | 0 | Normal | F |
| Rem025 | 70 | 10 | 20 | 0 | Normal | M |
| Rem026 | 42 | 40 | 18 | 0 | Normal | M |
| Rem027 | 42 | 30 | 7 | 21 | IGH/MALT1 | M |
| Rem028 | 81 | 10 | 9 | 0 | API2/MALT1 | M |
| Rem029 | 81 | 10 | 9 | 0 | API2/MALT1 | M |
| Rem030 | 72 | 10 | 18 | 0 | API2/MALT1 | F |
| Rem031 | 72 | 10 | 18 | 0 | API2/MALT1 | F |
| Rem032 | 25 | 50 | 25 | 0 | Normal | M |
| Rem033 | 81 | 10 | 9 | 0 | API2/MALT1 | M |
| Rem034 | 81 | 10 | 9 | 0 | Normal | F |
| Rem036 | 32 | 20 | 8 | 40 | API2/MALT1 | M |
| ID . | % Tumor . | % Lung . | % T cells . | % Follicles . | FISH . | Sex . |
|---|---|---|---|---|---|---|
| Rem001 | 64 | 20 | 16 | 0 | API2/MALT1 | M |
| Rem002 | 81 | 10 | 9 | 0 | Normal | M |
| Rem003 | 56 | 20 | 24 | 0 | Aneuploid | F |
| Rem004 | 72 | 20 | 8 | 0 | Normal | F |
| Rem005 | 24 | 60 | 16 | 0 | Aneuploid | M |
| Rem006 | 16 | 60 | 24 | 0 | Normal | F |
| Rem007 | 24 | 20 | 56 | 0 | Aneuploid | F |
| Rem008 | 48 | 40 | 12 | 0 | API2/MALT1 | M |
| Rem009 | 35 | 50 | 15 | 0 | Normal | M |
| Rem010 | 40 | 50 | 10 | 0 | Aneuploid | F |
| Rem011 | 24 | 40 | 18 | 18 | Normal | F |
| Rem012 | 32 | 20 | 16 | 32 | Normal | F |
| Rem013 | 24 | 40 | 12 | 24 | Normal | F |
| Rem014 | 24 | 70 | 6 | 0 | IGH/MALT1 | F |
| Rem015 | 28 | 60 | 12 | 0 | Normal | F |
| Rem016 | 64 | 20 | 16 | 0 | Normal | F |
| Rem017 | 40 | 50 | 10 | 0 | Aneuploid | F |
| Rem018 | 28 | 60 | 12 | 0 | Aneuploid | F |
| Rem019 | 56 | 30 | 14 | 0 | API2/MALT1 | F |
| Rem020 | 48 | 40 | 12 | 0 | Aneuploid | M |
| Rem021 | 15 | 50 | 35 | 0 | API2/MALT1 | F |
| Rem022 | 21 | 30 | 14 | 35 | Normal | M |
| Rem023 | 63 | 30 | 7 | 0 | IGH/MALT1 | M |
| Rem024 | 25 | 50 | 25 | 0 | Normal | F |
| Rem025 | 70 | 10 | 20 | 0 | Normal | M |
| Rem026 | 42 | 40 | 18 | 0 | Normal | M |
| Rem027 | 42 | 30 | 7 | 21 | IGH/MALT1 | M |
| Rem028 | 81 | 10 | 9 | 0 | API2/MALT1 | M |
| Rem029 | 81 | 10 | 9 | 0 | API2/MALT1 | M |
| Rem030 | 72 | 10 | 18 | 0 | API2/MALT1 | F |
| Rem031 | 72 | 10 | 18 | 0 | API2/MALT1 | F |
| Rem032 | 25 | 50 | 25 | 0 | Normal | M |
| Rem033 | 81 | 10 | 9 | 0 | API2/MALT1 | M |
| Rem034 | 81 | 10 | 9 | 0 | Normal | F |
| Rem036 | 32 | 20 | 8 | 40 | API2/MALT1 | M |
The tumor and lung content was estimated on frozen sections of the samples used for the gene expression profiling.
M indicates male; and F, female.
Gene expression profiling
RNA was extracted from frozen tissue using the PerfectPure RNA Tissue Kit (5 PRIME, Gaithersburg, MD), and GEPs were generated using the Affymetrix U133 plus 2.0 microarrays covering more than 47 000 transcripts according to the manufacturer's instructions (Affymetrix, Santa Clara, CA). All microarray data have been deposited with Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE13314.
Other gene expression datasets
For comparative studies, we obtained datasets from previously published, publicly available sources. Gene expression profiles of the following lymphoid malignancies performed using Affymetrix U133 microarrays were used: Burkitt lymphoma (BL), atypical Burkitt lymphoma (aBL), diffuse large B-cell lymphoma (DLBCL) of the germinal center (GC) and activated B-cell (ABC) subtypes,16 angioimmunoblastic T-cell lymphoma (AITL), anaplastic large cell lymphoma (ALCL), peripheral T-cell lymphoma unspecified (PTCL),17 Waldenstrom macroglobulinemia (WM), multiple myeloma (MM), and chronic lymphocytic leukemia (CLL).18 At the time of the analysis, no publicly accessible dataset using the same platforms were available for follicular lymphoma. We also used gene expression profiles of normal tissues such as plasma cells (NPC), peripheral blood B cells (BC),18 CD4-positive T cells (CD4 TC), CD8-positive T cells (CD8 TC), DR plus activated T cells (DR + TC), DR-T cells (DR-TC),17 and normal lung tissues19 (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Gene expression analysis
Raw gene expression data from Affymetrix CEL files were uploaded onto GeneSpring GX 7.3.1. (Agilent Technologies, Santa Clara, CA) and Robust Multi-Array (RMA) normalization performed. The normalized data were then log-transformed and median-centered for analysis.
For comparison with other lymphoid malignancies and normal cellular counterparts, variably expressed genes across the MALT lymphoma dataset were first identified by Welch analysis of variance (ANOVA) using variance computed by applying the Cross-Gene Error Model (CGEM) based on Deviation from one available within GeneSpring. This method overcomes the lack of replicates and variance associated with the individual samples and is similar in principle to variance filtering. The 1457 probes passing the significance filter of a corrected P value of 10−9 after multiple testing corrections using the Benjamini and Hochberg methods were used to cluster all the samples using the hierarchical agglomerative algorithm. Pearson correlation coefficient and centroid linkage were used as similarity and linkage methods, respectively. The functional categories of gene clusters were assessed according to gene ontology (GO) annotations.
To assess the relationship of MALT lymphoma cells to those of different lymph node compartments (germinal center, marginal zone, and mantle zone)20 and B-cell subsets (memory B cells),10 we extracted published signatures of these compartments and assessed the expression of genes constituting these signatures in the B-cell lymphomas included in our analysis (MALT, DLBCL, BL, and WM).
To identify genes specifically over- or underexpressed in MALT lymphoma, we performed ANOVA with multiple testing correction across the different diagnostic categories with post hoc pairwise analysis using the Tukey statistics. Probes differentially expressed at a corrected P value of .05 between the pairing of MALT with every other individual diagnosis were then intersected to obtain the uniquely expressed gene set.
For assessment of molecular heterogeneity within the MALT lymphomas, variably expressed genes were used for unsupervised clustering of the MALT lymphomas using the aforementioned algorithms. Differentially expressed probes between MALT lymphomas with and without MALT1 translocations t(14;18) and t(11;18) were identified using the Student t test with multiple testing corrections. For the analysis of spiked gene expression, we used the following set of sequential filters: genes with variable expression by ANOVA with CGEM and multiple testing correction at corrected P value of .05; present flag in at least 5 samples; raw expression value greater than 5000 in at least one sample.
Immunohistochemistry
Tissue microarrays (TMA) were constructed using paraffin-embedded tissue from all 35 specimens. A hematoxylin and eosin (H&E) section from each block was examined by a pathologist (E.D.R.), and the areas for TMA sampling were marked on the corresponding paraffin block. For each TMA, a 2.0-mm cylindrical core was removed from each donor paraffin block and transferred to a recipient paraffin block at defined array positions. Large diameter cores were used to preserve tumor morphology and architecture. Immunoperoxidase stains were applied to 5-μm sections of TMA using antibodies directed against MMP7 (clone ID-2; Millipore, Billerica, MA), SIGLEC-6 (polyclonal; Abcam, Cambridge, MA), RARA (polyclonal; Santa Cruz Biotechnology, Santa Cruz, CA), RGS-13 (polyclonal; Abcam), MALT1 (clone EP603Y; Epitomics, Burlingame, CA), CD20 (clone L26; Dako, Carpinteria, CA), CD3 (clone PS1; Novocastra Laboratories, Newcastle, United Kingdom), CD138 (clone MI15; Dako), and κ and λ immunoglobulin light chains (polyclonal; Dako). Immunophenotyping was also performed on frozen sections of each MALT lymphoma using antibodies directed against CD20 (clone L26; Dako), CD3 (clone Leu4; BD Biosciences, San Jose, CA), and CD138 (clone MI15; Dako). Tonsil was used for titrating the antibodies and as controls for all antibodies. Immunohistochemical assays were performed on all cases.
FISH
Interphase FISH was performed previously on nuclei isolated from all cases according to previously published methods.4 These cases were screened for translocations involving MALT1 using a breakapart (BAP) probe (Vysis, Downers Grove, IL). Cases that showed separation of the MALT1 BAP probe were further investigated using dual fusion (D-FISH) DNA probes for API2-MALT1 and IGH-MALT1 (Vysis). All cases were screened for translocations involving IGH using an IGH BAP probe (Vysis), an IGH-BCL10 D-FISH probe (homebrew), and/or an IGH-MALT1 D-FISH probe (Vysis). One hundred consecutive qualifying interphase nuclei from different areas of the same slide were scored. Based on these previous FISH studies, 10 specimens had API2-MALT1, 3 specimens had IGH-MALT1, and 22 specimens had no abnormalities involving MALT1 (Table 1).
FISH was also performed on paraffin sections of the TMA using a centromere-specific α-satellite (CEN) probe for chromosome 3 (Vysis) and a BAP for RARA (Vysis). A minimum of 50 cells were scored per case, and a minimum of 20 abnormal cells were required for that sample to be considered abnormal.4,21
Statistical analysis
The χ2 test was used to assess the association between the molecular subtypes with clinical and pathologic features of the tumors. The Student 2-tailed t test P values are reported. A P value less than .05 was considered significant.
Results
GEP of MALT lymphoma in relation to other B-cell lymphomas and normal tissues and cells
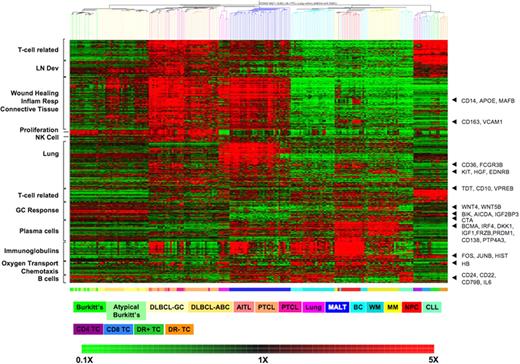
Using the 1457 probe sets that pass our statistical filter for unsupervised clustering of the samples, most unique diagnosis and cell types form distinct clusters (Figure 1). Cases of BL and aBL do not have distinct GEP and form a mixed cluster. DLCBL form a cluster with most of the ABC subtype clustered together. The AITL, ALCL, and PTCL samples are also intermixed, suggesting they are indistinguishable by global GEP. These results are consistent with the original studies from which these samples are obtained.17 The similarity in the expression profiles of WM with CLL and BC and the clustering of some MM and WM (in which plasma cells predominated over B cells) with NPCs was also consistent with our previous published study.18 The anchoring of gene clusters to known tissue or cell-type markers such as CD22, CD24, and CD79a in B cells; CD138 and PRDM1 in NPC and MM; AICDA (AID) in germinal center tumors (DLBCL); T-cell markers in T cells and different T-cell lymphomas; lack of GC signature in MALT lymphoma, and lack of proliferation signature in lung tissue, suggests that combining these disparate datasets faithfully reproduces the expected molecular signatures.
Clustering of different lymphoid malignancies and normal tissues by hierarchical clustering. Enriched GO biologic process for each gene clusters are indicated on the left. The top T cell–associated cluster of genes are overexpressed in MALT and normal and malignant T cells, but not other B-cell tumors or normal lung. Genes of interest anchoring these gene expression profiles are indicated on the right. The colored bar below the heatmap represents the different tissue types, and the legend key is just below. On the heatmap, red represented up-regulated genes and green down-regulated genes, with the scale represented at the bottom of the figure. The gene expression profiling datasets for normal lung, plasma cells and lymphocytes, and lymphoid neoplasms other than MALT lymphoma were obtained from previously published, publicly available sources as described in “Other gene expression datasets.”
Clustering of different lymphoid malignancies and normal tissues by hierarchical clustering. Enriched GO biologic process for each gene clusters are indicated on the left. The top T cell–associated cluster of genes are overexpressed in MALT and normal and malignant T cells, but not other B-cell tumors or normal lung. Genes of interest anchoring these gene expression profiles are indicated on the right. The colored bar below the heatmap represents the different tissue types, and the legend key is just below. On the heatmap, red represented up-regulated genes and green down-regulated genes, with the scale represented at the bottom of the figure. The gene expression profiling datasets for normal lung, plasma cells and lymphocytes, and lymphoid neoplasms other than MALT lymphoma were obtained from previously published, publicly available sources as described in “Other gene expression datasets.”
This, therefore, serves as a useful reference set to understand the molecular profile of MALT lymphoma. As our MALT lymphoma cases are derived from lung tissue, we have also included normal lung tissues in the dataset to identify lung-related signature. As expected, the molecular signature of MALT lymphoma is characterized by B-cell markers similar to other B-cell malignancies and normal B cells and to a lesser extent by plasma cell and immunoglobulin signature similar to plasma cell neoplasms and normal plasma cells (Figure 1). Consistent with their low-grade morphology and indolent behavior, they show decreased expression of proliferation genes, in particular compared with BL and DLBCL.
Significantly, MALT lymphomas express at high levels many of the genes highly expressed in T-cell lymphomas and normal T cells. As some of these genes are also expressed in DLBCL and to some extent by normal lung tissues and are absent in purified normal B- and T-cell subsets, they most likely reflect the stromal cells, connective tissue, and inflammatory cells seen in nonpurified tissue biopsy when tumor replacement is incomplete. The relatively lower expression of these genes in BL cases, where there is usually complete tumor infiltration, is consistent with this assumption. However, other genes in this group are overexpressed by MALT lymphomas, normal T cells, and T-cell lymphomas, but are relatively underexpressed in normal lung tissue and other B-cell malignancies (see Table S2 for a list of genes). These genes are associated with wound response, inflammatory response, and connective tissue as assessed by GO (Table S3).
Many of the lung MALT lymphomas also share a signature with normal lung tissues that is associated with lung development. The MALT cases that do not express this signature correspond to the tumors with the highest degree of tumor involvement with minimal lung tissue in the biopsy specimen (data not shown).
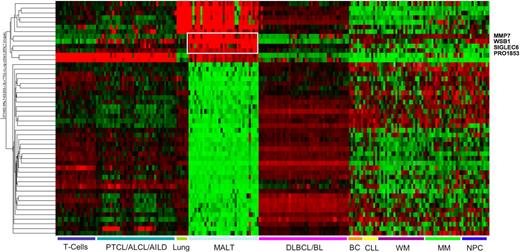
Fifty probes were differentially expressed between MALT lymphoma and all the other samples, 13 of which showed overexpression in MALT lymphoma (see Table S4). However, some of these were also highly expressed in T cells and normal lung tissue. Only 4, MMP7, SIGLEC6, WSB1, and PRO1853, are specifically overexpressed in MALT lymphoma. Thirty-seven genes including WNT1, MYCL1, and TCL1B are underexpressed in MALT lymphomas (Figure 2).
Heatmap representing the expression of genes with significantly different expression in MALT lymphoma compared with all the other lymphomas or normal tissues. The 4 genes overexpressed exclusively in MALT lymphoma are highlighted. The scaling of the heatmap is similar to Figure 1. The gene expression profiling datasets for normal lung and plasma cells and lymphoid neoplasms other than MALT lymphoma were obtained from previously published, publicly available sources as described in “Other gene expression datasets.”
Heatmap representing the expression of genes with significantly different expression in MALT lymphoma compared with all the other lymphomas or normal tissues. The 4 genes overexpressed exclusively in MALT lymphoma are highlighted. The scaling of the heatmap is similar to Figure 1. The gene expression profiling datasets for normal lung and plasma cells and lymphoid neoplasms other than MALT lymphoma were obtained from previously published, publicly available sources as described in “Other gene expression datasets.”
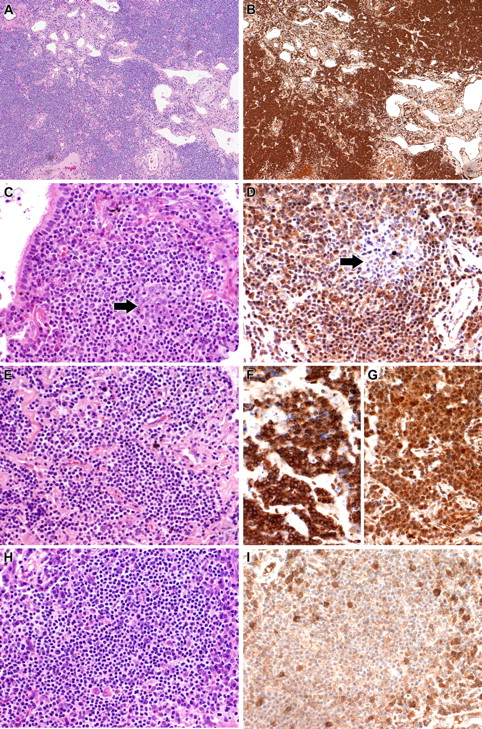
We validated 2 of the overexpressed transcripts for which antibodies for immunohistochemistry (IHC) are available, MMP7 and SIGLEC6. In tonsil, MMP7 was expressed by germinal center B cells, marginal zone B cells, and endothelial cells. Consistent with gene expression results, MMP7 was uniformly strongly expressed by all MALT lymphoma samples (Figure 3) independent of mRNA expression. In tonsil, SIGLEC6 was expressed weakly by germinal center B cells and the squamous epithelium. In MALT lymphomas, SIGLEC6 was expressed in all MALT lymphoma cases with varying degrees of intensity (Figure 3). Eight of the 11 cases showing highest level expression by GEP had strong expression of SIGLEC6 by IHC.
Validation of gene expression profiling by IHC. (A,B) Case 5 showing strong cytoplasmic expression of MMP7. (A) H&E; (B) MMP7. (C,D) Case 11 showing strong cytoplasmic expression of SIGLEC6 by the tumor cells. A reactive follicle ( ) included in the section is negative for SIGLEC6. (C) H&E; (D) SIGLEC6. (E-G) Case 14 with IGH/MALT1 translocation and spiked expression of RARA showing strong staining for MALT1 and RARA. (E) H&E; (F) MALT1 immunostain; (G) RARA. (H,I) Case 7, which lacked spiked RARA expression by gene expression profiling, is also negative for RARA by IHC. Plasma cells seen scattered through the tumor express RARA. (H) H&E; (I) RARA. All photographs were taken with a DP70 Olympus camera (Olympus, Tokyo, Japan) using an Olympus BX51 microscope (Olympus); images were acquired using DP Controller 2002 (Olympus) and processed using Adobe Photoshop version 7.0 (Adobe Systems, San Jose, CA). Original magnifications: ×100 (A,B), ×400 (C-I).
) included in the section is negative for SIGLEC6. (C) H&E; (D) SIGLEC6. (E-G) Case 14 with IGH/MALT1 translocation and spiked expression of RARA showing strong staining for MALT1 and RARA. (E) H&E; (F) MALT1 immunostain; (G) RARA. (H,I) Case 7, which lacked spiked RARA expression by gene expression profiling, is also negative for RARA by IHC. Plasma cells seen scattered through the tumor express RARA. (H) H&E; (I) RARA. All photographs were taken with a DP70 Olympus camera (Olympus, Tokyo, Japan) using an Olympus BX51 microscope (Olympus); images were acquired using DP Controller 2002 (Olympus) and processed using Adobe Photoshop version 7.0 (Adobe Systems, San Jose, CA). Original magnifications: ×100 (A,B), ×400 (C-I).
Validation of gene expression profiling by IHC. (A,B) Case 5 showing strong cytoplasmic expression of MMP7. (A) H&E; (B) MMP7. (C,D) Case 11 showing strong cytoplasmic expression of SIGLEC6 by the tumor cells. A reactive follicle ( ) included in the section is negative for SIGLEC6. (C) H&E; (D) SIGLEC6. (E-G) Case 14 with IGH/MALT1 translocation and spiked expression of RARA showing strong staining for MALT1 and RARA. (E) H&E; (F) MALT1 immunostain; (G) RARA. (H,I) Case 7, which lacked spiked RARA expression by gene expression profiling, is also negative for RARA by IHC. Plasma cells seen scattered through the tumor express RARA. (H) H&E; (I) RARA. All photographs were taken with a DP70 Olympus camera (Olympus, Tokyo, Japan) using an Olympus BX51 microscope (Olympus); images were acquired using DP Controller 2002 (Olympus) and processed using Adobe Photoshop version 7.0 (Adobe Systems, San Jose, CA). Original magnifications: ×100 (A,B), ×400 (C-I).
) included in the section is negative for SIGLEC6. (C) H&E; (D) SIGLEC6. (E-G) Case 14 with IGH/MALT1 translocation and spiked expression of RARA showing strong staining for MALT1 and RARA. (E) H&E; (F) MALT1 immunostain; (G) RARA. (H,I) Case 7, which lacked spiked RARA expression by gene expression profiling, is also negative for RARA by IHC. Plasma cells seen scattered through the tumor express RARA. (H) H&E; (I) RARA. All photographs were taken with a DP70 Olympus camera (Olympus, Tokyo, Japan) using an Olympus BX51 microscope (Olympus); images were acquired using DP Controller 2002 (Olympus) and processed using Adobe Photoshop version 7.0 (Adobe Systems, San Jose, CA). Original magnifications: ×100 (A,B), ×400 (C-I).
Cell of origin of MALT lymphoma
Comparison of B-cell lymphoma GEP with that of normal B-cell subsets showed that MALT lymphomas express genes associated with marginal zone B cells and memory B cells, with some overlap with mantle zone B cells, and underexpress genes associated with germinal center B cells. For example, germinal center differentiation markers CD10, BCL6, and AICDA were not expressed in MALT lymphoma but were expressed in DLBCL and BL, whereas genes such as CCL5, CCL6, CCR6, CXCR6, IFNGR1, and IL27RA that form the marginal zone/memory B-cell signature were overexpressed in MALT lymphoma but not expressed in DLBCL and BL. WM share some of the memory B-cell signature, partially overlapping with MALT lymphoma (Figure 4).
Expression of genes representing different B-cell compartment in different B-cell lymphomas. The scaling of the heatmap is the same as Figure 1. The gene expression profiling datasets for B-cell subsets were obtained from previously published, publicly available sources as described in “Other gene expression datasets.”
Expression of genes representing different B-cell compartment in different B-cell lymphomas. The scaling of the heatmap is the same as Figure 1. The gene expression profiling datasets for B-cell subsets were obtained from previously published, publicly available sources as described in “Other gene expression datasets.”
Molecular consequences of MALT1 translocations
Four hundred sixty-four probe sets were differentially expressed between MALT lymphomas with and without MALT1 translocations [t(11;18)/API2-MALT1, n = 10 and t(14;18)/IGH-MALT1, n = 3], with 290 up-regulated and 174 down-regulated in translocated samples. The most significantly up-regulated genes include MALT1, as expected, and also TRAF4 and TNFSF7; the most significantly down-regulated genes include CD64, IL6R, EPHA4, and XBP1 (see Table S5). The cases with t(14;18)(IGH/MALT1) had the highest expression of MALT1, consistent with higher transcription mediated by the IGH enhancer on chromosome 14 (Figure S1A). One case, t(11;18)(API2/MALT1)–positive by FISH, clustered with cases without MALT1 translocations. This tumor sample had a very low tumor fraction (20%), most likely accounting for the lack of signature associated with MALT1 translocations.
When the genes are analyzed in the context of interacting networks (see Document S1 “Network analysis”), the genes overexpressed in the MALT1 translocation-positive cases form a NF-KB network as expected. However, this is only part of a larger and more complex network comprising chemokine signaling mediated through the JAK-STAT and SRC pathways (Figure S1B). This is in agreement with the GO analysis, which showed that the genes up-regulated in tumors with MALT1 translocations were enriched for immune activation, NF-KB pathways, apoptosis, and protein kinase activity (Table S6). On the other hand, genes underexpressed in MALT1 translocation–positive cases are enriched for plasma cell–related genes and form a network involving JNK (Figure S1C). This is also in agreement with the GO analysis, which showed that genes down-regulated in tumors with MALT1 translocations were enriched for protein synthesis, MAPKKK, and JNK pathways (Table S7).
Molecular heterogeneity of pulmonary MALT lymphoma
Five hundred five genes were variably expressed among the MALT lymphoma samples. Initial clustering analysis showed that lung-related genes were highly expressed in samples with low tumor (and high normal lung) content, and genes on the Y chromosome were highly expressed in male patients (Figure S2). We therefore removed these 140 genes and repeated the unsupervised clustering (Figure 5). Most of the samples with MALT1 translocations (11 of 13) clustered together driven by a polyclonal immunoglobulin gene cluster that was associated with expression of plasma cell markers such as CD138 by GEP and the presence of a modest number of primarily polytypic plasma cells on histologic examination. Another cluster of samples (n = 7), all but 2 of which lacked MALT1 translocations, expressed CD138 but not the polyclonal immunoglobulin signature. Some of these patients had monoclonal immunoglobulins detected in their serum (data not shown), and in comparing the morphology and immunophenotype of the specimens that expressed CD138 but not the polyclonal immunoglobulin signature (n = 7) to those that did not (n = 28), the former group contained more prominent plasma cells (mean, 17% vs 5.5%) that were more likely to be light chain–restricted (50% vs 33%).
Unsupervised clustering of MALT lymphomas based on genes that are variably expressed across these tumors. The scaling of the heatmap is same as Figure 1.
Unsupervised clustering of MALT lymphomas based on genes that are variably expressed across these tumors. The scaling of the heatmap is same as Figure 1.
Analysis of genes with spiked expression in lung MALT lymphoma
Thirty probes representing 24 genes had spiked expression in at least one tumor sample (Table 2). The presence of spiked expression of duplicate probes as well as spiked expression of MALT1 in all 3 t(14;18)/(IGH/MALT1)–positive cases due to juxtaposition of the MALT1 gene on chromosome 18 to the IGH enhancer on chromosome 14 validates our approach. All 3 t(14;18)/(IGH/MALT1)–positive cases also showed strong cytoplasmic MALT1 expression by IHC (Figure 3). Interestingly, most of the spiked genes are more highly expressed than MALT1 is in the t(14;18)/IGH-MALT1 fusion–positive cases.
Probes and genes with spiked expression
| Probe ID . | Chromosome location . | Gene name . | Description . | Cases* . |
|---|---|---|---|---|
| 206121_at | 1p13 | AMPD1 | Adenosine monophosphate deaminase 1 (isoform M) | 2 |
| 205780_at | 22q13.31 | BIK | BCL2-interacting killer (apoptosis-inducing) | 5 |
| 208451_s_at | 6p21.3 | C4A /// C4B | Complement component 4A (Rodgers blood group) /// complement component 4B (Childo blood group) | 3 |
| 214428_x_at | 6p21.3 | C4A /// C4B | Complement component 4A (Rodgers blood group) /// complement component 4B (Childo blood group) | 3 |
| 226424_at | 19p13 | CAPS | Calcyphosine | 1 |
| 204606_at | 9p13 | CCL21 | Chemokine (C-C motif) ligand 21 | 4 |
| 218876_at | 16q22.1 | CGI-38 | Brain-specific protein | 7 |
| 208168_s_at | 1q31–32 | CHIT1 | Chitinase 1 (chitotriosidase) | 3 |
| 205229_s_at | 14q12-q13 | COCH | Coagulation factor C homolog, cochlin | 2 |
| 1554242_a_at | 14q12-q13 | COCH | Coagulation factor C homolog, cochlin | 2 |
| 229721_x_at | 22q11.3 | DERL3 | Der1-like domain family, member 3 | 3 |
| 219517_at | 15q15.3 | ELL3 | Elongation factor RNA polymerase II-like 3 | 2 |
| 219118_at | 12q13 | FKBP11 | FK506 binding protein 11, 19 kDa | 9 |
| 222067_x_at | 6p21.3 | HIST1H2BD | Histone 1, H2bd | 2 |
| 201656_at | 2q31.1 | ITGA6 | Integrin, α 6 | 4 |
| 216430_x_at | 15q14-q15 | SCGB2A2 | Secretoglobin, family 2A, member 2 | 9 |
| 205414_s_at | 17p12 | KIAA0672 | KIAA0672 gene product | 1 |
| 210018_x_at | 18q21 | MALT1 | Mucosa-associated lymphoid tissue lymphoma translocation gene 1 | 5 |
| 210017_at | 18q21 | MALT1 | Mucosa-associated lymphoid tissue lymphoma translocation gene 1 | 4 |
| 221286_s_at | 5q23-q31 | PACAP | Proapoptotic caspase adaptor protein | 6 |
| 201923_at | Xp22.11 | PRDX4 | Peroxiredoxin 4 | 3 |
| 223062_s_at | 9q21.2 | PSAT1 | Phosphoserine aminotransferase 1 | 5 |
| 206574_s_at | 8q24.3 | PTP4A3 | Protein tyrosine phosphatase type IVA, member 3 | 1 |
| 209695_at | 8q24.3 | PTP4A3 | Protein tyrosine phosphatase type IVA, member 3 | 1 |
| 1565358_at | 17q21 | RARA | Retinoic acid receptor, α | 11 |
| 210258_at | 1q31.2 | RGS13 | Regulator of G-protein signaling 13 | 11 |
| 1568752_s_at | 1q31.2 | RGS13 | Regulator of G-protein signaling 13 | 11 |
| 244887_at | 1q31.2 | RGS13 | Regulator of G-protein signaling 13 | 10 |
| 205725_at | 11q12.3-q13.1 | SCGB1A1 | Secretoglobin, family 1A, member 1 (uteroglobin) | 7 |
| 227952_at | 4p16.3 | ZNF595 | Zinc finger protein 595 | 7 |
| Probe ID . | Chromosome location . | Gene name . | Description . | Cases* . |
|---|---|---|---|---|
| 206121_at | 1p13 | AMPD1 | Adenosine monophosphate deaminase 1 (isoform M) | 2 |
| 205780_at | 22q13.31 | BIK | BCL2-interacting killer (apoptosis-inducing) | 5 |
| 208451_s_at | 6p21.3 | C4A /// C4B | Complement component 4A (Rodgers blood group) /// complement component 4B (Childo blood group) | 3 |
| 214428_x_at | 6p21.3 | C4A /// C4B | Complement component 4A (Rodgers blood group) /// complement component 4B (Childo blood group) | 3 |
| 226424_at | 19p13 | CAPS | Calcyphosine | 1 |
| 204606_at | 9p13 | CCL21 | Chemokine (C-C motif) ligand 21 | 4 |
| 218876_at | 16q22.1 | CGI-38 | Brain-specific protein | 7 |
| 208168_s_at | 1q31–32 | CHIT1 | Chitinase 1 (chitotriosidase) | 3 |
| 205229_s_at | 14q12-q13 | COCH | Coagulation factor C homolog, cochlin | 2 |
| 1554242_a_at | 14q12-q13 | COCH | Coagulation factor C homolog, cochlin | 2 |
| 229721_x_at | 22q11.3 | DERL3 | Der1-like domain family, member 3 | 3 |
| 219517_at | 15q15.3 | ELL3 | Elongation factor RNA polymerase II-like 3 | 2 |
| 219118_at | 12q13 | FKBP11 | FK506 binding protein 11, 19 kDa | 9 |
| 222067_x_at | 6p21.3 | HIST1H2BD | Histone 1, H2bd | 2 |
| 201656_at | 2q31.1 | ITGA6 | Integrin, α 6 | 4 |
| 216430_x_at | 15q14-q15 | SCGB2A2 | Secretoglobin, family 2A, member 2 | 9 |
| 205414_s_at | 17p12 | KIAA0672 | KIAA0672 gene product | 1 |
| 210018_x_at | 18q21 | MALT1 | Mucosa-associated lymphoid tissue lymphoma translocation gene 1 | 5 |
| 210017_at | 18q21 | MALT1 | Mucosa-associated lymphoid tissue lymphoma translocation gene 1 | 4 |
| 221286_s_at | 5q23-q31 | PACAP | Proapoptotic caspase adaptor protein | 6 |
| 201923_at | Xp22.11 | PRDX4 | Peroxiredoxin 4 | 3 |
| 223062_s_at | 9q21.2 | PSAT1 | Phosphoserine aminotransferase 1 | 5 |
| 206574_s_at | 8q24.3 | PTP4A3 | Protein tyrosine phosphatase type IVA, member 3 | 1 |
| 209695_at | 8q24.3 | PTP4A3 | Protein tyrosine phosphatase type IVA, member 3 | 1 |
| 1565358_at | 17q21 | RARA | Retinoic acid receptor, α | 11 |
| 210258_at | 1q31.2 | RGS13 | Regulator of G-protein signaling 13 | 11 |
| 1568752_s_at | 1q31.2 | RGS13 | Regulator of G-protein signaling 13 | 11 |
| 244887_at | 1q31.2 | RGS13 | Regulator of G-protein signaling 13 | 10 |
| 205725_at | 11q12.3-q13.1 | SCGB1A1 | Secretoglobin, family 1A, member 1 (uteroglobin) | 7 |
| 227952_at | 4p16.3 | ZNF595 | Zinc finger protein 595 | 7 |
Number of cases showing spiked expression of a given gene.
The genes with the most recurrently spiked expression are RARA and RGS13. By interphase FISH, all cases lacked a translocation involving RARA, and all cases except the t(14;18)/IGH-MALT1 fusion–positive cases lacked an IGH translocation. Therefore, other mechanisms must be involved in the spiked expression of RARA. By IHC in normal tonsil tissue, RARA was expressed by most lymphocytes whereas RGS13 expression was limited to germinal center B cells. In MALT lymphomas, there was variable expression of RARA and 7 of the 11 cases with spiked expression of RARA by GEP expressed high level of the protein by IHC (Figure 3). In contrast, RGS13 was weakly expressed by the neoplastic lymphocytes but strongly expressed by reactive germinal centers trapped within the neoplastic infiltrate (Figure 3).
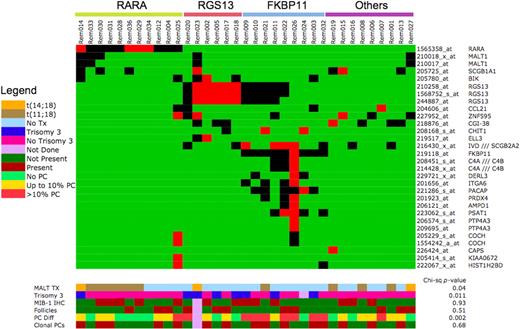
Interestingly, there was segregation of these spiked genes according to MALT1 translocation status. Spiked expression of RARA is seen mainly in tumors with MALT1 translocations (RARA group, n = 11), and spiked expression of RGS13, FKBP11, CCL21, PTP4A3, and ITGA6 mainly in those without (Figure 6). In addition, those tumors with spiked expression of RGS13 can be further divided into those with and without spiked expression of FKBP11 (FKBP11 and RGS13 groups, respectively). When these genes are analyzed in the context of interacting networks, the same segregation was observed with 2 networks, one involving G-protein signaling (RGS13 and PTP4A3) and receptor signaling (ITGA6) and the other involving NF-KB signaling (MALT1) and apoptosis pathway (BLK), intersecting through C-SRC (Figure S3).
Heatmap representing genes with spiked expression in MALT lymphoma. The color scale for gene expression is set to highlight cases with high expression of these genes. Genes that are expressed less than 4-fold higher than median are indicated in green, those that are expressed between 4- to 10-fold higher than median are indicated in black, whereas those that are expressed at greater than 10-fold higher than median are in red. The legend for the color bars indicating different pathologic features is presented in the figure.
Heatmap representing genes with spiked expression in MALT lymphoma. The color scale for gene expression is set to highlight cases with high expression of these genes. Genes that are expressed less than 4-fold higher than median are indicated in green, those that are expressed between 4- to 10-fold higher than median are indicated in black, whereas those that are expressed at greater than 10-fold higher than median are in red. The legend for the color bars indicating different pathologic features is presented in the figure.
We further dissected the gene expression profiles to understand the functional context of the molecular differences between the RARA, FKBP11, and RGS13 groups using gene set enrichment analysis (GSEA; see Document S1). Interestingly, the RGS13 group (n = 6) is enriched for genes located on chromosome 3, genes shown to be associated with proliferation, cell cycle, and poor prognosis in several tumor types (myeloma, breast cancer), and genes that are transcriptional targets of PAX5 (Table S8). The FKBP11 group (n = 9) is enriched for gene sets associated with plasma cell differentiation and genes that are transcriptional targets of XBP1 (Table S8). When we correlated these molecularly defined subgroups with available pathologic and genetic information, we found that the RARA group is enriched for MALT1 translocations (7/11 cases), the RGS13 group is enriched for trisomy 3 (4/6 cases) and prominent reactive follicles, and all cases but one of the FKBP11 group have increased plasma cells (8/9 cases), more than half (5/8 cases) of which are monotypic (Figure 6). In tumors with clonal plasma cells, both the B cells and plasma cells expressed the same light chain.
Discussion
This is the first comprehensive GEP study of MALT lymphoma. The only other study we are aware of was based on a cDNA platform using a limited number of genes and focused on comparison of low and high grade components of MALT lymphoma rather than the biology.22 Our study revealed several important insights into the biology of MALT lymphoma that may have diagnostic and therapeutic applications.
In contrast to other B-cell malignancies, MALT lymphomas have a prominent T-cell signature. The GO analysis of T-cell related genes overexpressed in lung MALT lymphomas show they are enriched for genes involved in T cell–mediated immune response and response to pathogens supporting an infectious or autoimmune etiology driven by T-cell help. This is consistent with the hypothesis that chronic antigenic stimulation either through infection or autoantigens in autoimmune conditions, leading to a T-dependent immune response is integral to MALT lymphoma pathogenesis.23 Although many infections and autoimmune conditions have been associated with MALT lymphomas affecting other sites, no infective agents or predisposing autoimmune conditions have been implicated for lung MALT lymphoma. It is therefore important to search for potential etiologic agents, in particular for infectious agents, using advanced technologies that have become available.24
Although very few genes are uniquely expressed by MALT lymphomas, MMP7 and SIGLEC6 were highly expressed in MALT lymphoma in comparison to other lymphoid malignancies, lung tissues, and normal lymphoid cells included in the study. MMP7 and SIGLEC6 expression was verified in MALT lymphoma at the protein level using IHC, with MMP7 expression being particularly uniformly strong in all cases. Although these markers may be developed as diagnostic tests for MALT lymphoma, the comparative GEP analysis was limited to diffuse large B-cell lymphoma, Burkitt lymphoma, and lymphoplasmacytic lymphoma categories. Further studies on larger set of clinical specimens will be required to establish daily diagnostic use. Nevertheless these findings may have important biologic consequences. Matrix metalloproteinases (MMPs) promote tumor progression through multiple mechanisms including extracellular matrix degradation, but also through signaling function relating to control of apoptosis, angiogenesis, and innate immunity.25 MMP7 is up-regulated through combined activation of transcription factors PEA3, c-JUN, β-catenin, and LEF-1 all downstream of classical oncogenes,26 and the MMP7 protein (also known as matrilysin) is secreted by tumor cells. MMP7 decreases cancer cell apoptosis by cleaving and shedding FAS ligand and cleaving proheparin-binding epidermal growth factor (pro-HB-EGF) to generate mature HB-EGF, which promotes cell survival by stimulating ERBB4.27 Of note, MMP7 has been mainly implicated in solid tumors and not hematologic malignancies. SIGLECs (sialic acid binding immunoglobulin-like lectins) are I-type lectins characterized by an N-terminal V-set immunoglobulin domain that mediates sialic acid binding followed by varying numbers of C2-set immunoglobulin domains. SIGLEC6 is involved in regulating cellular activation within the immune system.28 Functional studies of a related SIGLEC, SIGLEC7, have shown that it is an inhibitory NK-cell receptor,29 and activation leads to reduced cell growth30 and prevention of dendritic cell development.31 In addition, MMP7 and SIGLEC6 may be important in the homing of these lymphoid cells to the MALT. Whether these proteins have a role in the pathogenesis of MALT lymphoma will need to be clarified. If they do, targeting them may represent a MALT-specific therapeutic approach. In the case of MMP7, inhibitors are already available.32
Based on histologic features, immunophenotype and evidence for somatic hypermutation and antigen selection in the immunoglobulin heavy chain genes, it has been long speculated that MALT lymphomas are of marginal zone B-cell origin and have functional characteristics of memory B cells.33,34 Our analysis confirms the marginal zone and memory B-cell origin of MALT lymphoma at the global GEP level. MALT lymphomas lacked expression of genes highly expressed in germinal center B cells, but showed up-regulation of genes overexpressed by marginal zone and/or memory B cells. Interestingly, a subset of genes up-regulated in mantle zone B cells, such as CCL20 or CD36, were also expressed by MALT lymphomas. This is likely to reflect signal from residual mantle zones found in many MALT lymphomas.
Gastric MALT lymphomas with MALT1 translocations fail to respond to Helicobacter pylori eradication, suggesting that these translocations confer autonomous growth negating the need for continued H pylori–mediated help. Therefore, identification of pathways altered by MALT1 translocations is necessary to develop alternative targeted management strategies. In vitro studies have shown that the downstream effect of MALT1 translocations is the constitutive activation of the canonical NF-KB signaling pathway. However, this view is linear and limited. The use of global transcription profiling and network analysis allows us to analyze possible interacting networks that are affected as a result of these translocations. As expected, the NF-KB signaling network was identified among genes up-regulated in tumors with these translocations. However, interacting with the NF-KB network are larger networks involving the JAK-STAT networks and chemokine signaling via G-coupled proteins. These pathways are therefore possible sources of therapeutic intervention either individually or in concert.
The main heterogeneity within MALT lymphomas is driven by the presence or absence of translocations involving MALT1. Interestingly, there is a strong association between samples lacking these translocations and expression of plasma cell–related and polyclonal immunoglobulin genes. Increased plasma cells, which are sometimes monoclonal, are observed in some histologic specimens of MALT lymphoma. We showed for the first time that a plasmacytic component is predominantly seen in tumors without t(11;18)/API2-MALT1 or t(14;18)/IGH/MALT1. This is consistent with our findings that genes relatively down-regulated in tumors with these translocations are enriched for plasma cell–related genes and genes involved in protein synthesis in both the GO as well as the network analysis. This observation is also consistent with the concept that these translocations negate the need for an immunologically mediated oncogenic process, whereas plasma cells form part of the infiltrate together with T cells that presumably are required to maintain the oncogenic process in tumors without translocations. We also showed that in some tumors, more frequently the MALT1 translocation-negative cases, the plasma cells are clonal and share the same light chain restriction as the malignant B-cell clone, suggesting that in some MALT lymphomas, the clonal B cells are capable of terminal differentiation into plasma cells akin to the malignant B cells in WM.
The heterogeneity among MALT lymphomas can be further dissected by the expression pattern of genes with spiked expression. Tumors segregate based on the spiked expression of RARA, RGS13, or FKBP11. In silico functional analysis using GSEA reveals unique phenotypic associations for each molecular subgroup, which were verified on the histologic specimens. The differentiating characteristics include genetics (MALT1 translocations and trisomy 3) and histologic features (plasma cell percentage and clonality). As heterogeneity within MALT is anchored by deregulated gene expression, these may represent good therapeutic targets for each subtype.
Investigation by FISH showed that the spiked expression of RARA is not due to genetic alterations of the gene locus by translocations or amplifications. The spiked expression of RARA may therefore be due to juxtaposition of the gene to other strong regulatory element other than promoter elements on IgH, as have been shown in prostate cancer recently,15,35 or transmechanism.
RARA encodes the retinoic acid receptor α and is best known as one of the translocation partners in the t(15;17) in acute promyelocytic leukemia. Retinoic acid (RA) through RARA signaling plays an important role in hematopoiesis including regulation of lymphopoiesis.36 Interestingly, RA inhibits CD40- and interleukin-4 (IL-4)–induced proliferation of mantle cell lymphoma cell lines.37 As CD40-mediated signals are also implicated in proliferation of MALT lymphoma cells,22 targeting RARA with RA may be a potential therapeutic strategy in MALT lymphoma.
RGS13 is a member of the regulator G-protein signaling protein family.38 It is expressed by germinal center B cells and regulates B-cell responsiveness to chemokines such as CXCL12 and CXCL13.39 In mast cells, RGS13 modulates the signaling from Fc epsilon receptor I (FcERI).40 Interestingly, FcERI signaling in mast cells uses BCL10 and MALT1 pathway to activate NF-KB.41 Whether RGS13 has a regulatory role in BCL10/MALT1 pathway and NF-KB activation in B cells and lymphomas remains unknown.
To maximize the uniformity of our dataset, our analysis is restricted to primary MALT lymphomas arising from a single anatomic site. However, we believe our observations are applicable to MALT lymphoma affecting all extranodal sites, as we were able to dissect out signatures relating to the anatomical site of the biopsy specimen. It is generally thought that the pathophysiology and biology of MALT are similar regardless of involved sites.
In conclusion, we have identified new potential diagnostic markers, novel deregulated pathways, and potential new therapeutic targets. In addition, subgroups with distinct pathologic features and anchored by unique patterns of spike gene expression were identified. In the process, the importance of T cell-mediated chemokine signaling and NF-KB activation is highlighted.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by a University of Iowa/Mayo Clinic Lymphoma SPORE grant from the National Cancer Institute/National Institutes of Health (NCI/NIH; CA097274).
National Institutes of Health
Authorship
Contribution: W.J.C. analyzed data and wrote the paper; E.D.R. designed and performed research, analyzed data, and wrote the paper; R.F. and P.L.B. contributed vital analytical tools and analyzed data; J.A.V. performed research; P.J.K. designed research and analyzed data; and A.D. designed and performed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ahmet Dogan, Department of Laboratory Medicine and Pathology, Mayo Clinic, 200 First Street SW, Rochester, MN 55905; e-mail: dogan.ahmet@mayo.edu.
References
Author notes
*W.J.C. and E.D.R. contributed equally to this work.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal