Abstract
Waldenstrom macroglobulinemia (WM) is an incurable low-grade lymphoma characterized by bone marrow (BM) involvement of IgM secreting lymphoplasmacytic cells. The induction of unfolded protein response (UPR) genes (“physiologic” UPR) enables cells to differentiate into professional secretory cells capable of production of high amounts of endoplasmic reticulum (ER)–processed proteins, such as immunoglobulins. Ultimately, the initially cytoprotective UPR triggers an apoptotic cascade if ER stress is not corrected, called proapoptotic/terminal UPR. We show that WM cells inherently express the physiologic UPR machinery compared with normal BM cells, and that increased ER stress leads to proapoptotic/terminal UPR in WM cells. We therefore examined tunicamycin, ER stress inducer, for potential antitumor effects in WM. Tunicamycin induced significant cytotoxicity, apoptosis and cell-cycle arrest, and inhibited DNA synthesis in WM cell lines and primary BM CD19+ cells from patients with WM with an inhibitory concentration (IC50) of 0.5 μg/mL to 1 μg/mL, but not in healthy donor cells. Importantly, coculture of WM cells in the context of the BM microenvironment did not inhibit tunicamycin-induced cytotoxicity. Finally, we demonstrate that ER stress inducer synergizes with other agents used in the treatment of WM. These preclinical studies provide a framework for further evaluation of ER stress inducing agents as therapeutic agents in WM.
Introduction
The ER is the site where integral proteins and secretory proteins are folded into their tertiary structure, and multimeric proteins, such as immunoglobulins, are assembled.1,2 To survive under endoplasmic reticulum (ER) stress, eukaryotic cells have a self-protective mechanism against ER stress, termed the unfolded protein response (UPR).3,4 The specific induction of UPR genes enables cells to differentiate into professional secretory cells capable of tolerating the constitutive production of high amounts of ER-processed proteins, a process termed the “physiologic” UPR. The UPR maintains the quality of newly synthesized secretory and transmembrane proteins such as immunoglobulins, and is distinct from the “ER stress” or “terminal/proapoptotic” UPR, which is induced by nutrient deprivation or chemical agents that cause severe or prolonged ER stress, and triggers an apoptotic cascade called proapoptotic/terminal UPR.5-7 Several studies reveal a critical role for UPR activation for tumor cell resistance to hypoxia and tumor growth promotion, and suggest that the UPR may be an attractive target for antitumor modalities. It is therefore reasonable to assume that manipulation of ER stress might enhance the efficacy of chemotherapeutic drugs and provide new anticancer targets. So far, data support the potential of drugs that inhibit the normal functions and homeostasis of the ER in treatment of malignancies, and has lead to the development of heat shock protein 90 inhibitors.8,9 Interestingly, another novel agent, the proteasome inhibitor bortezomib, has been shown to target the ER function, by inducing components of the proapoptotic/terminal UPR.10
Waldenstrom macroglobulinemia is a low-grade lymphoma characterized by the presence of lymphoplasmacytic cells in the bone marrow (BM) and a serum monoclonal IgM in the circulation.11-13 Although indolent, it remains incurable with a median overall survival of 5 to 6 years. There is therefore an urgent need for the rational development of novel agents that target aberrant molecular pathways in WM.
We hypothezised that by increasing ER stress by use of tunicamycin, this would lead to proapoptotic/terminal UPR and therefore set the first step of a novel targeted therapy in WM. In this study, we show that WM cells inherently expressed the physiologic UPR machinery, and that increased ER stress leads to proapoptotic/terminal UPR in WM cells. Tunicamycin also induced significant antitumor activity on WM cells in vitro that provide the framework for further evaluation of ER stress inducing agents as therapeutic agents in WM.
Methods
Cells
The WM cell lines BCWM.114 and WSU-WM (from Dr Al Khatib, Wayne State University, Detroit, MI) and IgM secreting low-grade lymphoma cell lines MEC1 (DMSZ, Braunschweig, Germany) and RL (ATCC, Manassas, VA) were used in this study. All cell lines were cultured in RPMI-1640 containing 10% fetal bovine serum (FBS; Sigma-Aldrich, St Louis, MO), 2 μM l-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (GIBCO, Grand Island, NY).
Primary WM cells were obtained from BM samples using CD19+ microbead selection (Miltenyi Biotec, Auburn, CA) with over 90% purity, as confirmed by flow cytometric analysis with monoclonal antibody reactive to human CD20-PE (BD Biosciences, San Jose, CA). Peripheral blood and BM CD19+-selected cells were obtained from healthy volunteers and used in comparison for some experiments (All Cells, Berkeley, CA). Patient samples were obtained after approval from the Dana-Farber Cancer Institute Institutional Review Board. Informed consent was obtained from all patients in accordance with the Declaration of Helsinki protocol.
Reagents
Tunicamycin is an ER-stress inducer that inhibits N-linked glycosylation of lipids and tyrosine incorporation, which ultimately inhibit protein synthesis (Sigma-Aldrich). Bortezomib was obtained from Millennium Pharmaceuticals (Cambridge, MA). Dexamethasone and fludarabine were purchased from Sigma-Aldrich. The following cytokines, CD40L, BLyS, IL-6, and IGF1, were purchased from R&D Systems (Minneapolis, MN). Salubrinal was purchased from Axxora (San Diego, CA). The pan-caspase inhibitor Z-VAD-fmk was purchased from Promega (Madison, WI).
Cytotoxicity and DNA synthesis assays
Effect of tunicamycin on paracrine WM cell growth in the context of the BM microenvironment
To evaluate growth stimulation and signaling in WM cells adherent to bone marrow stromal cells (BMSCs), 3 × 104 BCWM.1 cells were cultured in BMSC-coated (ratio 3:1) 96-well plates for 48 hours in the presence or absence of tunicamycin in RPMI media reconstituted. BMSCs were cultured in Dulbecco modified Eagle medium (DMEM) reconstituted overnight for 24 hours before addition to BCWM.1 cells. [3H]-thymidine uptake was added for the last 8 hours and growth and proliferation were studied by measurement of DNA synthesis at 48 hours as described.16 In this experiment, the growth of both the BMSCs and tumor cells was measured, although as shown in the control conditions the growth of BMSCs is very slow and therefore does not affect the study of the growth of tumor cells.
Flow cytometric analysis
Cell-cycle analysis was profiled by flow cytometry using propidium iodide (PI) staining (Sigma-Aldrich), as described.15 Apoptosis was quantitated using Apo2.7 analysis (Beckman Coulter, Fullerton, CA), as described.15 Flow cytometric analysis was determined using an Epics flow cytometer (Coulter Immunology, Hialeah, FL).
Effect of tunicamycin on IgM secretion by BCWM.1 cells
Culture supernatant was harvested after overnight and 24 hours, and IgM concentration in the supernatant was determined by enzyme-linked immunosorbent assay (ELISA; Bethyl, Montgomery, TX), following manufacturer recommendations.
Colony-forming cell (CFC) assay
CFC assays were processed with nonadherent mononuclear fraction from BM of healthy volunteers using commercially available methylcellulose for human colony-forming cell assays (MethoCult; StemCell Technologies, Vancouver, BC), as described.15 Erythroid burst-forming units (BFU-Es), granulocyte macrophage–colony-forming units (CFU-GMs), macrophage colony-forming units (CFU-Ms), and granulocyte, erythroid, macrophage, megakaryocyte–colony-forming units (CFU-GEMMs) were counted at days 14 to 16.
Immunoblotting
BCWM.1 WM cells were lysed using lysis buffer (Cell Signaling Technology, Beverly, MA). The antibodies used for immunoblotting included: -p27kip-1, -p21waf-1/cip-1, -CDK2, -p-CDK2 (thr160), -CDK4, -CDK6, p53, -p-Rb (ser807/811), -HSP90, -HSP70, -p-HSP27, -HSP27, -caspase-8, -caspase-9, -caspase-3, -PARP, -Mcl-1, -p-BCL2 (Cell Signaling Technology); -ATF6 (Abcam, Cambridge, MA); -p-PERK (BioLegend, San Diego, CA); -α-tubulin, -EDEM, -GADD153/CHOP, -GRP78/BiP (Santa Cruz Biotechnology, Santa Cruz, CA); and caspase-4 and caspase-12 (BD Biosciences).
Quantitative RT-PCR analysis
Expression of human PERK, GADD34, CHOP/GADD153, Atf6, GRP78/BiP, Xbp1, IRE1α, and EDEM transcripts were determined using real-time quantitative reverse transcriptase–polymerase chain reaction (qPCR) based on TaqMan fluorescence methodology and expression of Xbp1 splicing was studied using SYBR green technique, following manufacturer protocols (Applied Biosystems, Foster City, CA). Applied Biosystems designed the TaqMan primer-probe set for those genes (Assays on Demand Gene Expression Product, PN4331182); PERK: Hs 00178128_ml; GADD34: Hs 00169585_ml; Chop/GADD153: Hs 00358796_gl; ATF6: Hs 00232586_ml; GRP78/BiP: Hs 00607129_gh; Xbp1: Hs 00231936_ml; IRE1α: Hs 00176385_ml; EDEM: Hs 00206467_ml. The primers used for Xbp1 splicing were previously published.3,17 Human 18S rRNA (Applied Biosystems, PN430832) was used as the endogenous control gene. PCRs were performed in the 7500 Real-Time PCR System (Applied Biosystems). All reactions were performed in triplicate and included a negative control. The relative level of target gene was normalized to the endogenous reference gene (18S rRNA) and determined using the ΔΔCT method.
Statistical analysis
Statistical significance of mean differences was determined using the Mann-Whitney U test. The interaction between tunicamycin with other therapy was analyzed using the CalcuSyn software program (Biosoft, Ferguson, MO) to determine whether the combinations were synergistic (CI < 0.8).15 The program calculates the combination index (CI) and determines the affected fraction (Fa) of cells for each combination, and provides isobologram that illustrates the effect of each combination.
Results
Baseline expression of “prosurvival/physiologic” UPR genes is constitutively elevated in patients with WM
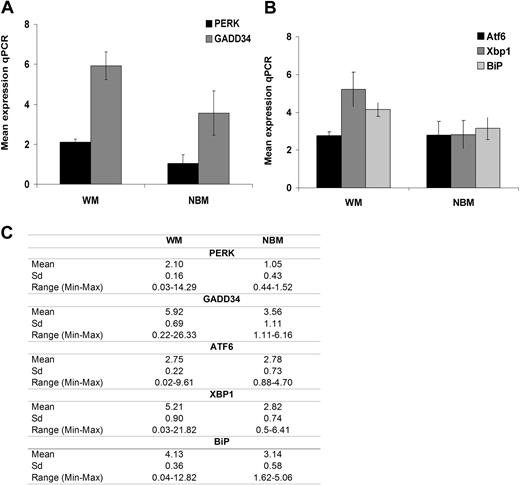
We first sought to determine the baseline level of prosurvival/physiologic UPR genes in WM cells. We studied UPR gene expression in CD19+ BM–selected cells from 16 patients with WM and compared these expressions with CD19+ BM–selected cells from 4 healthy donors (NBM). PERK (PKR-like ER kinase) is an ER transmembrane protein kinase that phosphorylates the translation initiation factor 2 in response to ER stress, leading to general translational attenuation. This translational control provides an efficient mechanism to reduce the burden of newly synthesized proteins to be folded by the ER.18,19 GADD34 (growth arrest and DNA damage inducible protein) transcription is induced to reverse the translational attenuation. As shown in Figure 1A, expression of PERK (P = .055) and GADD34 (P = .03) transcript expression was increased in patients with WM compared with NBM samples.
Baseline physiologic/prosurvival UPR gene expression in WM disease. UPR gene expression in BM CD19+ WM cells (N = 16) was compared with BM CD19+ cells from healthy donors (NBM, N = 4) using relative quantitative RT-PCR (qPCR). Panels show mean gene expression as a normalized ratio over ribosomal 18S. (A) Expression of genes of the PERK pathway, including PERK and GADD34. (B) Expression of genes of the ATF6 pathway, including ATF6, GRP78/BiP, and Xbp1. All data represent mean plus or minus SD of triplicate experiments. (C) Panel that summarizes the mean values for the transcript levels with SD and range.
Baseline physiologic/prosurvival UPR gene expression in WM disease. UPR gene expression in BM CD19+ WM cells (N = 16) was compared with BM CD19+ cells from healthy donors (NBM, N = 4) using relative quantitative RT-PCR (qPCR). Panels show mean gene expression as a normalized ratio over ribosomal 18S. (A) Expression of genes of the PERK pathway, including PERK and GADD34. (B) Expression of genes of the ATF6 pathway, including ATF6, GRP78/BiP, and Xbp1. All data represent mean plus or minus SD of triplicate experiments. (C) Panel that summarizes the mean values for the transcript levels with SD and range.
This increased expression of prosurvival/physiologic UPR genes was also found in the ATF6 (activating transcription factor 6) pathway that controls the transcriptional induction of ER resident molecular chaperones to increase the folding capacity of the ER, including BiP (immunoglobulin heavy chain–binding protein)/GRP78 (glucose regulated protein) and Xbp1 (X-box binding protein). Although ATF6 gene expression was similar in WM compared with NBM CD19+ cells, expression of Xbp1 (P = .03) and BiP (P = .047) genes was higher in WM than in NBM primary CD19+ cells (Figure 1B). The mean values with standard deviation (SD) and range are provided in Figure 1C.
WM cells demonstrate low levels of ER stress/proapoptotic terminal UPR gene expression at baseline
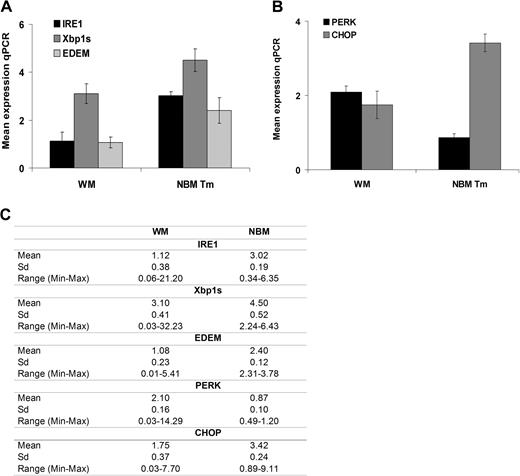
Under prolonged “stress,” the IRE1 (endoplasmic reticulum-to-nucleus signaling) pathway is activated and as a result, the initial cytoprotective UPR triggers an apoptotic cascade and facilitates cell death.20,21 This result in Xbp1 mRNA undergoes splicing (Xbp1s) at an IRE1α-specific cleavage site,17,22,23 while downstream EDEM regulates the extraction of misfolded proteins into the cytosol for proteasome-mediated destruction.24,25 As shown in Figure 2A, IRE1 (P = .03), Xbp1s (P = .045), and EDEM (P = .04) gene expression was lower in patients with WM than in tunicamycin-treated NBM samples. In addition, in response to prolonged ER stress, PERK induces cell-cycle arrest during G1 phase, and induction of GADD153/CHOP (C/EBP homologous protein), a proapoptotic transcription factor that potentiates apoptosis expression. We also observed significantly lower expression of CHOP in patient samples versus tunicamycin-treated NBM samples (P = .025; Figure 2B). The mean values with SD and range are provided in Figure 2C.
ER-stress/proapoptotic terminal UPR gene expression in WM. UPR gene expression was compared in BM CD19+ WM cells (N = 16) to BM CD19+ cells from healthy donors (NBM, N = 4) treated with tunicamycin (Tm; 10 μg/mL for 3 hours) using relative quantitative RT-PCR (qPCR). Panels show mean gene expression as a normalized ratio over ribosomal 18S. (A) Expression of genes of the IRE1 pathway, including IRE1, Xbp-1s, and EDEM. (B) Expression of genes of the PERK pathway, including PERK and GADD153/CHOP. All data represent mean plus or minus SD of triplicate experiments. (C) Panel that summarizes the mean values for the transcript levels with SD and range.
ER-stress/proapoptotic terminal UPR gene expression in WM. UPR gene expression was compared in BM CD19+ WM cells (N = 16) to BM CD19+ cells from healthy donors (NBM, N = 4) treated with tunicamycin (Tm; 10 μg/mL for 3 hours) using relative quantitative RT-PCR (qPCR). Panels show mean gene expression as a normalized ratio over ribosomal 18S. (A) Expression of genes of the IRE1 pathway, including IRE1, Xbp-1s, and EDEM. (B) Expression of genes of the PERK pathway, including PERK and GADD153/CHOP. All data represent mean plus or minus SD of triplicate experiments. (C) Panel that summarizes the mean values for the transcript levels with SD and range.
Taken together, those results confirm that constitutive increased expression of prosurvival/physiologic UPR genes, but not ER stress/proapoptotic terminal UPR components, are expressed in WM cells. These results correlate with some clinical presentation of WM, such as the large amounts of immunoglobulin secreted and that tumor cells are not undergoing cell death.26
Tunicamycin induces ER stress and promotes proapoptotic/terminal UPR gene expression in WM cells
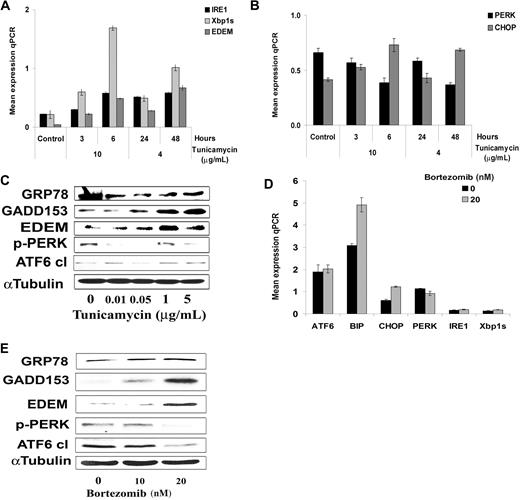
We next investigated whether sustained ER stress, using the known ER-stress inducer tunicamycin, resulted in prolonged protein synthesis inhibition and induction of proapoptotic/terminal UPR gene expression. We first explored ER stress gene expression in BCWM.1 cells treated with tunicamycin at different dose and time points. As showed in Figure 3A,B, tunicamycin (4 μg/mL at 24 and 48 hours, and 10 μg/mL at 3 and 6 hours) induced significant expression of genes that characterized the ER stress/“terminal” UPR in a time- and dose-dependent manner, such as IRE1, Xbp1s, EDEM, and CHOP. We then confirmed those results at the protein level in BCWM.1 cells, and found that BiP, CHOP, and EDEM protein expression increased in a dose-dependent manner at 6 hours, while PERK phosphorylation decreased at the same time point, and ATF6 expression did not vary much in that time point (Figure 3C). These results confirm that tunicamycin induces ER-stress and consequently proapoptotic/terminal UPR gene expression in BCWM.1 cells.
Tunicamycin induces terminal/proapoptotic ER stress. (A,B) Time- and dose-dependent induction of gene expression using relative quantitative RT-PCR (qPCR) in BCWM.1 cells treated with tunicamycin, 4 μg/mL for 24 and 48 hours and 10 μg/mL for 3 and 6 hours, respectively. All data represent mean plus or minus SD of triplicate experiments. (A) Expression of genes of the IRE1 pathway, including IRE1, Xbp-1s, and EDEM. (B) Expression of genes of the PERK pathway, including PERK and GADD153/CHOP. (C) Dose-dependent induction of terminal/proapoptotic ER stress proteins. BCWM.1 cells were cultured with tunicamycin (0.1 μg/mL to 5 μg/mL) for 6 hours. Whole-cell lysates were subjecting to Western blotting using anti-GADD153/CHOP, -GRP78/BiP, -ATF6, -p-PERK, -EDEM, and α-tubulin antibodies. (D,E) Bortezomib-induced ER stress in WM disease. (D) We studied expression at the transcriptional level of ATF6, GRP78/BiP, PERK, GADD153/CHOP, IRE1, and Xbp-1s following treatment of BCWM.1cells for 6 hours with 20 ng/mL bortezomib. (E) Induction of terminal/proapoptotic ER stress proteins was also studied in BCWM.1 cells cultured with 20 ng/mL bortezomib for 6 hours. Whole-cell lysates were subjected to Western blotting using anti-GADD153/CHOP, -GRP78/BiP, -ATF6, -p-PERK, -EDEM, and α-tubulin antibodies.
Tunicamycin induces terminal/proapoptotic ER stress. (A,B) Time- and dose-dependent induction of gene expression using relative quantitative RT-PCR (qPCR) in BCWM.1 cells treated with tunicamycin, 4 μg/mL for 24 and 48 hours and 10 μg/mL for 3 and 6 hours, respectively. All data represent mean plus or minus SD of triplicate experiments. (A) Expression of genes of the IRE1 pathway, including IRE1, Xbp-1s, and EDEM. (B) Expression of genes of the PERK pathway, including PERK and GADD153/CHOP. (C) Dose-dependent induction of terminal/proapoptotic ER stress proteins. BCWM.1 cells were cultured with tunicamycin (0.1 μg/mL to 5 μg/mL) for 6 hours. Whole-cell lysates were subjecting to Western blotting using anti-GADD153/CHOP, -GRP78/BiP, -ATF6, -p-PERK, -EDEM, and α-tubulin antibodies. (D,E) Bortezomib-induced ER stress in WM disease. (D) We studied expression at the transcriptional level of ATF6, GRP78/BiP, PERK, GADD153/CHOP, IRE1, and Xbp-1s following treatment of BCWM.1cells for 6 hours with 20 ng/mL bortezomib. (E) Induction of terminal/proapoptotic ER stress proteins was also studied in BCWM.1 cells cultured with 20 ng/mL bortezomib for 6 hours. Whole-cell lysates were subjected to Western blotting using anti-GADD153/CHOP, -GRP78/BiP, -ATF6, -p-PERK, -EDEM, and α-tubulin antibodies.
Since bortezomib is active in WM,27,28 and exerts its activity, at least in part, through induction of ER stress in myeloma,10,29 we next evaluated whether bortezomib (10 ng/mL and 20 ng/mL, 6 hours) also displayed its activity through induction of ER stress in WM. The results confirmed that bortezomib (20 ng/mL) induced ER stress gene expression in BCWM.1 cells at 6 hours (eg, genes that characterized the ER stress proapoptotic “terminal” UPR, IRE1, Xbp1s, and CHOP), similarly to tunicamycin (Figure 3D). We next studied the ER stress pathways at the protein level in BCWM.1 cells treated with bortezomib. We found that BiP, CHOP, and EDEM protein expression increased in a dose-dependent manner at 6 hours, while PERK phosphorylation along with cleaved ATF6 decreased in the same time point (Figure 3E).
Tunicamycin decreases proliferation and induces cytotoxicity of WM cells in a time- and dose-dependent fashion
We next investigated whether sustained ER stress using tunicamycin resulted in prolonged protein synthesis inhibition and decreased survival and induction of cell death. BCWM.1 cells were cultured in the presence of tunicamycin (0.01 μg/mL to 5 μg/mL) for 24 and 48 hours. Tunicamycin inhibited proliferation with an IC50 that varies from 0.05 to 0.1 μg/mL for BCWM.1 cells (Figure 4A) and WM-WSU WM cells and other IgM-secreting cell lines, RL and MEC1 (data not shown). Tunicamycin also induced cytotoxicity at 48 hours with an IC50 of 0.1 μg/mL in all cell lines tested including BCWM.1 cells (Figure 4B). The cytotoxic effect of tunicamycin was confirmed in primary tumor cells from patients with WM with an IC50 that varies from 0.05 μg/mL to 0.1 μg/mL for all primary samples tested (Figure 4C). In contrast, tunicamycin did not trigger cytotoxicity in peripheral blood CD19+ cells from 3 healthy volunteers at doses ranging from 0.01 μg/mL to 5 μg/mL (Figure 4D), and did not alter development of hematopoietic progenitor cells (data not shown). Salubrinal is a known ER stress protector that dephosphorylates eukaryotic translation initiation factor 2 subunit a (eIF2a).30,31 We next sought to determine whether tunicamycin induced cytotoxicity in the presence of ER stress protection. We observed that salubrinal did not rescue cells from tunicamycin-induced ER stress–mediated cytotoxicity (Figure 4E). We next studied whether IgM secretion in BCWM.1 culture supernatant decreases with tunicamycin treatment. As shown in Figure 4F, IgM secretion was decreased after 12 hours and 24 hours following treatment with tunicamycin at dose and time points (0.05 μg/mL and 0.1 μg/mL) that failed to induce cell death.
Tunicamycin induces cytotoxicity and decrease in proliferation. (A) Thymidine uptake assay. BCWM.1 WM cells were cultured with tunicamycin (0.01 μg/mL to 5 μg/mL) for 48 hours. (B) BCWM.1 cells were cultured with tunicamycin (0.01 μg/mL to 5 μg/mL) for 24 hours (△) and 48 hours (■), then cytotoxicity was studied. (C) Freshly isolated BM CD19+ cells from 3 patients with WM were cultured with tunicamycin (0.01 μg/mL to 1 μg/mL) for 48 hours. (D) Absence of cytotoxicity was observed on freshly isolated peripheral blood CD19+ from 3 healthy donors cultured with tunicamycin (0.01 μg/mL to 5 μg/mL) for 48 hours. Cytotoxicity was assessed by the MTT assay (B-D). (E) Salubrinal (50 μM and 75 μM) do not protect BCWM.1 cells from tunicamycin-induced (0.1 μg/mL and 1 μg/mL) cytotoxicity at 48 hours. (F) ELISA of BCWM.1 supernatant. IgM secretion was decreased after 12 hours and 24 hours of treatment with tunicamycin (0.05 μg/mL and 0.1 μg/mL).
Tunicamycin induces cytotoxicity and decrease in proliferation. (A) Thymidine uptake assay. BCWM.1 WM cells were cultured with tunicamycin (0.01 μg/mL to 5 μg/mL) for 48 hours. (B) BCWM.1 cells were cultured with tunicamycin (0.01 μg/mL to 5 μg/mL) for 24 hours (△) and 48 hours (■), then cytotoxicity was studied. (C) Freshly isolated BM CD19+ cells from 3 patients with WM were cultured with tunicamycin (0.01 μg/mL to 1 μg/mL) for 48 hours. (D) Absence of cytotoxicity was observed on freshly isolated peripheral blood CD19+ from 3 healthy donors cultured with tunicamycin (0.01 μg/mL to 5 μg/mL) for 48 hours. Cytotoxicity was assessed by the MTT assay (B-D). (E) Salubrinal (50 μM and 75 μM) do not protect BCWM.1 cells from tunicamycin-induced (0.1 μg/mL and 1 μg/mL) cytotoxicity at 48 hours. (F) ELISA of BCWM.1 supernatant. IgM secretion was decreased after 12 hours and 24 hours of treatment with tunicamycin (0.05 μg/mL and 0.1 μg/mL).
These results demonstrate that tunicamycin triggers significant cytotoxicity in WM and IgM-secreting cell lines, as well as in primary patient WM cells, and without toxicity to normal B cells. Moreover, tunicamycin decreased IgM secretion in WM cells.
Neither growth factors nor adherence to bone marrow stromal cells (BMSCs) protect against tunicamycin-induced WM cell cytotoxicity
Since the BM microenvironment confers tumor cell growth and drug resistance,15,16 we next studied whether tunicamycin could overcome the growth advantage and drug resistance conferred by the BM microenvironment to WM cells. BCWM.1 cells were cultured with tunicamycin (0.1 μg/mL to 1 μg/mL), in the presence or absence of BMSCs. Adherence of BCWM.1 cells to BMSCs triggered a 1.2-fold increase in the [3H]-thymidine uptake, and tunicamycin inhibited BCWM.1 cell growth in the context of the BM microenvironment in a dose-dependent fashion (P < .001; Figure 5A), confirming that tunicamycin retains significant antitumor activity even in the BM milieu.
Growth factors and coculture with BMSCs do not protect against tunicamycin-induced BCWM.1 cell cytotoxicity. (A) BCWM.1 cells were cultured with control media, and with tunicamycin (0.5 μg/mL to 1 μg/mL) for 48 hours, in the presence or absence of BMSCs. Cell proliferation was assessed using the [3H]-thymidine uptake assay. (B) BCWM.1 cells were cultured with control media in the presence or absence of IL-6 (25 ng/mL), CD40L (3 ng/mL), or BLyS (100 ng/mL), and treated with tunicamycin (0.1 μg/mL and 1 μg/mL) for 48 hours. Cytotoxicity was assessed by the MTT assay. All data represent mean plus or minus SD of triplicate experiments (A,B).
Growth factors and coculture with BMSCs do not protect against tunicamycin-induced BCWM.1 cell cytotoxicity. (A) BCWM.1 cells were cultured with control media, and with tunicamycin (0.5 μg/mL to 1 μg/mL) for 48 hours, in the presence or absence of BMSCs. Cell proliferation was assessed using the [3H]-thymidine uptake assay. (B) BCWM.1 cells were cultured with control media in the presence or absence of IL-6 (25 ng/mL), CD40L (3 ng/mL), or BLyS (100 ng/mL), and treated with tunicamycin (0.1 μg/mL and 1 μg/mL) for 48 hours. Cytotoxicity was assessed by the MTT assay. All data represent mean plus or minus SD of triplicate experiments (A,B).
Previous studies have demonstrated that IL-6, an important factor in development promoting plasmacytoid lymphocyte growth, is up-regulated in WM.32,33 Other cytokines and growth factors appear to promote WM tumor cell growth, such as BLyS (B lymphocyte stimulator)34 and CD40L (CD154).35 As shown in Figure 5B, IL-6 (100 ng/mL), BLyS (3 ng/mL), and CD40L (25 ng/mL) induced modest proliferation of BCWM.1 cells in about 10% to 25% increase at 48 hours, that was blocked by tunicamycin (0.1 μg/mL and 0.5 μg/mL). These results suggest that tunicamycin overcomes growth and drug resistance triggered by direct interaction with the BM microenvironment and by cytokines, such as IL-6, BLyS, and CD40L.
Tunicamycin induces caspase-dependent WM cell apoptosis
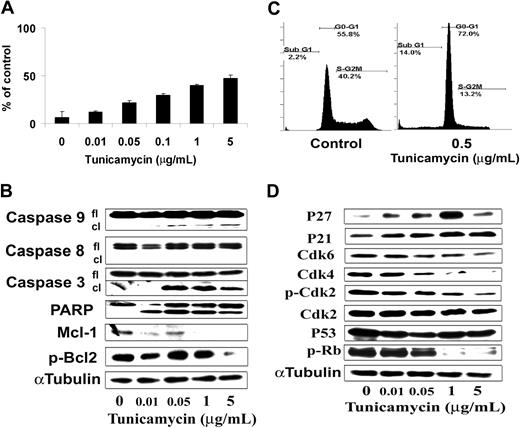
We next characterized the molecular mechanisms whereby tunicamycin induced cytotoxicity in BCWM.1 WM cells. Prolonged ER stress can be responsible for the activation of apoptosis through a mitochondrial-dependent and -independent pathway. We demonstrated that tunicamycin induced significant apoptosis as evidenced by positivity of Apo2.7 or propidium iodide staining by flow cytometric analysis. As shown in Figure 6A, tunicamycin (0.01 μg/mL to 5 μg/mL) induced dose-dependent apoptosis in BCWM.1- and IgM-secreting cell lines, with 0.5 μg/mL tunicamycin inducing 14% and 30% cell death in BCWM.1 cells at 24 hours and 48 hours, respectively (Figure 6A,C). The pan-caspase inhibitor Z-VAD-fmk (50 μM and 100 μM) did not inhibit tunicamycin-induced (0.1 μM and 1 μM) cytotoxicity at 48 hours (data not shown).36 To determine the mechanism of tunicamycin-induced apoptosis, we investigated the effect of tunicamycin on BCWM.1 cells by immunoblotting. The mitochondrial-independent pathway is thought to occur through initiator caspase-12. Activation of caspase-12 appears to be triggered only by various stimuli that activate ER stress. Caspase-12, once activated, directly cleaves caspase-9, activating it without the need for cytochrome c or Apaf-1, which in turn cleaves the effector caspase-3.37-39 Tunicamycin (0.05 μg/mL to 5 μg/mL) induced cleavage of caspase-9, caspases-3, and PARP in a dose-dependent fashion after overnight treatment (Figure 6B), but not of caspase-8, caspase-12, or caspase-4. We also noticed a decrease in the antiapoptotic molecules, p-BCL2 and Mcl-1 (Figure 6B). SAPK/JNK MAPK caspase-dependent cell apoptosis has already been demonstrated in B-cell malignancies40 and after ER stress induction.31 Tunicamycin-induced BCWM.1 cell apoptosis was accompanied with slight activation of SAPK/JNK at 6 hours using immunoblotting, and cytotoxicity triggered by tunicamycin (0.01 μg/mL and 0.1 μg/mL), which was blocked by the JNK1/2 inhibitor SP600125 (10 μM and 20 μM) at 48 hours (data not shown).
Tunicamycin induces apoptosis and cell-cycle arrest. (A) BCWM.1 cells were cultured with tunicamycin (0.01 μg/mL to 5 μg/mL) for 48 hours. Then the percentage of cells undergoing apoptosis was studied using positivity of Apo2.7 staining by flow cytometry. (B) Tunicamycin induces BCWM.1 WM cell apoptosis at 10 hours. BCWM.1 cells were cultured with tunicamycin (0.01 μg/mL to 5 μg/mL) then whole-cell lysates were subjecting to Western blotting using anti–caspase-9, –caspase-8, –caspase-3, -PARP, Mcl-1, p-Bcl2, and α-tubulin antibodies. (C) BCWM.1 cells were cultured without (control) and with tunicamycin (0.5 μg/mL) for 24 hours. Cell cycle was then studied using PI staining by flow cytometry. Percentages indicate cells in sub-G1 phase, G1 phase, and G2/M phase for 0.5 μg/mL tunicamycin. (D) BCWM.1 cells were cultured for 6 hours. Whole-cell lysates were subjecting to Western blotting using anti-CDK4, -CDK6, -CDK2, –p-CDK2, -p27, -p21, -p53, –p-Rb, and α-tubulin antibodies. All results represent mean plus or minus SD of triplicate experiments (A,C).
Tunicamycin induces apoptosis and cell-cycle arrest. (A) BCWM.1 cells were cultured with tunicamycin (0.01 μg/mL to 5 μg/mL) for 48 hours. Then the percentage of cells undergoing apoptosis was studied using positivity of Apo2.7 staining by flow cytometry. (B) Tunicamycin induces BCWM.1 WM cell apoptosis at 10 hours. BCWM.1 cells were cultured with tunicamycin (0.01 μg/mL to 5 μg/mL) then whole-cell lysates were subjecting to Western blotting using anti–caspase-9, –caspase-8, –caspase-3, -PARP, Mcl-1, p-Bcl2, and α-tubulin antibodies. (C) BCWM.1 cells were cultured without (control) and with tunicamycin (0.5 μg/mL) for 24 hours. Cell cycle was then studied using PI staining by flow cytometry. Percentages indicate cells in sub-G1 phase, G1 phase, and G2/M phase for 0.5 μg/mL tunicamycin. (D) BCWM.1 cells were cultured for 6 hours. Whole-cell lysates were subjecting to Western blotting using anti-CDK4, -CDK6, -CDK2, –p-CDK2, -p27, -p21, -p53, –p-Rb, and α-tubulin antibodies. All results represent mean plus or minus SD of triplicate experiments (A,C).
In addition, as shown in Figure 6C, tunicamycin (0.5 μg/mL) induced a G0/G1 arrest on cell-cycle analysis at 24 hours. We then studied by immunoblot the molecular pathways of cell-cycle G0/G1 arrest on BCWM.1 cells after overnight treatment with tunicamycin (0.5 μg/mL). Tunicamycin induced increased expression of p53 tumor suppressor protein as well as an increase of the expression of the cyclin-dependent kinase inhibitors p21waf-1/cip-1 and p27kip-1, in a dose-dependent fashion. These proteins are known to regulate cyclin-dependent kinase 2 (CDK2), CDK4, and CDK6,41 important for the G1-to-S transition of cell cycle through inactivation of retinoblastoma tumor suppressor protein (Rb).42 We observed that tunicamycin induced a decrease in CDK2 expression and phosphorylation on thr160 and an activation of Rb protein as shown by a decrease in Rb phosphorylation on ser807/811 on BCWM.1 cells (Figure 6D), and inhibition of cell-cycle progression.
Tunicamycin enhances cytotoxicity of other agents in WM disease
Guidelines on WM treatment either at relapse or frontline consider fludarabine and dexamethasone as major therapies in WM.13,43 However, there is a need to define the best combinations including those drugs and new agents since response rates, especially complete response rate, are limited when those agents are used alone. Since bortezomib is active in WM,27,28 we next evaluated the effects of tunicamycin in combination with bortezomib, as well as other drugs (fludarabine and dexamethasone), achievable on BCWM.1 cells.
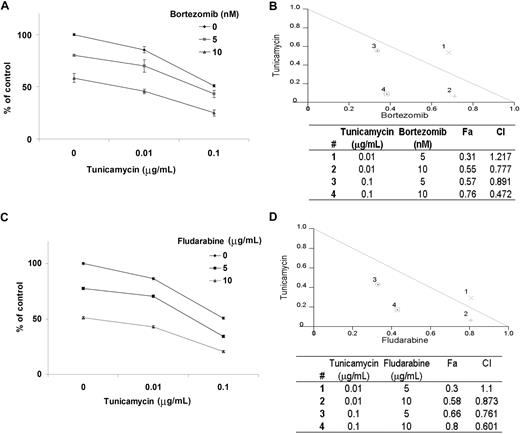
BCWM.1 cells were cultured for 48 hours with bortezomib (5 nM and 10 nM), tunicamycin (0.01 μg/mL to 0.1 μg/mL), or the combination, and cytotoxicity was measured using the MTT assay at 48 hours. As shown in Figure 7A, bortezomib-induced cytotoxicity was significantly increased by tunicamycin in a dose-dependent fashion. Tunicamycin 0.01 μg/mL and 0.1 μg/mL induced 25% and 50% cytotoxicity, which was augmented to 55% (CI = 0.7) and 76% (CI = 0.4) by bortezomib 10 nM, respectively, consistent with a synergistic effect (Figure 7B). Similarly, we studied the combination of tunicamycin (0.01 μg/mL and 0.1 μg/mL) with fludarabine (5 μg/mL and 10 μg/mL) in BCWM.1 cells at 48 hours using the MTT assay. As shown in Figure 7C, fludarabine 5 μg/mL and 10 μg/mL increased tunicamycin-induced (0.1 μg/mL) cytotoxicity to 66% (CI = 0.7) and 80% (CI = 0.6), also indicating synergism (Figure 7D). Lastly, we studied the effect of dexamethasone (50 nM and 100 nM) either alone or in combination with tunicamycin (0.01 μg/mL and 0.1 μg/mL), and observed no synergy with the combination with dexamethasone at 48 hours (data not shown).
Tunicamycin-induced cytotoxicity is enhanced in combination with other therapy. Cytotoxicity was measured using the MTT assay. All data represent mean plus or minus SD of triplicate experiments. (A,C) BCWM.1 cells were cultured with tunicamycin (0.01 μg/mL and 0.1 μg/mL) in the absence and presence of bortezomib (5 nM and 10 nM; A) and fludarabine (5 μg/mL and 10 μg/mL; C). (B,D) Normalized isobologram produced by the software Calcusyn. Values below the threshold line represent synergistic combination. Table shows affected fractions (Fa) and combination indexes (CI) with tunicamycin and other therapy. Numbers (#) correspond to those provided in the normalized isobologram. (B) BCWM.1 cells were cultured with either tunicamycin, bortezomib, or the combination for 48 hours. (D) BCWM.1 cells were cultured with tunicamycin, fludarabine, and the combination for 48 hours.
Tunicamycin-induced cytotoxicity is enhanced in combination with other therapy. Cytotoxicity was measured using the MTT assay. All data represent mean plus or minus SD of triplicate experiments. (A,C) BCWM.1 cells were cultured with tunicamycin (0.01 μg/mL and 0.1 μg/mL) in the absence and presence of bortezomib (5 nM and 10 nM; A) and fludarabine (5 μg/mL and 10 μg/mL; C). (B,D) Normalized isobologram produced by the software Calcusyn. Values below the threshold line represent synergistic combination. Table shows affected fractions (Fa) and combination indexes (CI) with tunicamycin and other therapy. Numbers (#) correspond to those provided in the normalized isobologram. (B) BCWM.1 cells were cultured with either tunicamycin, bortezomib, or the combination for 48 hours. (D) BCWM.1 cells were cultured with tunicamycin, fludarabine, and the combination for 48 hours.
Discussion
In this study, we demonstrated constitutive increased expression of prosurvival/physiologic UPR components, but not terminal/proapoptotic ER-stress components, which were also reported in other Ig-related malignancies such as multiple myeloma, also characterized by an excess amount of immunoglobulins in the serum of the patients.10
The ER stress or terminal/proapoptotic UPR is induced by nutrient deprivation or chemical agents that cause severe or prolonged ER stress.5,6 We therefore hypothesized that increased ER stress using tunicamycin, a potent ER stress inducer, would lead to proapoptotic/terminal UPR and consequently induce cytotoxicity and cell death, and enhance the efficacy of chemotherapeutic drugs in WM cells. We next demonstrated that tunicamycin, which blocks N-linked protein glycosylation, induces ER stress gene and protein expression in BCWM.1 WM cells. Others have demonstrated that bortezomib, a proteasome inhibitor, is active in WM and leads to rapid induction of terminal UPR components and ER stress–induced apoptosis.10
We also demonstrated that tunicamycin inhibited proliferation, induced cytotoxicity and apoptosis in WM cell lines and patient samples through activation of caspase-9 followed by caspase-3 and PARP cleavage. We did not observe caspase-12 and -4 activation, and caspase-8 cleavage occurred but at a later time point. Importantly, tunicamycin had no effect on normal mononuclear cells or growth of bone marrow colonies, suggesting a favorable therapeutic index.
The role of the bone marrow microenvironment in regulation of growth and drug resistance of malignant cells has been evocated previously in WM.15 Previous studies in WM have demonstrated that cytokines such as IL-6,32,33 BLyS,34 and CD40L (CD154),35 which mimic the BM microenvironment, also promote growth, survival, and trafficking of human WM cells. We show in these studies that adherence of BCWM.1 cells to BMSCs as well as addition of these cytokines and growth factors did not block tunicamycin-induced cytotoxicity in the context of the bone marrow milieu.
The regulation of signaling pathways in malignant cells is complex, and therefore rationally designed combinations of novel agent are needed. Bortezomib is now considered as a major drug in the treatment of WM disease, but more importantly, it also induces ER stress and promotes terminal UPR components.10 Interestingly, while tunicamycin promotes terminal UPR components by accumulation of misfolded proteins through blockage of N-linked protein glycosylation, bortezomib, a proteasome inhibitor, blocks the proteasome-mediated destruction of misfolded proteins extracted into the cytosol for degradation by the cytosolic 26S proteasome.44 We observed that the combination of 2 ER stress inducers, bortezomib and tunicamycin, induced synergistic cytotoxicity on WM cells. However, similar synergism was also observed with the combination of tunicamycin and fludarabine, implying that other mechanisms might be responsible for the synergism observed with tunicamycin.
In summary, we demonstrated that the prosurvival/physiologic UPR is constitutively activated in WM cells and that tunicamycin, an ER stress inducer that promotes proapoptotic/terminal UPR induced apoptosis and growth inhibition in WM cells in vitro, without toxicity in healthy donor cells. In addition, tunicamycin was able to overcome drug resistance and inhibit proliferation of tumor cells even in the bone marrow microenvironment. Finally, the combination of tunicamycin with bortezomib and fludarabine led to synergistic inhibition of survival in WM cells. These preclinical studies provide a framework for further evaluation of ER stress–inducing agents as therapeutic agents in WM.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
X.L. was supported by a grant from the Franco-American Fulbright Foundation. This work was supported in part by the International Waldenstrom Macroglobulinemia Foundation.
Authorship
Contribution: X.L., I.M.G., and S.T. designed the research and wrote the manuscript; and L.X., X.J., A.S., A.S.M., H.T.N., E.H., A.M.R., Z.R.H., A.W.H., D.D.S., S.A., K.O.C., J.S., B.C., C.P., and R.M. performed research and analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Steven P. Treon, Bing Program for Waldenstrom Macroglobulinemia, Dana-Farber Cancer Institute, 44 Binney Street, Boston, MA 02115; e-mail: steven_treon@dfci.harvard.edu.


![Figure 5. Growth factors and coculture with BMSCs do not protect against tunicamycin-induced BCWM.1 cell cytotoxicity. (A) BCWM.1 cells were cultured with control media, and with tunicamycin (0.5 μg/mL to 1 μg/mL) for 48 hours, in the presence or absence of BMSCs. Cell proliferation was assessed using the [3H]-thymidine uptake assay. (B) BCWM.1 cells were cultured with control media in the presence or absence of IL-6 (25 ng/mL), CD40L (3 ng/mL), or BLyS (100 ng/mL), and treated with tunicamycin (0.1 μg/mL and 1 μg/mL) for 48 hours. Cytotoxicity was assessed by the MTT assay. All data represent mean plus or minus SD of triplicate experiments (A,B).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/3/10.1182_blood-2007-10-116848/5/m_zh80040930010005.jpeg?Expires=1769164781&Signature=l3sj02heMI5jQoREHI1dnD~hyFRLYxh4NzQG6UwtJIc7r01-4Zg-eAgE8suNKsambhFyn-CbAHssLbNIow47VauMslpvwoa1m~JbrJtKuiGEMaHIW0bRgRk6xVcjYh-bdAPScprMYDJPbGqBU4oo-S~u0Nx0-s4mwjS-4~tfh1J9dwQ6V5sZwtRLXjRjn4a~ew2t9wfQ6Eok4QXYD-B6e6kdbryULKQeXRAaoYaxHf~mAvHmfCQl0sLxQqXe250V4FhyQ6-UPMty8HNlQVSlDH9S-vl0~G1bVjI0Q257~ANUS3AWW~gbj-7lJf4Pqw0ITx9EfzCIN55Rvao1ebR6wA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal