Abstract
KRAS is often mutated in human hematopoietic malignancies, including juvenile myelomonocytic leukemia (JMML) and T-cell lymphoblastic leukemia/lymphoma (TLL/L). However, the exact role and function of oncogenic KRAS mutations in the initiation and progression of JMML and TLL/L remain elusive. Here, we report the use of a mouse bone marrow transplantation model to study oncogenic Kras-induced leukemogenesis. We show that as the first genetic hit, oncogenic Kras mutations initiate both JMML and TLL/L, but with different efficiencies. Limiting dilution analyses indicated that an oncogenic Kras mutation alone is insufficient to produce frank malignancy. Instead, it cooperates with additional subsequent genetic event(s). Moreover, transplantation of highly purified hematopoietic stem cells (HSCs) and myeloid progenitors identified HSCs as the primary target for the oncogenic Kras mutation. Karyotypic analysis further indicated that secondary genetic hit(s) target lineage-specific progenitors rather than HSCs for terminal tumor transformation into leukemic stem cells. Thus, we propose the cellular mechanism underlying oncogenic Kras-induced leukemogenesis, with HSCs as the primary target by the oncogenic Kras mutations and lineage-committed progenitors as the final target for cancer stem cell transformation. Our model might be also applicable to other solid tumors harboring oncogenic Kras mutations.
Introduction
In the 1980s, RAS genes were first cloned as cellular homologs of the retroviral oncogenes v-Hras and v-Kras (reviewed in Barbacid1 ), which transform the infected cells and lead to sarcomas in rodents. Ras proteins belong to the superfamily of small GTPases, cycling between the active GTP-bound form and the inactive GDP-bound form.2,3 Conversion between these states occurs through intrinsic nucleotide exchange (from GDP-bound form to GTP-bound form) or GTP hydrolysis (from GTP-bound to GDP-bound form). These conversions are facilitated by numerous proteins, including Ras GEFs (guanine nucleotide exchange factors that promote the release of GDP and the association of GTP), and Ras GAPs (GTPase-activating proteins that accelerate GTP hydrolysis by Ras proteins; reviewed in Boguski and McCormick4 and Quilliam et al5 ).
Deregulated Ras signaling is prevalent in essentially all human cancers,6 and is achieved mainly through 2 mechanisms (reviewed in Shannon7 ). First, Ras proteins become aberrantly activated by the constitutive activation of a protein tyrosine kinase upstream of Ras (eg, cytokine receptors with constitutive tyrosine kinase activity and Bcr-Abl) or inactivation of proteins regulating the conversion between the Ras GTP-bound form and GDP-bound form (eg, Nf-1, a Ras-GAP). Second, acquired oncogenic RAS mutations disrupt Ras GTPase activities as well as their association with Ras GAPs, resulting in the accumulation of Ras proteins in their active GTP-bound form. Oncogenic mutations in the 3 RAS genes (Hras, Nras, and Kras) are identified in virtually all the human cancer types with various incidences and associations with specific RAS genes.6
Of the 3 Ras isoforms in mammals, Kras is the only essential Ras gene for normal mouse development.8-11 Oncogenic Kras mutations are prevalent in virtually all cancer types, making Kras one of the most frequently mutated genes in human cancers, including hematopoietic malignancies.6 In human patients with hematopoietic malignancies, oncogenic Kras mutations are often identified in myeloid and T-cell disorders, but rarely in B-cell disorders.6 Of all the myeloid disorders, a particular myeloproliferative disease, called juvenile myelomonocytic leukemia (JMML), shows a very tight connection to abnormal Ras signaling:12 approximately 30% of patients with JMML carry oncogenic mutations in the NRAS or KRAS gene; approximately 10% of patients carry mutations in the NF1 gene, which encodes a Ras GAP protein; and approximately 20% to 25% of patients carry mutations in the PTPN11 gene, which encodes a putative positive regulator of the Ras signaling pathway. Moreover, mouse models that are either deficient in NF1 or express oncogenic Kras at its endogenous level manifest some key features of this disease, such as overproduction of monocytes and hyperactivation of granulocyte-macrophage colony-stimulating factor (GM-CSF)–dependent signaling pathways.13-17
With respect to the T-cell disorders, the Nras or Kras gene is mutated in about 10% of patients with T-cell lymphoblastic leukemia/lymphoma (T-LL/L), and elevated levels of Ras signaling have been observed in about 50% of patients.18 Despite intensive investigation, several important issues regarding the exact role and function of oncogenic KRAS mutations in the initiation and progression of JMML and T-ALL/L remain elusive. For example, can oncogenic KRAS mutations initiate these disorders? If so, is it sufficient to produce frank leukemia on its own? What cell type(s) serve as its primary target(s)? And what cell type(s) eventually give rise to leukemic stem cells?
The results obtained in murine models using different conditional oncogenic Kras alleles remain controversial. When Guerra et al used a CMV-Cre transgene to induce widespread tissue expression of a conditional oncogenic Kras allele, tumorigenesis was largely restricted to the lung; no hematopoietic malignancies were identified.19 In contrast, when Braun et al and Chan et al used an Mx1-cre transgene to induce somatic expression of a different oncogenic Kras allele, tumorigenesis was observed in multiple tissues, and the induced mice developed a myeloproliferative disease.13,14 However, it remains unclear whether this disease is solely caused by cell-autonomous expression of oncogenic Kras in the hematopoietic compartment or if it occurs in combination with somatic oncogenic Kras expression in other cell types. The observation that this myeloproliferative disease is not transplantable into secondary recipients further supports this concern.14
To clearly define the cellular mechanism underlying oncogenic Kras-induced leukemogenesis, we established a mouse bone marrow transplantation model. We transplanted unfractionated bone marrow cells expressing oncogenic Kras from its endogenous locus into lethally irradiated mice. Depending on the donor cell dosage, 20% to 100% of the recipient mice developed malignancies in T cells and myeloid cells, including T-ALL/L and JMML. Limiting dilution analyses showed that the oncogenic Kras mutation alone is not sufficient to produce frank malignancy. We fractionated bone marrow cells into hematopoietic stem cells (HSCs) and myeloid progenitors and subsequently identified HSCs as the primary target of the oncogenic Kras mutation. Moreover, we showed that secondary genetic hit(s) target lineage-specific progenitors rather than HSCs for terminal tumor transformation. These findings lead us to propose a model explaining oncogenic Kras-induced leukemogenesis, a model that might also be applicable to other solid tumors harboring oncogenic Kras mutations.
Methods
Mice
All mouse lines were maintained in a pure C57BL/6 genetic background. Mice bearing the conditional oncogenic Kras (Lox-stop-Lox [LSL] Kras) mutation were crossed to C57BL/6 mice for more than 10 generations. The Mx1-cre mice were purchased from The Jackson Laboratory (Bar Harbor, ME), where they had been crossed to C57BL/6 mice for 7 generations. They were continuously crossed to C57BL/6 mice for more than 3 generations at the Whitehead Institute/Massachusetts Institute of Technology (MIT) facility. These 2 mouse lines were crossed to each other to generate mice carrying both alleles (LSL Kras/+; Mx-1 cre/+). Genotyping of the adult mice was performed as described.20
To induce Cre expression, 4- to 6-week-old mice were injected intraperitoneally with 250 μg polyinosinic-polycytidylic acid (pI-pC; Sigma-Aldrich, St Louis, MO) every other day for 3 doses. The injected mice were monitored daily for evidence of disease. All animals were killed 1 week after the last injection for experiments described in this manuscript except for the ones that were killed when moribund before this time frame. All experiments were conducted with the ethical approval of Committee on Animal Care at the Massachusetts Institute of Technology, Cambridge.
Murine bone marrow transplantation
Adult C57BL/6 recipient mice (CD45.1+, 6-8 weeks old) were irradiated with a Cesium source for 2 doses of 5 Gy (500 rad) each, delivered 3 hours apart. For sublethal irradiation, mice were administered one dose of 6.5 Gy (650 rad). Bone marrow cells were harvested from the femurs and tibias of pI-pC–treated mice (CD45.2+) and mice that provided competitor/helper cells (CD45.1+). When transplanting with total bone marrow cells, live nucleated donor cells (CD45.2+) and competitor cells were counted in 3% acetic acid with methylene blue (StemCell Technologies, Vancouver, BC). The cells were washed and resuspended in phosphate-buffered saline (PBS)/5% mouse serum, and injected into the retro-orbital venous sinus of irradiated CD45.1+ recipients. When transplanting fractionated bone marrow cells, fractionated CD45.2+ donor cells were mixed with 250 000 live nucleated CD45.1+ helper cells and injected into individual irradiated CD45.1+ recipients. The mice that underwent transplantation were maintained on trimethoprim-sulfamethoxazole–treated water for 2 weeks. Blood was obtained from the retro-orbital venous sinus regularly from 2 weeks to 8 months after transplantation for flow cytometric analysis and combined blood count (CBC). CBC analyses were performed mainly at the Hematology Clinical Core Facility at Children's Hospital Boston. The frequency of lineage-specific malignancy-initiating cells was calculated using the L-cal program from StemCell Technologies.
Flow cytometric analysis
For peripheral blood samples, red blood cells were lysed in ammonium chloride solution (StemCell Technologies) prior to antibody staining. Cells isolated from bone marrow, spleen, and thymus were resuspended in PBS with 2% fetal bovine serum (FBS) and passed through 25-μm cell strainers to obtain single-cell suspensions prior to antibody staining. Double-negative thymocytes were purified as CD4−CD8−B220−TER119− Mac-1−Gr-1− cells using StemSep columns per the manufacturer's instructions (StemCell Technologies).
The following directly conjugated antibodies were purchased from BD Pharmingen (San Diego, CA): CD45.1 (A20), CD45.2 (104), B220 (6B2), CD19 (1D3), Thy1.2 (53-2.1), Mac-1 (M1/70), Gr-1 (RB6-8C5), CD4 (RM4-5), CD8 (53-6.7), CD25 (7D4), CD41 (MWReg30), CD43 (S7), and CD44 (IM7). Directly conjugated anti-mouse IgM was purchased from Jackson ImmunoResearch Laboratories (West Grove, PA).
Histopathology
Mouse organs were fixed in 10% neutral buffered formalin (Sigma-Aldrich) and further processed at the Histology Lab of Division of Comparative Medicine, MIT, including dehydration, paraffin embedding, sectioning, and staining with hematoxylin and eosin.
Monocytes were sorted from peripheral blood samples as Mac-1+ Gr-1− cells (W1). A total of 5000 to 20 000 sorted monocytes were centrifuged onto slides (Cytospin 3; Shandon [now Thermo Fisher Scientific], Waltham, MA) and air-dried. Cells were fixed in −20°C methanol and stained with May-Grunwald-Giemsa stains according to the manufacturer's recommendations (Sigma-Aldrich).
Colony assays
A total of 105 bone marrow cells were plated in duplicate in semisolid medium MethoCult M3234 or M3231 (StemCell Technologies) supplemented with various concentrations of purified mouse GM-CSF (a gift from Amgen, Thousand Oaks, CA) according to the manufacturer's protocol. The colonies were counted after 7 to 10 days in culture.
Megakaryocyte quantification
The quantification of bone marrow megakaryocytes was performed as previously described.21
Fractionation of bone marrow cells
To purify HSCs, total bone marrow cells were stained with directly conjugated antibodies against CD41, CD48, and CD150 (BioLegend, San Diego, CA) and sorted twice for purity essentially as described previously.22
Myeloid progenitors were purified as Sca-1−Lin−IL7Rα− cells23,24 using the StemSep system per manufacturer's instructions (StemCell Technologies). The following biotin-labeled antibodies were purchased from eBioscience (San Diego, CA): Gr-1 (RB6-8C5), B220 (RA3-6B2), and CD3 (145-2C11). The following biotin-labeled antibodies were purchased from BD Pharmingen: IL7Rα (B12-1), Sca-1 (E13-161.7), IgM (R6-60.2), CD19 (1D3), CD4 (GK1.5), CD8 (53-6.7), Mac-1 (M1/70), and TER119.
G-banding karyotyping
Thymic lymphoma cells were isolated from individual mice and cultured in vitro for 2 to 4 weeks essentially as described previously.25 Bone marrow myeloid cells were purified as CD45.1−TER119−CD3−CD4−CD8−B220− cells from individual mice with JMML using the StemSep system per the manufacturer's instructions. Purified myeloid cells were cultured in vitro for 1 to 2 weeks essentially as described previously.26 Metaphase spreads were prepared after 4 to 6 hours of colcemid treatment in culture (Sigma-Aldrich). Mouse G-banding analysis was performed by Cell Line Genetics (Madison, WI). For each sample, typically 18 to 40 cells were analyzed.
Results
Recipient mice that received transplants of bone marrow cells expressing oncogenic Kras from its endogenous locus developed hematopoietic malignancies in multiple lineages
We induced oncogenic K-ras expression from its endogenous locus in the compound LSL Kras/+; Mx-1 cre/+ mice (Kras G12D; “Methods”) by injecting pI-pC every other day for 3 doses. The injected mice rapidly developed a myeloproliferative disease, consistent with previous reports on tumorigenesis in similar mice.13,14 However, due to the mechanism of oncogenic Kras induction, which occurs in many somatic cell types and is mediated by the interferon response pathway,27 any interpretation of these severe myeloproliferative disorders is complicated by effects mediated by somatic interferon action as well as by oncogenic Kras expression in nonhematopoietic cells.
To address the cell-autonomous role of oncogenic Kras in hematopoiesis and leukemogenesis, we took the approach of bone marrow transplantation. We crossed both the conditional oncogenic Kras mice (LSL Kras/+) and the Mx1-cre mice (Mx-1 cre/+) into a pure C57BL/6 genetic background for more than10 generations. The compound LSL Kras/+; Mx-1 cre/+ mice were then generated by crossing together mice with these alleles. We refer to the LSL Kras/+; Mx-1 cre/+ as Kras G12D mice and the Mx-1 cre/+ mice as wild-type control mice. One week after the last injection of pI-pC, bone marrow cells were isolated from donor mice, and various numbers of donor cells were mixed together with syngeneic competitor/helper cells in a 1:1 ratio and transplanted into lethally irradiated mice. Because the donor cells and competitor/helper cells express different cell-surface markers (CD45.2 vs CD45.1), we can trace CD45.2+ donor-derived hematopoiesis after transplantation in recipient mice. Since none of the mice that received wild-type control bone marrow cells developed malignancies, all recipient mice with hematopoietic malignancies as described in the rest of this study refer to mice that received Kras G12D bone marrow cells.
Depending on the donor cell dosages, 40% to 100% of recipient mice developed hematopoietic malignancies in multiple lineages, including thymic T-cell lymphoblastic lymphoma (Figure 1), T-cell leukemia (Figure 2), and phenotypes closely resembling human juvenile myelomonocytic leukemia (JMML; Figure 3). Interestingly, none of the mice developed B-cell malignancies, and in most of the animals that we examined, B-cell development was apparently normal (Figure 4). This disease pattern is very similar to that observed in human patients with hematopoietic malignancies that express an oncogenic Ras protein (reviewed in Bos6 ). Just as patients with cancer display a broad range of phenotypes even when they are diagnosed with the same cancer type, in many instances the diseased mice listed under the same cancer category showed various phenotypes. Therefore, we report here only the “consensus” phenotypes, which were displayed by most of the diseased animals.
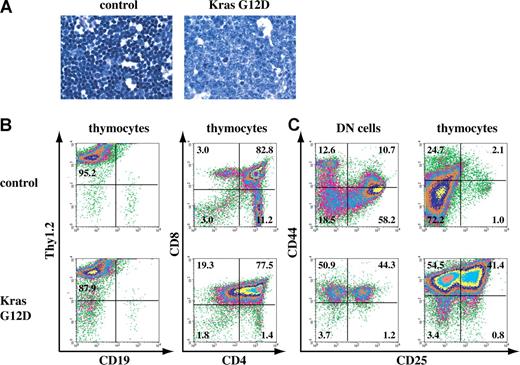
A fraction of recipient mice that received transplants of total bone marrow cells expressing oncogenic Kras from its endogenous locus developed thymic T-cell lymphoblastic lymphoma. (A) Representative histopathology (original magnifications of camera lenses used to take the picture listed in parentheses) from thymus (× 100) stained with hematoxylin and eosin (H&E) showed development of thymic lymphoma in mice that received transplants of bone marrow cells expressing oncogenic Kras G12D from its endogenous locus. (B,C) Flow cytometry analysis of thymocytes or purified DN cells isolated from the thymus of control mice or mice that received transplants of bone marrow cells expressing oncogenic Kras. These density plots were gated on live nucleated cells based on forward scatter and propidium iodide staining profiles. More than 98% of the cells analyzed here are donor-derived as judged by their CD45.2 expression. Representative data are shown. The percentages of cells in quadrants of interest are indicated.
A fraction of recipient mice that received transplants of total bone marrow cells expressing oncogenic Kras from its endogenous locus developed thymic T-cell lymphoblastic lymphoma. (A) Representative histopathology (original magnifications of camera lenses used to take the picture listed in parentheses) from thymus (× 100) stained with hematoxylin and eosin (H&E) showed development of thymic lymphoma in mice that received transplants of bone marrow cells expressing oncogenic Kras G12D from its endogenous locus. (B,C) Flow cytometry analysis of thymocytes or purified DN cells isolated from the thymus of control mice or mice that received transplants of bone marrow cells expressing oncogenic Kras. These density plots were gated on live nucleated cells based on forward scatter and propidium iodide staining profiles. More than 98% of the cells analyzed here are donor-derived as judged by their CD45.2 expression. Representative data are shown. The percentages of cells in quadrants of interest are indicated.
A fraction of recipient mice that received transplants of total bone marrow cells expressing oncogenic Kras from its endogenous locus developed T-cell leukemia. Flow cytometric analysis of peripheral blood (PB), bone marrow (BM), and spleen (SP) cells isolated from control mice or mice that received transplants of bone marrow cells expressing oncogenic Kras G12D from its endogenous locus. These density plots were gated on live nucleated cells based on forward scatter and propidium iodide staining profiles. More than 98% of cells analyzed here are donor-derived as judged by their CD45.2 expression. Representative data are shown. The percentages of cells in quadrants of interest are indicated.
A fraction of recipient mice that received transplants of total bone marrow cells expressing oncogenic Kras from its endogenous locus developed T-cell leukemia. Flow cytometric analysis of peripheral blood (PB), bone marrow (BM), and spleen (SP) cells isolated from control mice or mice that received transplants of bone marrow cells expressing oncogenic Kras G12D from its endogenous locus. These density plots were gated on live nucleated cells based on forward scatter and propidium iodide staining profiles. More than 98% of cells analyzed here are donor-derived as judged by their CD45.2 expression. Representative data are shown. The percentages of cells in quadrants of interest are indicated.
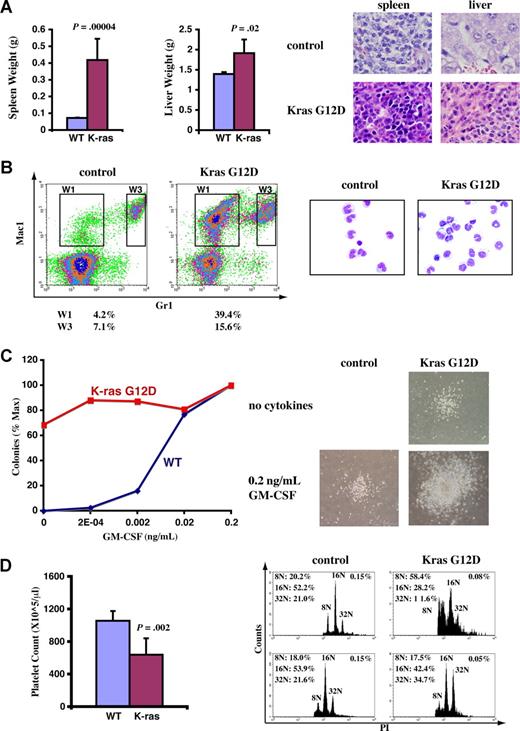
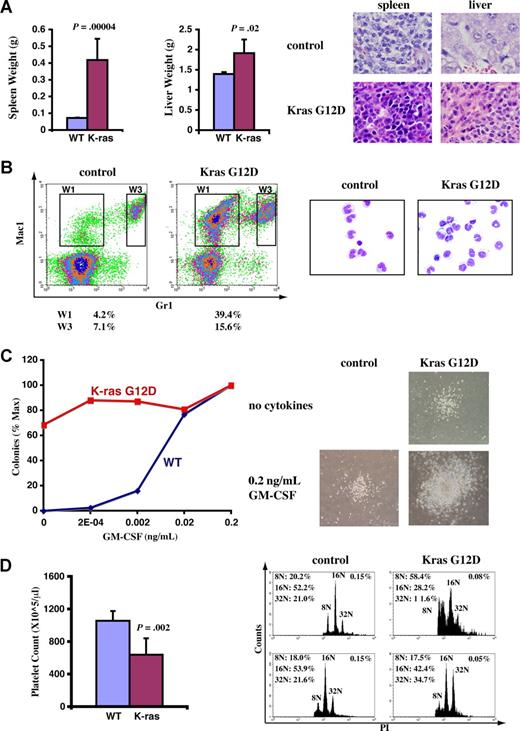
A fraction of recipient mice that received transplants of total bone marrow cells expressing oncogenic Kras from its endogenous locus developed JMML-like phenotypes. Diseased and control mice were killed approximately 3 months after transplantation when diseased mice became moribund. (A) Diseased mice showed enlarged spleen and liver. Representative histologic H&E sections from spleen (×60) and liver (×60) showed an extensive infiltration of myelomonocytic cells in the splenic and hepatic parenchymas of the recipient mice that received transplants of bone marrow cells expressing oncogenic Kras G12D (original magnifications of camera lenses used to take the pictures listed in parentheses). (B) Flow cytometric analysis of peripheral blood samples from control mice or mice that received transplants of bone marrow cells expressing oncogenic Kras. These density plots were gated on donor-derived, live nucleated cells based on forward scatter, propidium iodide staining, and CD45.2 expression profiles. Representative data are shown. The percentages of cells in regions of interest are indicated. W1 cells were sorted, centrifuged on slides, and stained with May-Grunwald-Giemsa stains to confirm their monocyte identity (right). (C) A total of 105 bone marrow cells were plated in duplicate in semisolid medium with or without GM-CSF. The data were presented as percentages of the maximum number of colonies formed in culture with 0.2 ng/mL GM-CSF. The experiments were repeated independently using cells from at least 3 mice, and representative results obtained from one mouse are shown here. Photomicrographs (original magnification ×20) show wild-type and mutant Kras G12D myeloid progenitor colonies grown in different concentrations of GM-CSF. (D) Platelet counts in peripheral blood and bone marrow (BM) megakaryocyte analysis. CD41+ megakaryocytes from control BM and Kras G12D BM were analyzed for DNA content. Only mature megakaryocytes with 8N and greater ploidy are shown. The percentages of mature megakaryocytes are indicated. (A,D) Student t test was performed. The data are presented as averages plus SD.
A fraction of recipient mice that received transplants of total bone marrow cells expressing oncogenic Kras from its endogenous locus developed JMML-like phenotypes. Diseased and control mice were killed approximately 3 months after transplantation when diseased mice became moribund. (A) Diseased mice showed enlarged spleen and liver. Representative histologic H&E sections from spleen (×60) and liver (×60) showed an extensive infiltration of myelomonocytic cells in the splenic and hepatic parenchymas of the recipient mice that received transplants of bone marrow cells expressing oncogenic Kras G12D (original magnifications of camera lenses used to take the pictures listed in parentheses). (B) Flow cytometric analysis of peripheral blood samples from control mice or mice that received transplants of bone marrow cells expressing oncogenic Kras. These density plots were gated on donor-derived, live nucleated cells based on forward scatter, propidium iodide staining, and CD45.2 expression profiles. Representative data are shown. The percentages of cells in regions of interest are indicated. W1 cells were sorted, centrifuged on slides, and stained with May-Grunwald-Giemsa stains to confirm their monocyte identity (right). (C) A total of 105 bone marrow cells were plated in duplicate in semisolid medium with or without GM-CSF. The data were presented as percentages of the maximum number of colonies formed in culture with 0.2 ng/mL GM-CSF. The experiments were repeated independently using cells from at least 3 mice, and representative results obtained from one mouse are shown here. Photomicrographs (original magnification ×20) show wild-type and mutant Kras G12D myeloid progenitor colonies grown in different concentrations of GM-CSF. (D) Platelet counts in peripheral blood and bone marrow (BM) megakaryocyte analysis. CD41+ megakaryocytes from control BM and Kras G12D BM were analyzed for DNA content. Only mature megakaryocytes with 8N and greater ploidy are shown. The percentages of mature megakaryocytes are indicated. (A,D) Student t test was performed. The data are presented as averages plus SD.
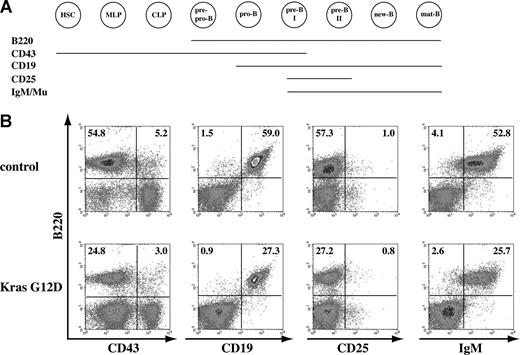
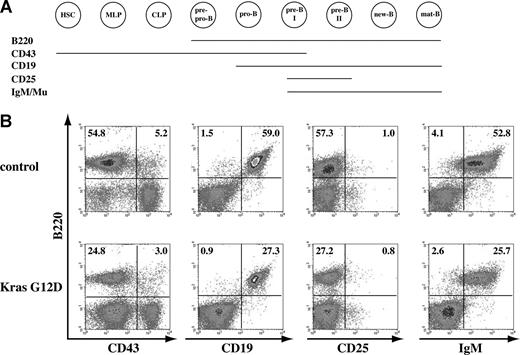
B-cell development is apparently normal in recipient mice that received transplants of total bone marrow cells expressing oncogenic Kras. (A) Schematic illustration of different cell-surface markers associated with different B-cell developmental stages. (B) Flow cytometric analysis of spleen cells isolated from control mice or mice that received transplants of bone marrow cells expressing oncogenic Kras. These density plots were gated on donor-derived, live nucleated cells based on forward scatter, propidium iodide staining, and CD45.2 (donor cell marker) expression profiles. At least 10 mice with T-cell malignancy and/or JMML were examined, and representative data from one mouse are shown here. The percentages of cells in quadrants of interest are indicated.
B-cell development is apparently normal in recipient mice that received transplants of total bone marrow cells expressing oncogenic Kras. (A) Schematic illustration of different cell-surface markers associated with different B-cell developmental stages. (B) Flow cytometric analysis of spleen cells isolated from control mice or mice that received transplants of bone marrow cells expressing oncogenic Kras. These density plots were gated on donor-derived, live nucleated cells based on forward scatter, propidium iodide staining, and CD45.2 (donor cell marker) expression profiles. At least 10 mice with T-cell malignancy and/or JMML were examined, and representative data from one mouse are shown here. The percentages of cells in quadrants of interest are indicated.
Most recipient mice developed T-cell malignancies
To test how efficiently the oncogenic Kras mutations can induce hematopoietic malignancies, we transplanted 2.5 × 105 total bone marrow cells expressing oncogenic Kras along with same number of competitor cells into 52 lethally irradiated recipient mice (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). We found that 50% of recipient mice (26 of 52) developed clinically significant thymic hyperplasia/lymphomas (Figure 1). Compared with control thymus, which was full of mature T cells with dense nuclear staining, the lymphoma thymus was filled with more immature cells with diffuse nuclear staining (Figure 1A). To identify the cell origin of this thymic lymphoma, thymocytes were examined for their expression of Thy1.2 and CD19, markers for T cells and B cells, respectively (Figure 1B). Most of the cells were Thy1.2+ but CD19−, indicating that the thymic lymphoma originated from T cells. Most of these mice did not show detectable T-cell dominance in peripheral blood or bone marrow (data not shown) when they died or were humanely killed 3 to 6 months after transplantation (Figure S1).
We next examined the lymphoma development in the recipient mice (Figure 1B). In a normal thymus, T-cell development progresses from CD4, CD8 double-negative (DN) thymocytes to CD4, CD8 double-positive (DP) thymocytes and finally to mature CD4 or CD8 single-positive (SP) T cells.28 At the early stages of this T-cell lymphoma, the thymocytes mainly displayed a hyperplasia phenotype with an approximately 3- to 5-fold increase of cell mass but absence of neoangiogenesis. In more than 95% of the cases, the thymocytes started to lose CD4 SP cells (data not shown). The thymocytes at this stage could not propagate in vitro as established cell lines. At more advanced stages of T-cell lymphoma, thymus weights and thymocyte counts increased approximately 10-fold compared with controls, and neoangiogenesis was very prominent (data not shown). CD4 SP cells were almost absent, and the DP thymocytes gradually lost CD4 expression to become CD8 SP (Figure 1B). The thymocytes at this stage were readily propagated in vitro, and eventually all became CD8 SP (data not shown). This pattern of disease progression closely resembles the development of T-cell lymphoma identified in EβR/Rp53−/− mice.25
We then examined at which stage(s) the T-cell lymphoma disrupted thymic T-cell development. Given the immature lymphoblastic morphology of lymphoma thymocytes, we first examined the developmental status of the DN cells in the malignant thymus (Figure 1C). Depending on the status of CD44 and CD25 expression, normal DN thymocytes can be further divided into 4 successive subpopulations: CD44+CD25− (DN1), CD44+CD25+ (DN2), CD44−CD25+ (DN3), and CD44−CD25−(DN4). We purified DN cells from control and lymphoma thymus. The lymphoma DN thymocytes were primarily blocked at the DN1 and/or DN2 stages as evidenced by the accumulation of CD44+CD25− and CD44+CD25+ cells (Figure 1C; data not shown). Interestingly, when we examined the expression status of CD44 and CD25 on total thymocytes, unlike the control thymocytes that were mainly CD44−CD25−, the lymphoma thymocytes remained CD44+CD25− and/or CD44+CD25+. However, despite the abnormal differentiation of DN cells, the lymphoma DP and CD8 SP thymocytes managed to initiate a partial T-cell differentiation program because the expression of CD24 and CD127 was largely normal, whereas the expression of CD3, CD5, and CD69 was either absent or greatly reduced in many cases (data not shown; Table S1). These results suggested that T-cell lymphoma disrupted normal development of DN1 and/or DN2 cells, which could drive partial but abnormal T-cell differentiation. Alternatively, T-cell lymphoma disrupted normal development in mature T cells, which fail to down-regulate DN cell markers.
In addition to T-cell lymphoma, we found that approximately 10% of recipient mice (5 of 52) developed T-cell leukemia, either acute or chronic. Mice with acute T-cell leukemia showed T cells dominant in peripheral blood 4 weeks after transplantation; these mice died 1 to 2 weeks after diagnosis (Figure S1). Mice with chronic T-cell leukemia displayed the symptoms 3 to 4 months after transplantation and often were associated with thymic T-cell lymphoma. Under both of these conditions, T cells predominated in peripheral blood, bone marrow, and spleen (Figure 2). For example, in control mice, fewer than 5% of nucleated peripheral blood cells expressed the Thy1.2 antigen characteristic of T cells, whereas more than 85% of the cells in the Kras G12D mice expressed this antigen. We then examined T-cell development in affected bone marrow, which displayed a similar progressive feature as that in recipient mice with T-cell lymphoma; most thymocytes were DP, and many apparently had started to lose CD4 expression and gradually became CD8 SP cells (Figure 2).
A fraction of recipient mice developed a disease closely resembling human JMML, a type of myeloproliferative disease
In addition to T-cell malignancies, approximately 8% of recipient mice (4 of 52) developed phenotypes closely resembling those of patients with JMML. The diseased mice exhibited high monocyte levels in peripheral blood 6 to 8 weeks after transplantation; all of these mice died 6 to 8 weeks after diagnosis (Figure S1). Therefore, we killed these mice around 3 months after transplantation when they became moribund. The diseased mice showed marked splenomegaly and mild hepatomegaly (Figure 3A). On average, the spleens increased approximately 5-fold, and livers increased approximately 40% in weight compared with control mice. Both the splenic and hepatic parenchymas were heavily infiltrated by myelomonocytic cells, with profound involvement of the white pulps in the spleen and portal triad areas in the liver. The lobular architecture was completely effaced by the infiltrative tumor cells (Figure 3A). In peripheral blood, the total white blood cell counts showed only a moderate increase (∼ 40%-150% increase compared with controls; data not shown). However, monocyte overproduction was prominent (Figure 3B), as the monocyte compartment (Mac-1+ Gr-1−; W1) was enlarged more than 10-fold (∼ 4% of white blood cells in controls and ∼ 30%-60% in diseased mice). When total bone marrow cells isolated from these diseased mice were cultured in vitro, myeloid colonies formed spontaneously in the absence of cytokines, and the colony sizes increased dramatically (Figure 3C). The diseased mice showed decreased platelet counts in peripheral blood (Figure 3D), apparently caused by decreased megakaryocytes (defined as CD41+ cells with DNA content no less than 8N) in bone marrow21 and/or defective megakaryocyte differentiation (demonstrated as reduced DNA content profile; Figure 3D). Moreover, the diseased mice were anemic with higher reticulocyte counts (data not shown). All these phenotypes are characteristic of human patients with JMML.29,30
B-cell development is apparently normal in recipient mice
Bone marrow B-cell development is divided into several stages based on their cell-surface expression of B220, a marker expressed throughout B-cell development, in combination with other surface markers (Figure 4A; reviewed in Hardy and Hayakawa31 ). We examined B-cell development in bone marrow and spleen isolated from at least 10 recipient mice with hematopoietic malignancies. B-cell development was essentially normal in both tissues (Figure 4; data not shown). Due to the infiltration of other hematopoietic cells and extramedullary hematopoiesis, the B-cell compartment was often underrepresented in the spleens of recipient mice. However, B-cell development was apparently normal (9 of 10 mice; “Discussion”), and we did not observe a B-cell malignancy in any of the recipient mice that we examined (n = 10; Figure 4).
Oncogenic Kras mutation alone is not sufficient to produce hematopoietic malignancies in recipient mice
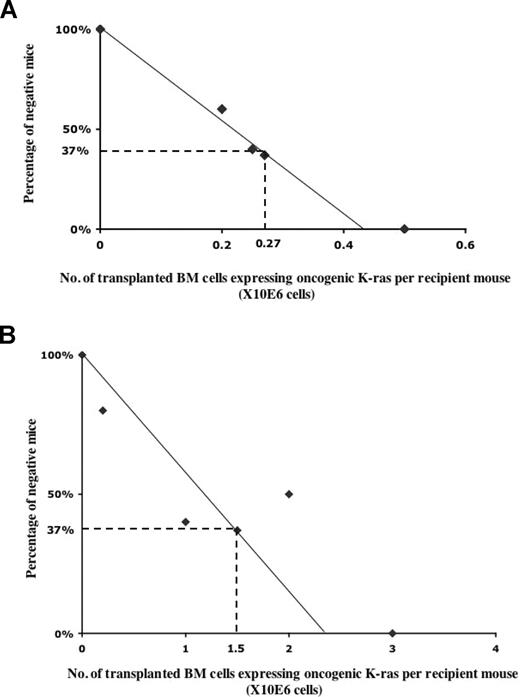
We noticed that not all of the recipient mice developed a hematopoietic malignancy, and that the fraction of diseased mice appeared to vary in proportion to the number of G12D donor cells transplanted. Thus, we used the limiting dilution method to determine directly the frequency of cells that initiate hematopoietic malignancies of different lineages (Figure 5). To this end, we transplanted different numbers of donor bone marrow cells expressing oncogenic Kras from its endogenous locus, together with congenic competitor/helper cells, in a 1:1 ratio into recipient mice (n was ≥ 10 at each data point). We determined the number of injected cells necessary to achieve a situation in which 37% of mice that received transplantations were free of cancer; the reciprocal of this value represents the frequency of malignancy-inducing cells in the injected cell population.
Calculation of the frequency of lineage-specific malignancy-initiating cells. Limiting dilution analysis of the frequency of T-cell malignancy-initiating cells (A) and JMML-initiating cells (B). Lethally irradiated CD45.1 recipient mice received transplants of various numbers of total bone marrow cells expressing oncogenic Kras together with competitor cells in a 1:1 ratio. At each data point, at least 10 recipient mice were used. Plotted is the percentage of recipient mice free of T-cell malignancy (A) or JMML (B) 6 to 8 months after transplantation. The frequency of malignancy-initiating cells was calculated as described in “Methods.”
Calculation of the frequency of lineage-specific malignancy-initiating cells. Limiting dilution analysis of the frequency of T-cell malignancy-initiating cells (A) and JMML-initiating cells (B). Lethally irradiated CD45.1 recipient mice received transplants of various numbers of total bone marrow cells expressing oncogenic Kras together with competitor cells in a 1:1 ratio. At each data point, at least 10 recipient mice were used. Plotted is the percentage of recipient mice free of T-cell malignancy (A) or JMML (B) 6 to 8 months after transplantation. The frequency of malignancy-initiating cells was calculated as described in “Methods.”
Because the recipient mice were susceptible to several distinct malignancies that occurred at dramatically different frequencies, we calculated the frequency of leukemia-initiating cells based on their specific lineages. Therefore, the negative mice scored in Figure 5 only represent the mice that do not carry the particular lineage-specific malignancy we scored. In some cases, particularly at intermediate cell doses, these negative mice do carry a malignancy in other unscored lineage(s).
We compared the frequency of normal hematopoietic stem cells to those of malignancy-inducing cells (Figure 5). The frequency of cells able to initiate a T-cell malignancy, including T-cell lymphoma and T-cell leukemia, was approximately 1 per 270 000 bone marrow cells, whereas the frequency of JMML-initiating cells was only 1 per 1.5 million bone marrow cells. These numbers are significantly lower than the frequency of normal hematopoietic stem cells, which is approximately 1 per 10 000 total bone marrow cells.32 We and others used several methods to establish that upon pI-pC induction, the recombination frequency of Mx1-cre at the endogenous Kras locus was consistently above 80% to 90% (data not shown).13,14 Therefore, the low frequencies of disease-initiating cells are not caused by the poor induction efficiency of oncogenic Kras expression. Rather, our data suggests that the oncogenic Kras mutation alone is insufficient to produce frank hematopoietic malignancies, and that oncogenic Kras needs to cooperate with rare, additional genetic event(s).
Oncogenic Kras targets hematopoietic stem cells to initiate lineage-specific hematopoietic malignancies
Many experiments showed that both HSCs and myeloid progenitors are targets for oncogenic transformation of normal cells into leukemic stem cells.33,34 Thus, we wanted to determine which population of cells serves as the primary target for an oncogenic Kras mutation. If the oncogenic Kras mutation must act on an HSC to initiate a malignancy, we would expect that transplantation of HSCs but not lymphoid or myeloid progenitors expressing oncogenic Kras would lead to T-cell leukemia/lymphoma and/or JMML. However, if lymphoid or myeloid progenitors serve as the target of an oncogenic Kras mutation for leukemogenesis, transplantation of such cells expressing oncogenic Kras would result in T-cell malignancy or JMML, respectively.
To this end, we fractionated total bone marrow cells into HSCs and myeloid progenitors. HSCs were purified, based on their characteristic expression pattern of SLAM receptors, as CD41−CD48−CD150+ cells.22 After sorting twice, the purity of HSCs was typically approximately 60% to 70% based on the reassessment of purified cells using the same markers. Based on limiting dilution experiments using wild-type cells, we calculated that the frequency of functional HSCs was one in 12 to 16 injected cells (data not shown). Myeloid progenitors, including common myeloid progenitors, granulocyte-macrophage progenitors, and erythroid-megakaryocyte progenitors, were purified as Lin−Sca-1−IL7Rα− cells.23,24 The purity of myeloid progenitors was typically greater than 70% based on their surface phenotype (data not shown). When 20 000 myeloid progenitors in which oncogenic Kras expression had been induced were transplanted into sublethally irradiated mice alone or into lethally irradiated mice together with 2.5 × 105 congenic helper cells, none of the 30 mice developed JMML-like phenotypes (data not shown). In contrast, when 20 HSCs in which oncogenic Kras expression had been induced were transplanted into lethally irradiated mice together with 2.5 × 105 helper cells, 22 of 36 mice developed various hematopoietic malignancies (Table 1). Of these 22 diseased mice, 1 developed both severe JMML and T-cell lymphoma, 2 developed severe JMML alone, and 19 developed T-cell lymphoma or leukemia alone. Our results indicate that as the first genetic hit, oncogenic Kras mutation targets HSCs to initiate hematopoietic malignancies.
Secondary genetic hit(s) target lineage-committed progenitors for the final tumor transformation
An interesting question is which population of cells are targets for the secondary genetic hits following oncogenic Kras mutations, as those cells are likely to be transformed into leukemic stem cells. The target could be HSCs or more lineage-committed progenitors. To distinguish between these 2 possibilities, we isolated and karyotyped thymic lymphoma T cells and bone marrow myeloid cells from the same mouse that received transplants of purified HSCs expressing oncogenic Kras from its endogenous locus and that developed with both T-cell lymphoma and JMML (Table 1). If the secondary genetic hit(s) target and transform HSCs, we would expect that both thymic T cells and bone marrow myeloid cells display the same or largely identical chromosomal abnormalities. However, if the secondary genetic hit(s) target more lineage-committed progenitors for the final tumor transformation, we would expect that these 2 cell types would display different karyotypes.
Mouse G-banding analysis on thymic lymphoma T cells showed that all cells tested carried the identical chromosomal abnormalities, including trisomy chromosomes 14 and 15 as well as gain of a fused chromosome. This indicates that the T-cell lymphoma was clonal (Figure 6A). In contrast, all the myeloid cells showed an apparently normal karyotype (Figure 6B). Further characterization of these 2 cell populations using a more sensitive CGH analysis identified completely nonoverlapping genomic lesions in both (Table S2). These results suggest that secondary genetic hit(s) arise in separate T cell–specific and myeloid cell–specific progenitors to generate T-cell lymphoma and JMML, respectively.
Malignant T cells and myeloid cells isolated from the same mouse displayed different karyotypes. Lymphoma T cells and malignant myeloid cells were isolated from the same animal that received a transplant of purified HSCs expressing oncogenic Kras from its endogenous locus and that developed both T-cell lymphoma and JMML (Table 1). Mouse chromosomal G-banding analysis was performed on these cells (“Methods”). A total of 18 thymic lymphoma cells were analyzed; all displayed the same karyotype. A representative image of a thymic lymphoma cell (A) showed trisomy chromosomes 14 and 15 (indicated by →) as well as gain of a fused mysterious chromosome (indicated by → labeled with marker). A total of 20 myeloid cells were analyzed; all showed apparently normal karyotype (B).
Malignant T cells and myeloid cells isolated from the same mouse displayed different karyotypes. Lymphoma T cells and malignant myeloid cells were isolated from the same animal that received a transplant of purified HSCs expressing oncogenic Kras from its endogenous locus and that developed both T-cell lymphoma and JMML (Table 1). Mouse chromosomal G-banding analysis was performed on these cells (“Methods”). A total of 18 thymic lymphoma cells were analyzed; all displayed the same karyotype. A representative image of a thymic lymphoma cell (A) showed trisomy chromosomes 14 and 15 (indicated by →) as well as gain of a fused mysterious chromosome (indicated by → labeled with marker). A total of 20 myeloid cells were analyzed; all showed apparently normal karyotype (B).
Oncogenic Kras-induced hematopoietic malignancies are transplantable into secondary recipients
To determine whether oncogenic Kras-induced hematopoietic malignancies are transplantable, we isolated bone marrow cells from individual diseased mice that had received transplants of HSCs expressing oncogenic Kras. For mice that developed a T-cell malignancy alone, whether T-cell leukemia or lymphoma, 1 to 5 × 106 total bone marrow cells were transplanted into lethally or sublethally irradiated secondary recipient mice. All secondary recipient mice developed acute T-cell leukemia and died within 4 weeks after transplantation. For the 2 mice that developed severe JMML alone, 1 died before we could harvest bone marrow cells. Therefore, we only performed a secondary transplantation from the other mouse; 5 × 106 total bone marrow cells were transplanted into lethally irradiated secondary recipient mice. All recipient mice developed symptoms similar to human acute myeloid leukemia and died within 2 weeks after transplantation. For the 1 mouse that developed both T-cell lymphoma and severe JMML, 5 × 106 total bone marrow cells were transplanted into lethally irradiated secondary recipient mice. Of the recipient mice, 40% developed acute T-cell leukemia and died within 4 to 6 weeks after transplantation. The rest of the mice developed both JMML and T-cell lymphoma and died within 3 months after transplantation. Taken together, our data shows that all of the oncogenic Kras-induced hematopoietic malignancies are transplantable into secondary recipients.
Discussion
In this paper, we define the cellular mechanism concerning induction of hematologic malignancies by oncogenic Kras expressed from its endogenous locus. First, we demonstrate that as the first genetic hit oncogenic Kras induces TLL and JMML in a cell-autonomous manner. Second, oncogenic Kras mutation is insufficient to induce frank hematopoietic malignancies. Instead, it cooperates with secondary genetic hit(s). Third, HSCs serve as the primary target by the oncogenic Kras mutation, but lineage-specific progenitors are the target cells for gene alterations that induce the final leukemic transformation.
Oncogenic Kras mutations initiate hematopoietic malignancies as the first genetic hit
Our result that somatic activation of oncogenic Kras expression from its endogenous promoter rapidly induces a myeloproliferative disease (MPD) with complete penetrance is in sharp contrast to Guerra et al's finding that no myeloid malignancies developed even after months of observation when they used a CMV-Cre transgene to activate a different oncogenic Kras allele.19 There are 2 possible reasons for these differences. First, CMV-Cre might not work as efficiently as Mx1-Cre in bone marrow cells. Second, the conditional oncogenic Kras allele constructed by Guerra et al carries an additional modification with an internal ribosomal entry site (IRES)–βgeo cassette inserted to the 3′ untranslated region (UTR) of the endogenous Kras locus. This modification might reduce the expression level of the endogenous Kras protein.
We find that transplanting total bone marrow cells expressing oncogenic Kras into lethally irradiated recipient mice leads to hematopoietic malignancies with various penetrances. This result is different from Chan et al' s observation that all transplant recipients remained healthy and disease-free after 120 days.14 The discrepancy is not caused by different experimental procedures, as we also performed bone marrow transplantation (BMT) into sublethally irradiated mice and obtained similar results as we reported here using a competitive BMT strategy. The difference is very likely caused by the different genetic backgrounds of our mice versus theirs. Their mice were maintained on a mixture of 129Sv/Jae, C57BL/6, and Balb/c genetic backgrounds, whereas our mice are in a pure C57BL/6 background. Therefore, despite the use of wild-type littermates as BMT recipients, their BMT experiments likely result in mutual rejections between host and graft cells, which might prevent leukemogenesis efficiently.
Although it appears that oncogenic Kras expression alone is sufficient to induce MPD (closely resembling to human JMML) in mice described in the original papers,13,14 the MPD phenotypes are complicated by systemic interferon-mediated responses, induced oncogenic Kras expression in nonhematopoietic cells, and the simultaneous expression of oncogenic Kras in 80% to 90% of myeloid cells. Thus, the initiation of MPD phenotypes with 100% penetrance in the primary mice might be a transient phenomenon attributable to microenvironmental factors and does not necessarily imply the long-term maintenance in a cell-autonomous manner. As we showed that the initiation and maintenance of JMML, the type of MPD closely related to the MPD initially described only reached 100% when large amounts of total bone marrow cells were transplanted (Figure 5B). When we transplanted purified HSCs expressing oncogenic Kras into recipient mice, only a small fraction of mice consistently developed JMML (Table 1). This is mainly due to the low frequency of JMML-initiating cells, which have to incorporate both oncogenic Kras mutation and rare secondary genetic hits (“Results”). It is also possible that either the MPD state or the pI-pC treatment hinders the engraftment of JMML-initiating cells and thus contributes to our results.
Oncogenic Kras mutation induces TLL efficiently
Oncogenic Kras initiates TLL very efficiently in our mouse model. There are 2 possible explanations for this. First, the oncogenic Kras mutation might provide unique proliferative/survival advantages to precursor T cells, as seen with several other cell types.13,14,17,35 We observed rapid expansion and recovery of the lymphoid compartment as early as 2 weeks after transplanting total bone marrow cells expressing oncogenic Kras. This is in sharp contrast to mice that received transplants of control cells, in which the myeloid compartment expanded and recovered ahead of the lymphoid compartment (data not shown). In addition, due to the important role of Ras signaling in inducing cytokine gene production (reviewed in Genot and Cantrell36 ), overproducing lymphocyte-stimulating cytokines might act as an autocrine in a positive feedback loop on the proliferation of lymphocytes expressing oncogenic Kras.
Second, oncogenic Kras might affect T-cell differentiation on its own, which precedes the final T-cell malignancy. Ras signaling is important for regulating T-cell functions and maintaining a normal immune system (reviewed in Scheele et al37 ). In recipient mice that developed JMML alone, the thymus size was often greatly reduced. However, even in the degenerated thymus that were absent of any malignancy, T-cell development displayed a similar partial differentiation pattern as that shown in thymic lymphoma T cells (data not shown), indicating that oncogenic Kras signaling affects T-cell differentiation.
We analyzed the clonality of 3 independent cell lines established from oncogenic Kras-induced primary T-cell lymphoma (Figures 6A,S2). One of the cell lines is monoclonal, consistent with our mouse G-banding analysis, whereas the other 2 cell lines are oligoclonal, with one clone dominant in the whole populations.
Interestingly, we did not identify any recipient mice that developed a B-cell malignancy. However, we did observe abnormal B-cell development in one particular mouse that received transplants of HSCs expressing oncogenic Kras and that developed both thymic T-cell lymphoma and JMML; B-cell development was partially blocked at the pro-B stage (data not shown). We speculate that if the mouse survived longer, it could have eventually developed a B-cell malignancy. Nevertheless, the absence of B-cell malignancies in our mouse model is consistent with the rare incidence of human patients with B-cell malignancies carrying oncogenic RAS mutations.6
The JMML-like phenotypes initiated by oncogenic Kras are more severe than those initiated by inactivation of Nf1
The JMML-like phenotypes induced by oncogenic Kras are much more severe than those initiated by Nf1 deficiency, as demonstrated by shorter disease latency, more aggressive phenotypes, and more importantly, presence of disease phenotypes when cotransplanted with wild-type competitor cells (Figure 5; data not shown).13,16 This indicates that oncogenic Kras is a much more potent inducer of JMML than Nf1 deficiency. There are 2 possible explanations for the difference in the phenotypic severity. First, the Ras-GTP level might be lower in cells deficient in Nf1 than in cells expressing oncogenic Kras from its endogenous promoter. Oncogenic Ras mutations impair both endogenous Ras GTPase activities and the association of Ras proteins with Ras GAP proteins, whereas NF1 abolishes its interaction with Ras proteins without affecting their GTPase activities. Conceivably, Nf1 inactivation is a less severe biochemical lesion than an oncogenic Ras mutation. In addition, in mammals NF1 is only 1 of more than 20 Ras GAP proteins. It is likely that its inactivation is partially compensated by other family members. Second, NF1 might be a GAP protein that preferentially acts on Hras and/or Nras, rather than Kras. In this scenario, although cells deficient in NF1 show elevated Ras GTP levels, the level of Kras GTP might remain similar to that in wild-type cells. The measurement of total Ras GTP levels as well as Kras GTP levels in bone marrow cells deficient in NF1 and in bone marrow cells expressing oncogenic Kras would distinguish between these 2 possibilities.
Role of oncogenic Kras mutations as the first genetic hit in leukemogenesis
Oncogenic Kras mutations are proposed to confer proliferative and/or survival advantage during leukemogenesis but not to affect cell differentiation.38 Based on our work and that of others, we think that this proposal is only partially correct and might be incomplete as well. First, oncogenic Kras mutations do confer proliferative and/or survival advantage to hematopoietic cells. In both fetal and adult stages, oncogenic Kras leads to hyperactivation of cytokine-dependent signaling pathways.17,35 Consequently, progenitors harboring oncogenic Kras mutations form much bigger colonies in vitro than do wild-type cells (Figure 3).13,14 Second, oncogenic Kras mutations do affect hematopoietic cell differentiation. Ineffective erythropoiesis is observed at both fetal and adult stages in vivo and in vitro.35,39,40 Third, oncogenic Kras mutations might affect genome stability directly or indirectly. We observed that in the absence of additional genetic changes, the activity/function of HSCs expressing oncogenic Kras exhausted much faster than that of wild-type HSCs (data not shown), suggesting that oncogenic Kras accelerates HSC aging probably by abnormally stimulating them into the cell cycle. This phenomenon has been reported in several other cases.26,41,42 At the progenitor cell level, we and others showed that oncogenic Kras stimulates extensive cell proliferation both in vivo and in vitro. Cell proliferation or cell division is an inherently error-prone process, with mutations arising naturally during DNA replication. Therefore, oncogenic Kras mutation might affect genome stability indirectly through stimulating active cell proliferation. On the other hand, we did observe random genomic changes (eg, random gain or loss of chromosomes) in the process of establishing cell lines from primary T-cell lymphomas harboring oncogenic Kras mutations (data not shown). The incidence of random genomic changes is significantly higher than that observed in other actively proliferating cell lines, indicating that oncogenic Kras mutations might have some direct effects on genome stability.
A model for oncogenic Kras-induced leukemogenesis
We find that expression of oncogenic Kras alone is not sufficient to generate frank malignancies; it has to cooperate with additional subsequent genetic change(s) (Figure 5). Indeed, we identified additional genomic lesions in all the cell lines established from primary leukemia/lymphoma by mouse G-banding analysis and/or CGH analysis (data not shown). Therefore, it was not a surprise to find that HSCs rather than myeloid progenitors serve as the primary target of oncogenic Kras mutation, as this allows progeny cells to incorporate and select cooperative additional changes over an extended period of time. This is not always the case for other oncogenes. For example, a number of MLL fusion proteins are sufficient to induce myeloid leukemia in both HSCs and myeloid progenitors.33,34
Due to the low frequency of common lymphoid progenitors (CLPs),43 we were unable to test directly whether CLPs expressing oncogenic Kras would be sufficient to induce T-cell malignancies. Given the fact that some T cells have self-renewal capability, it is possible that both HSCs and CLPs expressing oncogenic Kras are able to induce T-cell malignancies.
Our further study on malignant T cells and myeloid cells isolated from the same diseased animal carrying hematopoietic malignancies in both cell types showed that T cells and myeloid cells had different karyotypes. Because we transplanted no more than 2 functional HSCs per recipient mouse (“Results”), the possibility that the secondary genetic hits target 2 HSCs to induce both T-cell and myeloid malignancies is extremely slim. It is more likely that, following an oncogenic Kras mutation, secondary genetic hit(s) target lineage-specific progenitors to generate lineage-specific malignancies. In another words, in oncogenic Kras–induced T-cell lymphoma/leukemia and JMML, it is very likely that leukemic stem cells arise from lineage-committed progenitors rather than HSCs themselves.
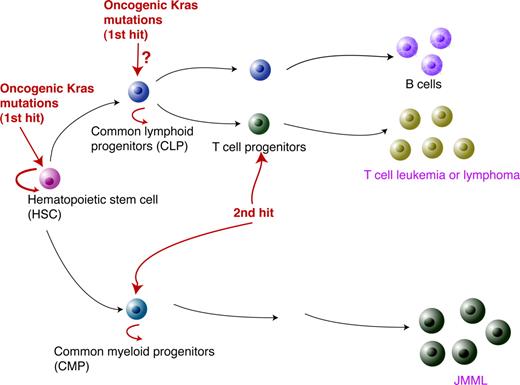
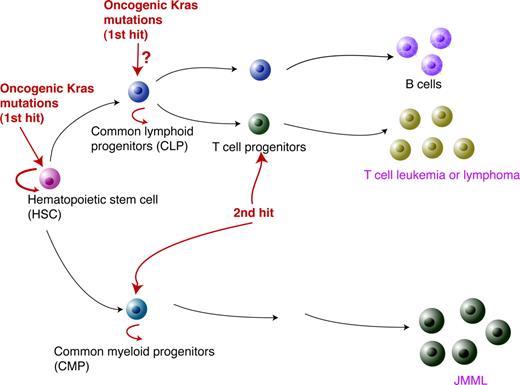
Based on our data, we propose the following model to explain oncogenic Kras-induced leukemogenesis (Figure 7). As the first genetic hit to initiate leukemogenesis, the oncogenic Kras mutation has to occur in HSCs. While the hematopoietic cells carrying this mutation keep proliferating and differentiating, secondary genetic hits occur in more linage-committed lymphoid or myeloid progenitors. In cooperation with the oncogenic Kras mutation, this leads in T cells to T-cell malignancies or in the myeloid compartment to JMML.
A model for oncogenic Kras-induced leukemogenesis. As the first genetic hit in leukemogenesis, oncogenic Kras mutations have to occur in HSCs. However, this event alone is insufficient to induce lineage-specific malignancies. The second hit is likely to occur in lineage-specific progenitors to initiate leukemogenesis.
A model for oncogenic Kras-induced leukemogenesis. As the first genetic hit in leukemogenesis, oncogenic Kras mutations have to occur in HSCs. However, this event alone is insufficient to induce lineage-specific malignancies. The second hit is likely to occur in lineage-specific progenitors to initiate leukemogenesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Drs David A. Tuveson and Tyler E. Jacks for generously providing us with conditional oncogenic Kras mice and Dr Fernando Camargo for kindly providing us with Mx1-cre mice. We are grateful to Drs Mark Kiel and Sean Morrison for their helpful suggestions of isolating HSCs based on SLAM receptor expression. We thank Drs Emery Bresnick, Qiang Chang, Norman Drinkwater, Shannon Kenny, and Bill Sugden for helpful discussion and critical comments on the manuscript. We are grateful to Dr Charles W. M. Roberts for generously providing us with the genomic southern probe to analyze TCRβ locus recombination and to Dr Jianzhu Chen for assessing the clonality of our lymphoma cell lines. We also thank Stacey Sullivan and Tony Chavarria for their help with mice.
This work was supported by a Howard Temin Award from the National Cancer Institute to J.Z. and National Institutes of Health grant PO1 HL 32262 to H.F.L.
National Institutes of Health
Authorship
Contribution: J.Z. designed and executed the experiments and wrote the manuscript; J.W. and Y.L. conducted the experiments; H.S. performed CGH analysis; K.H.Y. performed histopathology analysis; H.F.L. designed the experiments and wrote the manuscript; and M.F. performed histopathology analysis, designed the experiments, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jing Zhang, 1400 University Avenue, McArdle Lab/Room 417A, Madison, WI 53706; e-mail: zhang@oncology.wisc.edu.