Abstract
The human Mixed-Lineage-Leukemia-5 (MLL5) gene is located in a genomic region frequently deleted in patients with myeloid malignancies and encodes a widely expressed nuclear protein most closely related to MLL1, a Trithorax transcriptional regulator with established involvement in leukemogenesis. Although the physiologic function of MLL5 is completely unknown, domain structure and homology to transcriptional regulators with histone methyltransferase activity suggest a role in epigenetic gene regulation. To investigate physiologic functions of Mll5, we have generated a knockout mouse mutant using Cre/loxP technology. Adult homozygous Mll5-deficient mice are obtained at reduced frequency because of postnatal lethality. Surviving animals display a variety of abnormalities, including male infertility, retarded growth, and defects in multiple hematopoietic lineages. Interestingly, Mll5−/− mice die of sublethal whole-body irradiation but can be rescued with wild-type bone marrow grafts. Flow cytometric ana-lysis, bone marrow reconstitution, and in vivo BrdU-labeling experiments reveal numerical, functional, and cell-cycle defects in the lineage-negative Sca-1+, Kit+ (LSK) population, which contains short- and long-term hematopoietic stem cells. Together, these in vivo findings establish several nonredundant functions for Mll5, including an essential role in regulating proliferation and functional integrity of hematopoietic stem/progenitor cells.
Introduction
Loss of chromosome 7 or deletions on its long arm are common cytogenetic alterations in myeloid malignancies, which often indicate poor prognosis.1 Molecular mapping of a commonly deleted segment within chromosome band 7q22 has led to the identification of a hitherto unknown gene,2 which was shown to be expressed in all tissues analyzed and to encode a transcript with an open reading frame for 1858 amino acids. Over its entire length, the deduced protein was found 38% identical and 51% similar to MLL1, a common target of chromosomal translocations in human leukemias and the founding member of the mammalian MLL/trithorax family of transcriptional regulators.3 Based on homology and overall architecture of the protein product, the new gene was classified as the fifth MLL/trithorax member, and hence designated MLL5.2
MLL1 is the human ortholog of Drosophila trithorax. The Drosophila genome also harbors a homolog of MLL5, the functionally uncharacterized gene CG9007, encoding a predicted protein with virtually identical domain structure and 40% sequence similarity to MLL5.2 In the fly, Trithorax group proteins (Trx-G) are important for maintaining expression of Hox and other developmental genes through modification of chromatin. They are antagonized by proteins of the Polycomb group (Pc-G), which maintain repressed states. As part of large but distinct multicomponent protein complexes, Trx-G and Pc-G proteins control the maintenance, but not the initiation, of target gene expression and thus serve as epigenetic guardians of cell identity.4 Similar to Drosophila, mammalian Pc-G and Trx-G proteins act antagonistically on Hox genes,5 but there is also evidence for their involvement in cellular proliferation and tumorigenesis.6,7
A defining hallmark of MLL/Trx-G proteins is the concomitant presence of 2 distinct protein modules, the PHD and SET domains. PHD (plant homeodomain) is a conserved zinc finger motif that has been found in many eukaryotic proteins, notably in nuclear factors with chromatin regulatory functions.8,9 Increasing evidence suggests that a key function of PHD domains could be to tether partner proteins to chromatin by binding simultaneously to both.9,10 The second characteristic motif of MLL/Trx-G proteins, the SET domain (suppressor of variegation, enhancer of zeste, trithorax), has been recognized as an element that can confer lysine methyltransferase activity, and an increasing number of SET domain proteins have been shown to mediate methylation of specific lysine residues in histone tails.11,12 Epigenetic marking of transcriptional states thus seems a key function of SET domain proteins.
MLL5 contains single PHD and SET domains and lacks other obvious conserved sequence elements, exactly like its Drosophila homolog CG9007. Transfection studies in cell lines have demonstrated that MLL5 is a nuclear protein that forms speckled foci, and both up- and down-regulation of MLL5 transcripts inhibits cell cycle progression.13,14 No other information on potential functions of MLL5 is yet available, and murine Mll5 has not been described at all. Here we report the phenotype of adult mice that survive constitutive Mll5 deficiency. These animals exhibit several hematopoietic abnormalities. Most interestingly, homozygous Mll5−/− mice show increased sensitivity to ionizing radiation because of defects in the bone marrow stem/progenitor compartment. The observed phenotypes establish an important role of Mll5 in primitive hematopoietic cell function.
Methods
Additional methods are available online (Document S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Generation of Mll5-deficient mice
Relevant fragments of the mouse Mll5 gene were cloned from genomic DNA of E14.1 (129/Ola-derived) embryonic stem cells by polymerase chain reaction (PCR) using Expand Long Template PCR System (Roche Diagnostics, Mannheim, Germany). The complete sequence of the targeting vector (pVM-BK) can be obtained from H.J.F. on request. Mll5-deficient mice were generated from 2 independently targeted embryonic stem cell clones (87 and 315) following conventional methodology (Document S1). Animals heterozygous for the “knockin” allele (consisting of a loxP-flanked neomycin cassette positioned upstream of exon 3 and a third loxP site inserted downstream of exon 4) were intercrossed with animals of the ubiquitous deleter strain CMV-Cre15 to generate heterozygous knockout mice lacking the neomycin cassette, and exons 3 and 4 on one Mll5 allele. Mll5+/− heterozygotes were backcrossed 5 times with C57BL/6 partners before being intercrossed to generate homozygous Mll5−/− mice. All data presented in this study were obtained with offspring from heterozygous parents of the fifth C57BL/6 backcross. All animal experiments were approved by the Regierungsprasidium Tubingen Institutional Review Board.
Generation of polyclonal rabbit anti-Mll5 antisera
Peptide synthesis, coupling of peptides to Limulus hemocyanine as carrier, immunization of rabbits, and affinity purification of monospecific immunoglobulin Gs (IgGs) was done at BioGenes (Berlin, Germany). Polyclonal sera were raised successfully against amino acids 37 to 51 (KSNSFPHQLYTSSSH; epitope 1), 220 to 234 (VPKVTDKRRKKSGEK; epitope 2), and 1447 to 1458 (CPPSPHTENPPKSS; epitope 3) of the murine Mll5 protein, which are encoded by exons 2, 6, and 25, respectively.
Flow cytometry
A list of all antibodies and details of staining procedures are given in Document S1. Stained cells were analyzed and sorted on FACSCalibur or FACSAria cytometers (BD Biosciences, San Jose, CA) using CellQuest and DIVA software.
Peripheral blood cell counts
Blood was collected from the lateral tail vein of 10- to 14-week-old animals in ethylenediaminetetraacetic acid-coated vials (Sarstedt, Nürnbrecht, Germany). Samples were analyzed on an Advia 120 Hematology System (Bayer, Tarrytown, NY).
Bone marrow reconstitution experiments
Alymphoid (Rag2−/− γc−/−) recipients and congenic CD45.1 mice (B6.SJL-Ptprc(a)/BoAiTac) were purchased from Taconic Farms (Germantown, NY). Lin-negative donor cells were obtained by fluorescence-activated cell sorter (FACS)–sorting bone marrow (BM) cells negative for the following surface markers: CD3ϵ, CD4, CD8, CD11b, CD19, Gr1, NK1.1, and TER119. Donor and recipient mice were 9- to 12-week-old females. Noncompetitive reconstitution: 36 C57BL/6 mice received a lethal dose (2× 600 cGy; 3 hours between doses) of whole-body irradiation. Fourteen of the irradiated animals were reconstituted each with 3 × 104 FACS-sorted Lin-negative BM cells from 2 separate Mll5−/− donors (7 recipients per donor). Another 14 of the irradiated recipients received the same number of cells from respective wild-type (WT) littermates (again, 7 recipients per donor). The remaining 8 animals received phosphate-buffered saline (PBS) as control. Competitive reconstitution into (Rag2−/−γc−/−) or CD45.1-congenic recipients: FACS-sorted Lin-negative BM cells from Mll5−/− mice or WT littermates were thoroughly mixed with an equal number of Lin-negative competitor cells sorted from CD45.1 congenics. A total of 104 (for Rag2−/−γc−/− recipients) or 5 × 104 cells (for CD45.1-congenic recipients) were injected into the tail vein of each recipient irradiated either sublethally at 400 cGy (Rag2−/−γc−/− recipients) or lethally with a split dose (5 hours between doses) of 2 × 550 cGy (CD45.1 congenic recipients) using a 137Cs source. Recipients were given 1.1 g/L neomycin sulfate in drinking water for the first 12 weeks after injection. For analysis, mice were either bled from the tail vein or killed to obtain lymphoid organs. Contribution to hematopoietic compartments was determined using expression of CD45.1 and CD45.2 as marker.
Cell-cycle analysis
Mice were injected intraperitoneally with bromodeoxyuridine (BrdU; 1 mg of BrdU/animal; BD Biosciences) and killed 2 hours later. Lineage-negative BM cells were enriched using Dynabeads Sheep antirat IgG (Invitrogen, Dynal, Oslo, Norway), stained for surface markers to detect stem cells, and then fixed, permeabilized, and stained with 7-amino-actinomycin D and anti-BrdU antibody using the FITC BrdU Flow Kit (BD Biosciences) according to the manufacturer's protocol. For long-term incorporation, BrdU was administered orally (1 mg/mL in drinking water) for 7 days.
Statistical analysis
Results are presented as mean plus or minus SD for continuous variables and in absolute and relative frequencies for categorical variables. Statistical significance of differences between 2 groups was assessed using the 2-tailed unpaired t test, unless stated otherwise in the corresponding figure legends. P values less than .05 were considered significant.
Results
Homozygous inactivation of Mll5 results in partial lethality, reduced growth, and male sterility
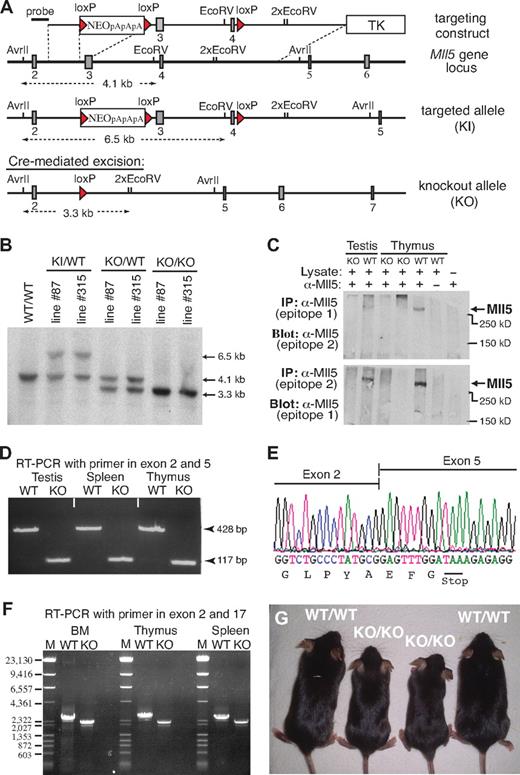
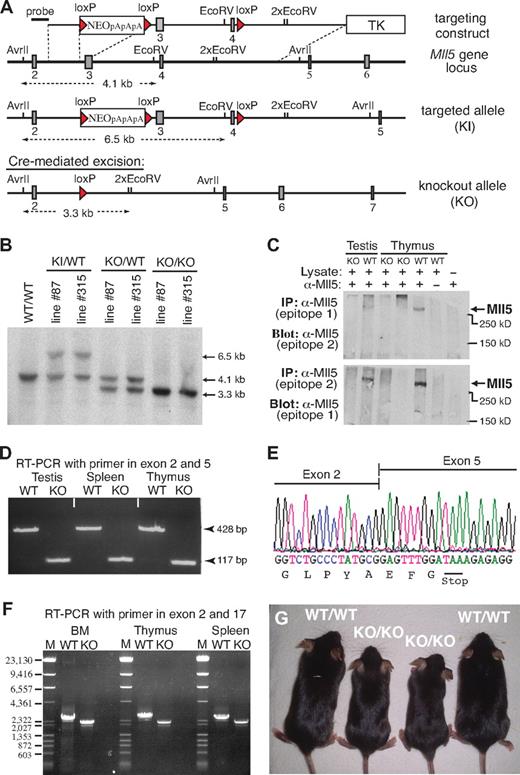
We identified the mouse Mll5 gene based on sequence homology with its recently described human ortholog.2,14 Like human MLL5, the mouse gene appears to be widely expressed, with transcripts being detectable in most, if not all, adult tissues (Document S1; Figure S1). To explore the physiologic role of this Trithorax-like gene, we generated 2 independent lines of Mll5-deficient mice by deleting exons 3 and 4 using Cre/loxP technology (Figure 1A). Southern blotting confirmed correct modification of Mll5 alleles in both mutant mouse lines (Mll5-87 and Mll5-315; Figure 1B). Excision of exons 3 and 4 was chosen as the most promising strategy to achieve complete inactivation of Mll5 because, together, both exons encode the unique PHD domain and loss of exons 3 and 4 shifts the reading frame in the Mll5 transcript, resulting in the formation of a translational stop codon after just 65 of 1868 amino acids (see discussion of RT-PCR and sequence analysis in the next paragraph).
Generation of Mll5-deficient mice. (A) Targeting strategy. Filled rectangles represent exons; red triangles, loxP sites. TK indicates thymidine kinase gene; NEO, neomycin resistance gene followed by 3 polyadenylation sites (pApApA); KI, knockin; KO, knockout. (B) Southern blot of AvrII/EcoRV-digested genomic DNA isolated from mice (tail biopsies) of the indicated lines and genotypes. The blot was hybridized with the external probe depicted in panel A. (C) Western blot of immunoprecipitated protein extracts from testis and thymus of WT and homozygous Mll5 knockout (KO) mice. Epitope 1 is encoded by exon 2 and epitope 2 by exon 6 of the Mll5 gene. (D) RT-PCR amplification of total RNA from testis, spleen, and thymus of an Mll5−/− mouse (KO) and a WT control. The PCR product of 117 bp demonstrates the absence of exons 3 and 4, encoding the PHD domain, and efficient splicing from exon 2 to exon 5 in knockout mice. (E) Sequence analysis of the splice junction in the 117-bp RT-PCR band shown in panel B. The nucleotide sequence confirms splicing between exon 2 and 5 in Mll5−/− mice and the resultant shift in the reading frame, which leads to the appearance of an in-frame stop codon (TAA). (F) RT-PCR amplification of total RNA from BM, thymus, and spleen of an Mll5−/− mouse (KO) and a WT control with primers located in exon 1 and exon 17. There is no evidence for splice products from exon 2 to any of the 12 exons after exon 5, essentially excluding the possibility of a mutated transcript with restored reading frame. For better resolution, all RT-PCR products were also analyzed by agarose gel electrophoresis after digestion with NcoI (data not shown), again with no evidence for additional splice products. The faint band immediately below the major RT-PCR product, present in WT and knockout tissues, corresponds to an in-frame splice variant lacking exon 16 (data not shown). The schematic structure of the expected mRNA in WT and Mll5−/− mice (KO) along with the location of exons, PCR primers, and sizes of PCR products is shown in Figure S2. (G) Postnatal growth defects in Mll5-deficient mice: 2 pairs of 6-week-old Mll5+/+ (WT/WT) and Mll5−/− (KO/KO) male littermates. See also Figure S3.
Generation of Mll5-deficient mice. (A) Targeting strategy. Filled rectangles represent exons; red triangles, loxP sites. TK indicates thymidine kinase gene; NEO, neomycin resistance gene followed by 3 polyadenylation sites (pApApA); KI, knockin; KO, knockout. (B) Southern blot of AvrII/EcoRV-digested genomic DNA isolated from mice (tail biopsies) of the indicated lines and genotypes. The blot was hybridized with the external probe depicted in panel A. (C) Western blot of immunoprecipitated protein extracts from testis and thymus of WT and homozygous Mll5 knockout (KO) mice. Epitope 1 is encoded by exon 2 and epitope 2 by exon 6 of the Mll5 gene. (D) RT-PCR amplification of total RNA from testis, spleen, and thymus of an Mll5−/− mouse (KO) and a WT control. The PCR product of 117 bp demonstrates the absence of exons 3 and 4, encoding the PHD domain, and efficient splicing from exon 2 to exon 5 in knockout mice. (E) Sequence analysis of the splice junction in the 117-bp RT-PCR band shown in panel B. The nucleotide sequence confirms splicing between exon 2 and 5 in Mll5−/− mice and the resultant shift in the reading frame, which leads to the appearance of an in-frame stop codon (TAA). (F) RT-PCR amplification of total RNA from BM, thymus, and spleen of an Mll5−/− mouse (KO) and a WT control with primers located in exon 1 and exon 17. There is no evidence for splice products from exon 2 to any of the 12 exons after exon 5, essentially excluding the possibility of a mutated transcript with restored reading frame. For better resolution, all RT-PCR products were also analyzed by agarose gel electrophoresis after digestion with NcoI (data not shown), again with no evidence for additional splice products. The faint band immediately below the major RT-PCR product, present in WT and knockout tissues, corresponds to an in-frame splice variant lacking exon 16 (data not shown). The schematic structure of the expected mRNA in WT and Mll5−/− mice (KO) along with the location of exons, PCR primers, and sizes of PCR products is shown in Figure S2. (G) Postnatal growth defects in Mll5-deficient mice: 2 pairs of 6-week-old Mll5+/+ (WT/WT) and Mll5−/− (KO/KO) male littermates. See also Figure S3.
To verify loss of Mll5 at the protein level, we raised polyclonal rabbit antisera against 3 distinct epitopes encoded by exon 2, 6, and 25, respectively. Although all 3 affinity-purified sera specifically recognized recombinant GFP-Mll5 fusion proteins in cell lines transiently transfected with appropriate expression constructs (data not shown), we failed to detect endogenous Mll5 protein by conventional Western blotting in any tissue tested (lung, testis, spleen). Inability to detect the endogenous protein by Western blotting has also been reported for human MLL5 and proposed to reflect low protein stability.14 Consistent with this hypothesis, using immunoprecipitation to enrich for Mll5, we succeeded to visualize a single band corresponding in size to full-length Mll5 (Figure 1C). Importantly, no such band was detectable in mice with a homozygous deletion of exons 3 and 4, although the epitopes recognized by our sera are located outside the deleted region. These findings are fully in line with reverse-transcribed polymerase chain reaction (RT-PCR) and sequence analysis data (Figure 1D-F; Figure S2), which confirm the absence of in-frame Mll5 transcripts in mutant mice, and thus demonstrate that we have generated a true Mll5-null mutant.
Mice heterozygous for the targeted allele were outwardly indistinguishable from WT littermates and were obtained at the expected Mendelian frequency in backcrosses with both C57BL/6 and 129/Ola partners. To reduce potential variability in phenotypes resulting from genetic background, all subsequent studies were done with offspring from parents of 5th backcross onto C57BL/6. Interestingly, the number of homozygous mutant offspring from Mll5+/− intercrosses was significantly reduced at the time of weaning, irrespective of gender (Table 1). Genotyping of embryos from timed matings between heterozygous parents did not reveal enhanced lethality at any stage of embryonic development. For instance, at embryonic day 18.5, that is, a few hours before birth, viable Mll5−/− fetuses were present at expected numbers (Table 2). A reduction in the percentage of homozygous Mll5-deficient pups became apparent only among newborns 1 day postpartum (Table 2). A further decline in the number of Mll5−/− pups was observed in the following few days (data not shown). Autopsy of recently deceased animals did not reveal any macroscopic defects that could account for the premature death of approximately 40% of Mll5−/− pups. Our genotype analyses of embryos and newborns thus demonstrate partial lethality among Mll5−/− offspring within the first days of postnatal life. Surviving animals from both independently generated knockout lines (Mll5-87 and Mll5-315) exhibited indistinguishable phenotypes and were used with respective littermate controls interchangeably for all subsequent analyses.
Mll5−/− pups surviving the first days after birth developed into apparently healthy adults. After weaning, no difference in viability between Mll5−/− mice and WT littermates was noticeable, at least during the 9- to 14-week period, after which most animals were killed for experiments. However, within litters, most Mll5−/− animals could be readily identified by their reduced size (Figure 1G). At 6 weeks of age, the average weight difference between Mll5−/− mutants and littermate controls was 16% for males and 12% for females (Figure S3A). Weight differences persisted during pubertal growth and adulthood. Consistent with the normal frequency of Mll5−/− mice before birth, fetuses at day 18.5 of gestation did not exhibit any genotype-specific weight differences (Figure S3B). In marked contrast to Mll1-mutant mice, which suffer from several bone malformations even when heterozygous,16 no skeletal defects were visible in Mll5-deficient embryos (Figure S3C). Taken together, our data suggest that Mll5 is not essential for embryonic development and that physiologically relevant deficits of Mll5 deficiency manifest only after birth.
Mating of Mll5−/− males with Mll5+/− or Mll5−/− females never resulted in pregnancies. To specifically assess male fertility, 9 young (6- to 8-week-old) Mll5−/− males were housed over a period of 7 to 12 months with 2 C57BL/6 females each. Whereas Mll5+/− controls were fertile, none of the homozygous Mll5-deficient males ever sired offspring. Histologic and molecular analyses of testis and epididymi revealed a severe defect in spermiogenesis, resulting in almost complete absence of mature sperm (Figure S4; Document S1). In contrast to mutant males, not all Mll5−/− females were completely infertile, but overall reproductive performance was severely impaired as well (data not shown).
Mice lacking Mll5 exhibit multiple hematopoietic defects
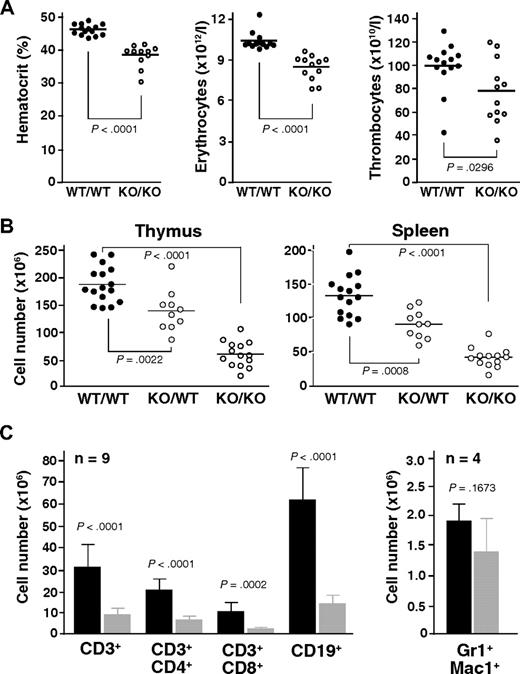
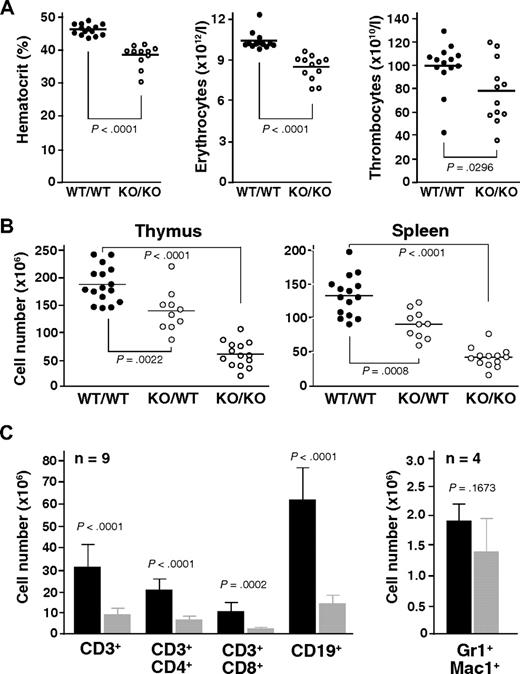
Automated analysis of peripheral blood revealed a consistently decreased hematocrit along with reduced erythrocyte counts in Mll5−/− mice (Figure 2A). Thrombocyte and leukocyte numbers were also decreased on average but varied much more between individual animals. The reductions in hematocrit and red blood cell counts were not associated with changes in erythrocyte, leukocyte, or thrombocyte morphology, as determined by manual examination of May-Grünwald-Giemsa-stained blood smears and automated measurements of mean cell volume (data not shown).
Mll5 mutant mice exhibit reduced blood cell counts and hypocellularity in spleen and thymus. (A) Result of automated blood analysis for hematocrit, erythrocyte, and thrombocyte numbers. (B) Total number of thymocytes and nucleated splenocytes in mice with the indicated genotypes. Only data from litters with at least 1 WT and 1 Mll5−/− animal are shown. (C) Total number of T cells (CD3+), B cells (CD19+), and myeloid cells (Gr1+Mac1+) in spleens of Mll5−/− mice (▩) and WT littermates (■).
Mll5 mutant mice exhibit reduced blood cell counts and hypocellularity in spleen and thymus. (A) Result of automated blood analysis for hematocrit, erythrocyte, and thrombocyte numbers. (B) Total number of thymocytes and nucleated splenocytes in mice with the indicated genotypes. Only data from litters with at least 1 WT and 1 Mll5−/− animal are shown. (C) Total number of T cells (CD3+), B cells (CD19+), and myeloid cells (Gr1+Mac1+) in spleens of Mll5−/− mice (▩) and WT littermates (■).
Gross inspection of internal organs revealed a specific reduction in the size of lymph nodes, thymus, and spleen in animals lacking Mll5, much more than can be accounted for by the general reduction in body weight (data not shown). Whereas the architecture of thymus and spleen was generally maintained (Figure S5), the number of thymocytes and splenocytes was strongly reduced, averaging less than 30% that of sex-matched WT littermates (Figure 2B). Interestingly, we also observed an intermediate reduction of total thymocyte and splenocyte numbers in heterozygous Mll5 mutant mice, when littermates of all 3 genotypes were compared, clearly indicating that Mll5 haploinsufficiency is not without phenotypic consequences. Flow cytometry of Mll5−/− splenocytes revealed strong reductions in the total number of B and T cells (Figure 2C). γδ T cells were present but also reduced (data not shown). Analysis of the T-cell receptor repertoire with a panel of anti-Vα and anti-Vβ antibodies did not reveal any significant differences in the usage of V elements, comparing peripheral T cells or single-positive thymocytes from Mll5−/− mice and WT controls (Figure S6), arguing against major defects in V(D)J recombination as explanation for the observed deficiency in mature T cells. The absolute number of Gr1/Mac1-positive myeloid splenocytes was on average only mildly reduced, suggesting that Mll5 deficiency may be more detrimental for cells of lymphoid descent (see “Discussion”).
Consistent with the observed peripheral T-cell deficiency, thymopoiesis was found markedly impaired in mutant mice. Flow cytometric analysis of Mll5−/− thymocytes with antibodies against CD4 and CD8 coreceptors revealed hypocellularity in all 4 developmental subsets, with CD4+CD8+ cells being affected the most (Figure 3A). We also probed for specific defects within the CD4−CD8− double-negative compartment, which contains the most immature thymocyte populations,17 by analyzing these cells for surface expression of CD44 and CD25 molecules. CD44 and CD25 mark discrete developmental subsets, progressing from CD44+CD25− (so-called double-negative 1 = DN1) to CD44+CD25+ (DN2) to CD44−CD25+ (DN3) and finally CD44−CD25− (DN4) maturational stages. Whereas hypocellularity was apparent in all 4 subsets, the most dramatic reduction in total numbers (average, 7.0-fold) was found in the DN2, and to a somewhat lesser extent (average, 3.6-fold) in the DN1 subset, ie, affecting the 2 earliest developmental stages (Figure 3B). Because the DN1 subpopulation is known to be heterogeneous with less than 50% of cells constituting true, mainstream thymocyte precursors, we included Kit (CD117) as an additional marker in the analysis.18 Interestingly, the most dramatic reduction (average, 16.9-fold) in total cell number was consistently found in the Kit-expressing subset, ie, the earliest thymocyte progenitor population (Figure 3C). In addition to the defects in T lymphopoiesis, flow cytometry of BM also revealed significant reductions in the cellularity of pro- and pre-B cell stages (Document S1; Figure S7), indicating perturbations in B-cell development as well.
Impaired lymphopoesis in the absence of Mll5. (A-C) Flow cytometric analysis of thymocytes from 10- to 14-week-old Mll5−/− mice (KO/KO) and WT littermates (WT/WT). (Left dot plots) Data from 1 representative staining. Total thymocyte numbers are given on top of the CD4/CD8 panels. (Right bar graphs) Total number of cells with the indicated immunophenotype, as average per animal plus or minus SD (n = 7).  represents Mll5−/− mice; ■, WT littermates. (D) Competitive BM reconstitution into alymphoid Rag2−/−γc−/− recipients. FACS-sorted Lin-negative BM cells (CD45.2) from Mll5−/− mice (KO) or WT littermates were mixed with an equal number of Lin-negative competitor cells (CD45.1) and injected into sublethally irradiated (400 cGy) recipients. Individual recipients reconstituted with Mll5−/− donor cells (○) or donor cells from WT littermates (●) are shown (data from 2 independent experiments combined).
represents Mll5−/− mice; ■, WT littermates. (D) Competitive BM reconstitution into alymphoid Rag2−/−γc−/− recipients. FACS-sorted Lin-negative BM cells (CD45.2) from Mll5−/− mice (KO) or WT littermates were mixed with an equal number of Lin-negative competitor cells (CD45.1) and injected into sublethally irradiated (400 cGy) recipients. Individual recipients reconstituted with Mll5−/− donor cells (○) or donor cells from WT littermates (●) are shown (data from 2 independent experiments combined).
Impaired lymphopoesis in the absence of Mll5. (A-C) Flow cytometric analysis of thymocytes from 10- to 14-week-old Mll5−/− mice (KO/KO) and WT littermates (WT/WT). (Left dot plots) Data from 1 representative staining. Total thymocyte numbers are given on top of the CD4/CD8 panels. (Right bar graphs) Total number of cells with the indicated immunophenotype, as average per animal plus or minus SD (n = 7).  represents Mll5−/− mice; ■, WT littermates. (D) Competitive BM reconstitution into alymphoid Rag2−/−γc−/− recipients. FACS-sorted Lin-negative BM cells (CD45.2) from Mll5−/− mice (KO) or WT littermates were mixed with an equal number of Lin-negative competitor cells (CD45.1) and injected into sublethally irradiated (400 cGy) recipients. Individual recipients reconstituted with Mll5−/− donor cells (○) or donor cells from WT littermates (●) are shown (data from 2 independent experiments combined).
represents Mll5−/− mice; ■, WT littermates. (D) Competitive BM reconstitution into alymphoid Rag2−/−γc−/− recipients. FACS-sorted Lin-negative BM cells (CD45.2) from Mll5−/− mice (KO) or WT littermates were mixed with an equal number of Lin-negative competitor cells (CD45.1) and injected into sublethally irradiated (400 cGy) recipients. Individual recipients reconstituted with Mll5−/− donor cells (○) or donor cells from WT littermates (●) are shown (data from 2 independent experiments combined).
To probe for functional deficits specifically in lymphopoiesis, we performed competitive reconstitution experiments using combined Rag-2 and cytokine receptor common γ subunit (Rag2−/− γc−/−) deficient mice as recipients. Because these animals are unable to generate their own T, B, and NK cells,19,20 they cannot reject BM grafts. In addition, all lymphocytes found in such animals after BM reconstitution are of donor origin. In 2 independent experiments, lineage-depleted BM cells from an Mll5−/− mouse or a WT littermate were mixed at a 1:1 ratio with lineage-depleted competitor cells derived from BM of congenic CD45.1+ mice, and injected intravenously into sublethally (400 cGy) irradiated Rag2−/−γc−/− recipients. When white blood cells from reconstituted animals were recovered at regular intervals and stained with antibodies against CD3 (pan T-cell marker), CD19 (pan B-cell marker), and CD45.2 (noncompetitor donor marker), a consistent disadvantage of Mll5−/− cells with regard to B- as well as T-lineage reconstitution was readily apparent (Figure 3D). In accordance with our cytofluorometric data, these findings document intrinsic developmental defects in B and T lymphopoiesis.
In addition to the profound defects in T- and B-cell development, we observed an approximately 40% reduction in the total number of Lin−/Kit+/Sca-1− BM cells, which include erythro-myeloid progenitors.21 Specifically, the total number of cells in populations generally considered as common myeloid, megakaryocyte-erythrocyte, and granulocyte-monocyte progenitors were reduced in the absence of Mll5 (Figure S8). However, we did not find evidence for additional or more specific perturbations in myelopoiesis in the BM of Mll5−/− mice, at least under steady-state conditions.
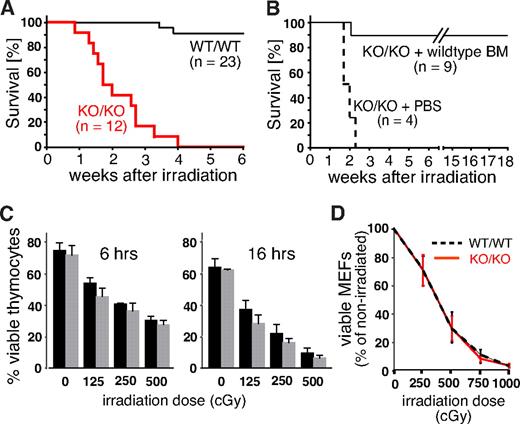
Mll5-deficient mice are hypersensitive to ionizing radiation
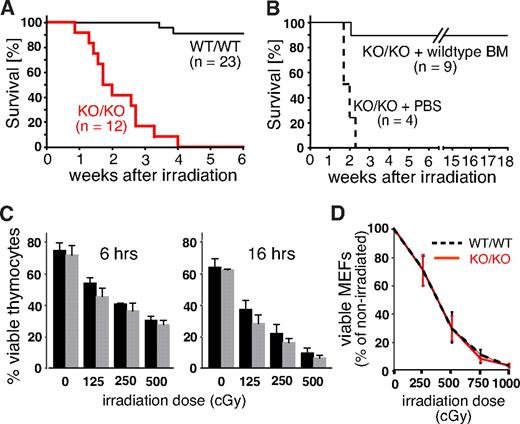
Growth retardation, impaired fertility, and defects in lymphopoiesis are, when combined, a phenotype often associated with genetic defects in the DNA repair machinery. To reveal potential defects in the response to DNA damage, we exposed Mll5−/− mice and control littermates to whole-body γ irradiation (WBI). At a sublethal dose of 750 cGy, only 2 of 23 WT animals died within the 6-week observation period. Strikingly, all 12 homozygous Mll5-deficient animals died of WBI at a median time of 13 days (Figure 4A). To test whether the increased radiation sensitivity of Mll5-deficient mice is solely the result of BM failure, or, alternatively, to cumulative systemic effects on other organs, we irradiated 13 age-matched Mll5−/− mice (750 cGy) and injected 9 of them intravenously with 104 lineage-depleted WT BM cells immediately after WBI. The remaining 4 animals received PBS as control. Whereas all 4 PBS-treated Mll5−/− mice died within 16 days, 8 of 9 BM-reconstituted mutants were rescued and survived beyond 18 weeks after irradiation (Figure 4B). Thus, sublethally irradiated Mll5−/− mice die specifically because of BM failure and not because of the sensitivity of other tissues to WBI.
Mll5−/− mice die of sublethal WBI because of BM failure. (A) Survival curves of Mll5−/− mice (KO/KO) and WT littermate controls (WT/WT) exposed to 750 cGy of WBI (P < .001; log rank test). (B) Survival curves of Mll5−/− mice reconstituted with Lin-negative WT C57BL/6 BM cells or PBS immediately after WBI with 750 cGy (P < .001; log rank test). (C) Similar radiation sensitivity of Mll5−/− ( ) and WT (■) thymocytes. The number of viable cells was determined 6 and 16 hours after irradiation by FACS using propidium iodide and annexin V staining. (D) There was no difference in radiation sensitivity between MEFs isolated from day 14.5 Mll5−/− (red solid line) and WT (black dashed line) embryos. The number of viable cells was determined 6 days after irradiation by trypan blue exclusion. Data in panels C and D summarize 3 independent experiments with cells from 3 different mice/embryos of each genotype.
) and WT (■) thymocytes. The number of viable cells was determined 6 and 16 hours after irradiation by FACS using propidium iodide and annexin V staining. (D) There was no difference in radiation sensitivity between MEFs isolated from day 14.5 Mll5−/− (red solid line) and WT (black dashed line) embryos. The number of viable cells was determined 6 days after irradiation by trypan blue exclusion. Data in panels C and D summarize 3 independent experiments with cells from 3 different mice/embryos of each genotype.
Mll5−/− mice die of sublethal WBI because of BM failure. (A) Survival curves of Mll5−/− mice (KO/KO) and WT littermate controls (WT/WT) exposed to 750 cGy of WBI (P < .001; log rank test). (B) Survival curves of Mll5−/− mice reconstituted with Lin-negative WT C57BL/6 BM cells or PBS immediately after WBI with 750 cGy (P < .001; log rank test). (C) Similar radiation sensitivity of Mll5−/− ( ) and WT (■) thymocytes. The number of viable cells was determined 6 and 16 hours after irradiation by FACS using propidium iodide and annexin V staining. (D) There was no difference in radiation sensitivity between MEFs isolated from day 14.5 Mll5−/− (red solid line) and WT (black dashed line) embryos. The number of viable cells was determined 6 days after irradiation by trypan blue exclusion. Data in panels C and D summarize 3 independent experiments with cells from 3 different mice/embryos of each genotype.
) and WT (■) thymocytes. The number of viable cells was determined 6 and 16 hours after irradiation by FACS using propidium iodide and annexin V staining. (D) There was no difference in radiation sensitivity between MEFs isolated from day 14.5 Mll5−/− (red solid line) and WT (black dashed line) embryos. The number of viable cells was determined 6 days after irradiation by trypan blue exclusion. Data in panels C and D summarize 3 independent experiments with cells from 3 different mice/embryos of each genotype.
Mll5 is expressed in a wide variety of different cell types, including thymocytes and embryonic fibroblasts (Figure S1C,D). Thymocytes are highly sensitive to DNA damage–induced cell death22,23 and thus could serve as an accessible cell population for molecular studies. Similarly, mouse embryonic fibroblasts (MEFs) are frequently used as primary cell lines to demonstrate and study ubiquitous DNA repair defects in specific mouse mutants. To test whether cells outside the BM also exhibit increased radiation sensitivity in the absence of Mll5, we thus compared survival of thymocytes and day 14.5 MEFs from WT and Mll5−/− mice/embryos after γ irradiation, but no striking differences were observed (Figure 4C,D). Taken together, these results strongly suggest that Mll5 plays a vital role in the response to DNA-damaging γ radiation, which, however, appears restricted to specific cell types, including the hematopoietic stem/progenitor population.
Phenotypic and functional defects in the hematopoietic stem/progenitor cell compartment of mice lacking Mll5
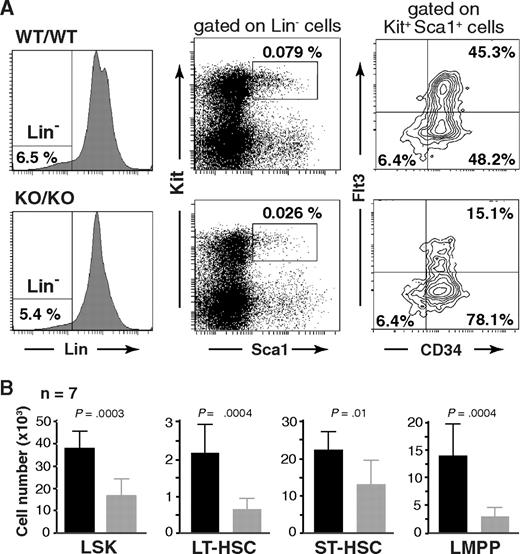
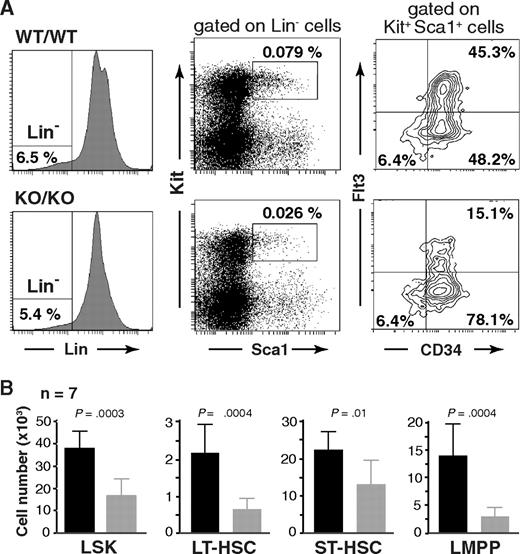
To investigate a potential role of Mll5 in hematopoietic stem cell (HSC) biology, we first determined the number and immunophenotype of lineage-negative (Lin−), Sca-1+, Kit+ cells, commonly referred to as the L(in)S(ca)K(it) population,24 which contains HSCs and early progenitors. Interestingly, whereas the overall expression profile of Sca-1 and Kit seemed unaltered, the total number of LSK cells was consistently reduced more than 2-fold in the absence of Mll5 (Figure 5). Further fractionation of LSK cells based on CD34 and Flt3 expression25-27 revealed a similar reduction in the total number of long-term HSCs (LT-HSC: CD34−Flt3−) and an approximately 4-fold reduction of lymphoid-primed multipotent progenitors (LMPP: CD34+Flt3+).28 The total number of LSK CD34+Flt3− cells, commonly considered as HSC with short-term reconstituting potential (ST-HSC), was least affected in Mll5−/− mice (on average, ∼ 25% reduced), which is reflected in an increased percentage of these cells relative to the other 2 LSK subpopulations (Figure 5A right panels). The reduced cellularity in the LMPP compartment is consistent with respect to the observed defects in lymphoid development, as reductions in early T- and B-cell progenitors have been reported to be associated with loss of Flt329 or its ligand.30
Phenotypic defects in the HSC compartment of Mll5−/− mice. (A) Reduced frequency of Lin−/Sca1+/Kit+ (LSK) cells (middle panels) and altered CD34/Flt3 subset composition (right panels) in Mll5−/− mice. (Left panels) Lineage gates. Plots from one representative pair of animals are shown. (B) Total number of cells in the respective HSC subpopulation, as average per animal (2 tibiae, 2 femora). LT-HSC, CD34−Flt3− LSK subset; ST-HSC, CD34+Flt3− LSK subset; LMPP, CD34+Flt3+ LSK subset.
Phenotypic defects in the HSC compartment of Mll5−/− mice. (A) Reduced frequency of Lin−/Sca1+/Kit+ (LSK) cells (middle panels) and altered CD34/Flt3 subset composition (right panels) in Mll5−/− mice. (Left panels) Lineage gates. Plots from one representative pair of animals are shown. (B) Total number of cells in the respective HSC subpopulation, as average per animal (2 tibiae, 2 femora). LT-HSC, CD34−Flt3− LSK subset; ST-HSC, CD34+Flt3− LSK subset; LMPP, CD34+Flt3+ LSK subset.
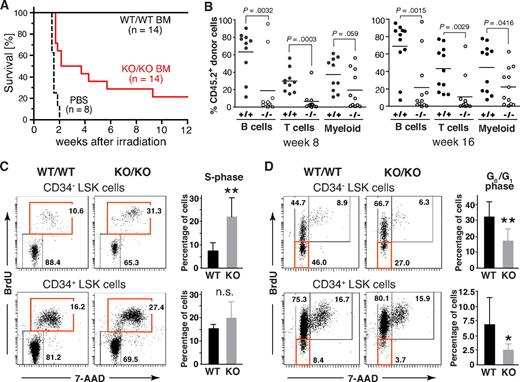
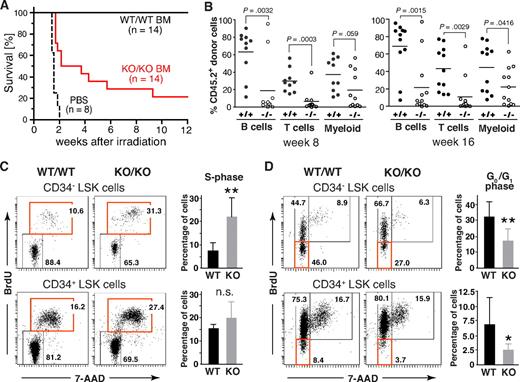
To assess the functional properties of HSCs from Mll5−/− mice, we first tested their ability to rescue lethally irradiated WT recipients. As expected, all mice, which received PBS as control, died within 2 weeks after WBI (12 Gy). Interestingly, whereas all 14 animals that had received 3 × 104 FACS-sorted Lin-negative BM cells from WT donors survived, 11 of 14 recipients reconstituted with the same number of Mll5−/− cells died (Figure 6A). These data demonstrate an intrinsic functional defect of Lin-negative BM cells lacking Mll5, which compromises their ability to efficiently rescue lethally irradiated recipients.
Phenotypic defects in the HSC compartment of Mll5−/− mice are associated with reduced reconstitution potential and increased cycling of CD34− LSK cells. (A) Inefficient rescue of lethally irradiated mice with Mll5−/− BM grafts. C57BL/6 females were lethally irradiated and grafted with FACS-sorted Lin-negative BM cells from either Mll5−/− mice (KO/KO) or WT littermates (WT/WT). Results are a composite of 2 independent experiments (P < .001; log rank test). (B) Competitive reconstitution. Lin-negative donor (CD45.2) BM cells were mixed with an equal number of Lin-negative CD45.1 competitor cells and injected into lethally irradiated CD45.1 mice. Peripheral blood was analyzed 8 (left) and 16 weeks (right) after reconstitution by FACS for donor-derived B lymphoid (CD19), T lymphoid (CD3), and myeloid (Gr1+/Mac1+) cells. Individual recipients reconstituted with Mll5−/− donor cells (○) or donor cells from WT littermates (●) are shown (data from 2 independent experiments combined). (C,D) Cell cycle analysis of LSK cells by in vivo BrdU labeling. (C) Increased cycling of CD34-negative LSK cells in the absence of Mll5 as assessed by 7-amino-actinomycin D/BrdU staining and FACS analysis 2 hours after intraperitoneal injection of BrdU into Mll5-deficient (KO/KO) mice and WT (WT/WT) littermate controls. (Left panels) Representative dot plots from 1 of 4 independent experiments. (Right bar graphs) Average percentage of BrdU-labeled cells in the CD34-negative (top) and CD34-positive (bottom) LSK compartment (**P < .05; n.s. indicates not significant; n = 4). (D) Reduced frequency of noncycling LSK cells in the absence of Mll5. BrdU was given orally to Mll5−/− (KO/KO) mice and WT littermate controls (WT/WT) for 7 days. BrdU incorporation in CD34− (top) and CD34+ LSK cells was evaluated by FACS analysis. (Left panels) Representative dot plots from 1 of 4 independent experiments. (Right bar graphs) Average percentage of noncycling (BrdU-negative) cells in the CD34-negative (top) and CD34-positive (bottom) LSK compartment (**P < .05, *P = .12; n = 4).
Phenotypic defects in the HSC compartment of Mll5−/− mice are associated with reduced reconstitution potential and increased cycling of CD34− LSK cells. (A) Inefficient rescue of lethally irradiated mice with Mll5−/− BM grafts. C57BL/6 females were lethally irradiated and grafted with FACS-sorted Lin-negative BM cells from either Mll5−/− mice (KO/KO) or WT littermates (WT/WT). Results are a composite of 2 independent experiments (P < .001; log rank test). (B) Competitive reconstitution. Lin-negative donor (CD45.2) BM cells were mixed with an equal number of Lin-negative CD45.1 competitor cells and injected into lethally irradiated CD45.1 mice. Peripheral blood was analyzed 8 (left) and 16 weeks (right) after reconstitution by FACS for donor-derived B lymphoid (CD19), T lymphoid (CD3), and myeloid (Gr1+/Mac1+) cells. Individual recipients reconstituted with Mll5−/− donor cells (○) or donor cells from WT littermates (●) are shown (data from 2 independent experiments combined). (C,D) Cell cycle analysis of LSK cells by in vivo BrdU labeling. (C) Increased cycling of CD34-negative LSK cells in the absence of Mll5 as assessed by 7-amino-actinomycin D/BrdU staining and FACS analysis 2 hours after intraperitoneal injection of BrdU into Mll5-deficient (KO/KO) mice and WT (WT/WT) littermate controls. (Left panels) Representative dot plots from 1 of 4 independent experiments. (Right bar graphs) Average percentage of BrdU-labeled cells in the CD34-negative (top) and CD34-positive (bottom) LSK compartment (**P < .05; n.s. indicates not significant; n = 4). (D) Reduced frequency of noncycling LSK cells in the absence of Mll5. BrdU was given orally to Mll5−/− (KO/KO) mice and WT littermate controls (WT/WT) for 7 days. BrdU incorporation in CD34− (top) and CD34+ LSK cells was evaluated by FACS analysis. (Left panels) Representative dot plots from 1 of 4 independent experiments. (Right bar graphs) Average percentage of noncycling (BrdU-negative) cells in the CD34-negative (top) and CD34-positive (bottom) LSK compartment (**P < .05, *P = .12; n = 4).
We also performed competitive stem cell reconstitution experiments, using allelic differences at the CD45 locus to distinguish donor from competitor/recipient cells. For these experiments, FACS-sorted Lin-negative BM cells from either Mll5−/− mice or WT littermates (CD45.2 background) were mixed at a 1:1 ratio with the equivalent cell population of WT C57BL/6.SJL mice (CD45.1 background) and injected intravenously into lethally irradiated C57BL/6.SJL recipients, which allow concomitant evaluation of both lymphoid and myeloid reconstitution, which had not been possible in Rag2−/−γc−/− recipients because of undefined CD45 background. Flow cytometric analysis of peripheral blood at 4, 8, 12, and 16 weeks after reconstitution revealed significant differences between WT and Mll5−/− donors. Although Lin-negative BM cells from both Mll5 genotypes were able to generate B cells (CD19+), T cells (CD3+), and granulocytes (Gr1+/Mac1+), even under competitive conditions, Mll5−/− BM cells exhibited a markedly decreased capacity to reconstitute B- and T-cell populations (Figure 6B), confirming our earlier data with Rag2−/−γc−/− recipients. Whereas the generation of Gr1+/Mac1+ myeloid cells was impaired as well, the effect was less striking. Of note, Mll5 deficiency did not influence the homing potential of sorted Lin-negative BM cell populations, as assessed in short-term transfer experiments with 2 different read-outs, that is, direct detection of fluorescently labeled donor cells by FACS 16 to 18 hours after transfer and quantification of homed progenitors in methylcellulose assays (Figure S9). Taken together, these findings highlight a nonredundant and cell-autonomous role of Mll5 in maintaining hematopoietic stem/progenitor cell functionality.
Impaired function of hematopoietic stem/progenitor cells is frequently associated with excessive cell cycling.31-34 To address a potential role for Mll5 in hematopoietic stem/progenitor cell proliferation, we performed in vivo BrdU labeling experiments. Interestingly, the percentage of LSK CD34− cells (LT-HSCs) that had incorporated BrdU over a 2-hour period was consistently increased approximately 3-fold in Mll5-deficient mice versus WT controls (Figure 6C). To evaluate the frequency of noncycling stem cells, BrdU was administered to mice orally for 7 days. Strikingly, the percentage of quiescent (G0/G1) cells in the LSK compartment was consistently reduced by 40% to 60% in the absence of Mll5 (Figure 6D). Taken together, our findings demonstrate that loss of Mll5 results in reduced numbers, impaired function, and excessive cycling of primitive hematopoietic cells.
Discussion
The candidate leukemia suppressor MLL5 is the latest addition to the mammalian Trithorax/MLL gene family.2,14 However, no information on physiologic functions of MLL5 or its murine ortholog has been available to date. Here we report the generation and characterization of mice lacking Mll5 constitutively. Heterozygous mutants are fully viable and fertile. When intercrossed, morphologically normal embryos of all 3 possible genotypes are found at Mendelian frequencies up to day 18.5 of gestation, indicating that Mll5 is not essential for embryonic development. However, soon after birth, approximately 40% of Mll5−/− pups die for as yet unknown reasons. Penetrance of postnatal lethality and severity of some other phenotypic traits may be influenced by genetic background (V.M. and H.J.F., unpublished data, October 5, 2008). To reduce genetic variability, all studies reported here were done with offspring from parents of fifth C57BL/6 backcross. Interestingly, Mll5−/− mice surviving postnatal lethality appear as healthy and active as WT littermates. To date, targeted null mutations have been described only for 2 of the 5 known Mll/Trithorax family members, including 3 independently mutated alleles for Mll116,35,36 and a null allele for Mll2.37 In all cases, homozygous mutation resulted in complete embryonic lethality. Mll5−/− mice are thus the first in which consequences of an Mll/Trithorax gene deficiency can be studied in an adult mammal.
Mll5−/− mice are mildly growth-retarded, display multiple hematopoietic defects, in particular in the lymphoid compartment, and reduced fertility, with males being completely sterile. This phenotype combination is often observed in mutants with defects in the homologous recombination pathway of DNA repair. Intriguingly, Mll5−/− mice proved hypersensitive to whole-body γ irradiation (WBI). To this point, the phenotype overlaps markedly with mouse mutants lacking Atm,38 53Bp1,39 or histone H2ax,40,41 important players in DNA double-strand break repair. However, in contrast to these latter mutants, Mll5 deficiency does not seem to sensitize all tissues. For instance, MEFs and thymocytes lacking Mll5 do not exhibit significantly increased sensitivity to γ-radiation or DNA interstrand cross-linking agents, like mitomycin C (V.M. and H.J.F., unpublished data, October 5, 2008). Furthermore, adoptive transfer of WT BM is sufficient to rescue Mll5−/− mice from death after WBI, also arguing against generalized DNA repair defects. Whether Mll5 participates in a pathway of DNA damage control that is confined to hematopoietic pro-genitor populations, as demonstrated recently for the transcriptional repressor Slug,42 remains to be determined.
Besides increased radiation sensitivity, Mll5−/− hematopoietic stem/progenitor cell populations exhibit several additional phenotypic and functional deficits. The total number of LSK cells, which contain HSCs and early progenitors, is consistently reduced by more than 50%. Within this population, all 3 cell subsets defined by CD34 and Flt3 expression are decreased in absolute numbers, with the strongest reductions in the HSC compartment with long-term reconstitution potential and the LMPP subset. However, these differences in cell numbers probably do not explain the observed functional deficits of Lin-negative BM cells from Mll5−/− mice. Poor performance in competitive reconstitution assays and, in particular, inefficient rescue of lethally irradiated mice by as many as 3 × 104 Lin-negative Mll5−/− BM cells argue for stem/progenitor defects on a per cell basis. Our data thus indicate a significant contribution of Mll5 to hematopoietic stem/progenitor cell homeostasis and function.
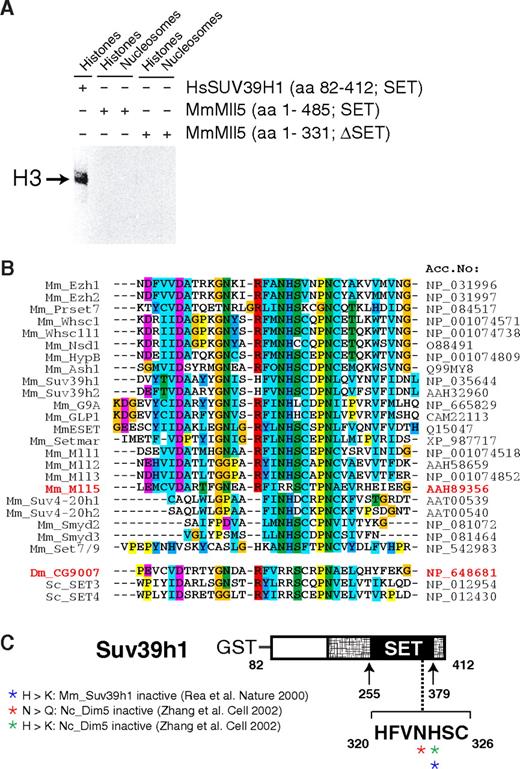
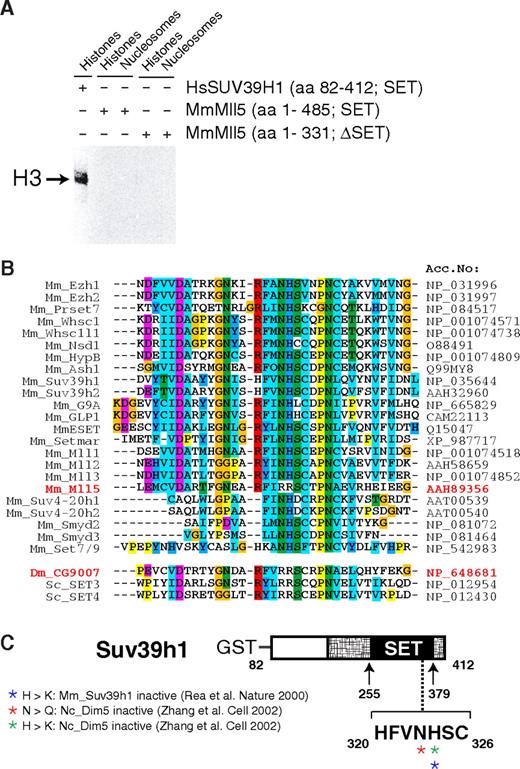
A critical role of Mll5 in primitive hematopoietic cells may not come as a total surprise. Mll1, its closest paralog, is essential for definitive hematopoiesis, as shown by in vitro differentiation of Mll1−/− embryonic stem cells43 and analysis of chimeric44 or, most recently, conditionally mutant mice.33,45 Like its Drosophila ortholog Trithorax, Mll1 regulates expression of several Hox genes, an activity that appears to be mediated, at least in part, by histone lysine methylation.5,16,46 Indeed, the SET domain of human MLL1 has been shown to methylate lysine at position 4 of histone H3.46,47 However, despite specific efforts, we did not find evidence that Mll5 mediates its effects by regulating Hox gene expression (Figure S10) or by methylating histones. For instance, we have compared the global histone methylation pattern in phenotypically affected tissues of Mll5−/− mice and WT littermates with a panel of 12 distinct antimethyl histone antibodies but did not detect significant differences (Figure S11). We were also unable to detect in vitro methyltransferase activity of recombinant Mll5 proteins when purified histones or recombinant histone octamers were used as substrates (Figure 7A). Indeed, alterations in critical amino acids within the SET domain strongly suggest that Mll5 may function in a very different manner from other known members of the Mll family (Figure 7B,C), in line with marked structural differences between Mll5 and its homologs.37 Mll5 therefore seems to fulfill unique functions, which obviously cannot be compensated for by Mll1-4 in our mouse mutants.
Mll5 is a SET domain protein that appears to lack histone methyltransferase activity. (A) Recombinant Mll5 does not reveal in vitro histone methyltransferase activity toward core histones, isolated from calf thymus, or to nucleosomes isolated from Saccharomyces cerevisiae. Recombinant Mll5 lacking its SET domain and HsSUV39H1 were used as negative and positive controls, respectively. (B) Alignment of the catalytic region within the SET domain of various mouse proteins and Drosophila and S cerevisiae MmMll5 homologs CG9007, SET3, and SET4, respectively. The 2 adjacent amino acids asparagine (N) and histidine (H), which are part of one of the most conserved sequence motifs in the SET domain (R-F/Y-I-N-H-X-C-X-P-N) and are invariant in all SET domains with established lysine methyltransferase activity, are altered to arginines in Mll5. The same substitutions are found in Drosophila CG9007 and S cerevisiae SET3 and SET4. Interestingly, whereas no information on the enzymatic activities of CG9007 and SET4 is available, specific efforts to detect histone methyltransferase activity for SET3 proved unsuccessful.48 Sequence alignment was performed using the Clustal-X program. Colors highlight conservation of different residues among the SET domain proteins. (C) Schematic diagram highlighting amino acids required for catalytic activity, as determined by mutational analyses of the MmSuv39h1 and NcDim5 HMTs. Mutation of the conserved asparagine (N) or histidine (H) amino acids into related (also basic!) residues glutamine (Q) or lysine (K) was sufficient to destroy histone methyltransferase activity of Dim5 and SUV39H1.49,50 Crystal structure analyses suggest that the conserved N-H residues are critical for binding the methyl-donor S-adenosyl-L-methionine (AdoMet).11 In MmMll5 and the fly and yeast orthologs, these critical residues are replaced by arginines. Taken together, these observations suggest that the SET domain of Mll5 lacks histone methyltransferase activity and might be involved in alternative activities, such as, for example, protein-protein interactions, a proven additional function of the Mll1 SET domain.51
Mll5 is a SET domain protein that appears to lack histone methyltransferase activity. (A) Recombinant Mll5 does not reveal in vitro histone methyltransferase activity toward core histones, isolated from calf thymus, or to nucleosomes isolated from Saccharomyces cerevisiae. Recombinant Mll5 lacking its SET domain and HsSUV39H1 were used as negative and positive controls, respectively. (B) Alignment of the catalytic region within the SET domain of various mouse proteins and Drosophila and S cerevisiae MmMll5 homologs CG9007, SET3, and SET4, respectively. The 2 adjacent amino acids asparagine (N) and histidine (H), which are part of one of the most conserved sequence motifs in the SET domain (R-F/Y-I-N-H-X-C-X-P-N) and are invariant in all SET domains with established lysine methyltransferase activity, are altered to arginines in Mll5. The same substitutions are found in Drosophila CG9007 and S cerevisiae SET3 and SET4. Interestingly, whereas no information on the enzymatic activities of CG9007 and SET4 is available, specific efforts to detect histone methyltransferase activity for SET3 proved unsuccessful.48 Sequence alignment was performed using the Clustal-X program. Colors highlight conservation of different residues among the SET domain proteins. (C) Schematic diagram highlighting amino acids required for catalytic activity, as determined by mutational analyses of the MmSuv39h1 and NcDim5 HMTs. Mutation of the conserved asparagine (N) or histidine (H) amino acids into related (also basic!) residues glutamine (Q) or lysine (K) was sufficient to destroy histone methyltransferase activity of Dim5 and SUV39H1.49,50 Crystal structure analyses suggest that the conserved N-H residues are critical for binding the methyl-donor S-adenosyl-L-methionine (AdoMet).11 In MmMll5 and the fly and yeast orthologs, these critical residues are replaced by arginines. Taken together, these observations suggest that the SET domain of Mll5 lacks histone methyltransferase activity and might be involved in alternative activities, such as, for example, protein-protein interactions, a proven additional function of the Mll1 SET domain.51
Clearly, additional experimentation will be required to unravel molecular mechanisms and to precisely define the particular aspect of hematopoietic stem/progenitor cell function that is compromised in the absence of Mll5. Our in vivo BrdU-labeling experiments and the resultant demonstration of increased cell cycling within the most primitive HSC subset (CD34− LSK cells) may provide a first important hint. Interestingly, increased proliferation of CD34− LSK cells has been found in several mouse mutants with similar phenotypic and functional defects in the HSC compartment, including Gfi−/−,32 Foxa3−/−,34 conditional Mll1−/−,33 and most recently Pbx1−/−31 mouse mutants. In some cases, increased cell cycling was interpreted to reflect an essential role of the respective gene in maintaining HSC quiescence and thus in preserving the stem cell pool.31,33,34 Although this is an attractive possibility to explain the observed defects in Mll5-deficient mice as well, we currently cannot exclude that enhanced proliferation is just a stress response of mutant cells to compensate for other defects in HSC function.
Human MLL5 was identified as a candidate leukemia suppressor gene in 7q22, a chromosomal region frequently deleted in patients with aggressive AML. To date, we have been unable to detect any form of spontaneous leukemia in our heterozygous or homozygous Mll5-deficient mice. However, this does not exclude an important role of Mll5 in tumor suppression. Most of the Mll5−/− animals surviving postnatal lethality were killed for experimentation by 3 to 4 months of age. The number of Mll5−/− animals monitored for up to 1 year, mainly to assess fertility defects, might not have been sufficient to detect malignancies with low incidence or long latency. In addition, genetic alterations in addition to loss of Mll5 may be required for clinical disease. Curiously, 7q22 deletions are particularly common in patients who develop leukemia as a second malignant neoplasm after chemotherapy with DNA alkylating drugs.1 It is tempting to speculate that loss of Mll5 might sensitize hematopoietic progenitor populations to DNA damaging agents. With the availability of constitutive and conditional (Document S1) Mll5-deficient mice, this hypothesis can now be tested.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank E. Warlich. M. Thorwirth, and A. Tasdogan for mouse genotyping, M. Habdank for help with qPCR analysis, I. Steinhauser for automated blood analysis, G. Terszowski for skillful operation of the FACSAria, and R. Syhachak for expert care of our mouse colony.
This work was supported by grants from Landesforschungsschwerpunkt: Mechanismen der Transformation lympho-hämatopoetischer Zellen and from Deutsche Forschungsgemeinschaft (FE-578/1-1). Research in the laboratory of A.H.F.M.P. was supported by the Novartis Research Foundation and the NoE network The Epigenome (LSHGCT-2004-503433).
Authorship
Contribution: V.M., B.M., U.B., F.Z., K.H., and O.W. conducted experiments; C.B. did blastocyst injections; K.D. and H.D. identified human MLL5 and provided the cDNA sequence; A.H.F.M.P. provided essential reagents; V.M., H.-R.R., P. S.-C., A.H.F.M.P., and H.J.F. designed the experiments; and H.J.F. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hans Jörg Fehling, Albert-Einstein-Allee 11, D-89081 Ulm, Germany; e-mail: joerg.fehling@uni-ulm.de.



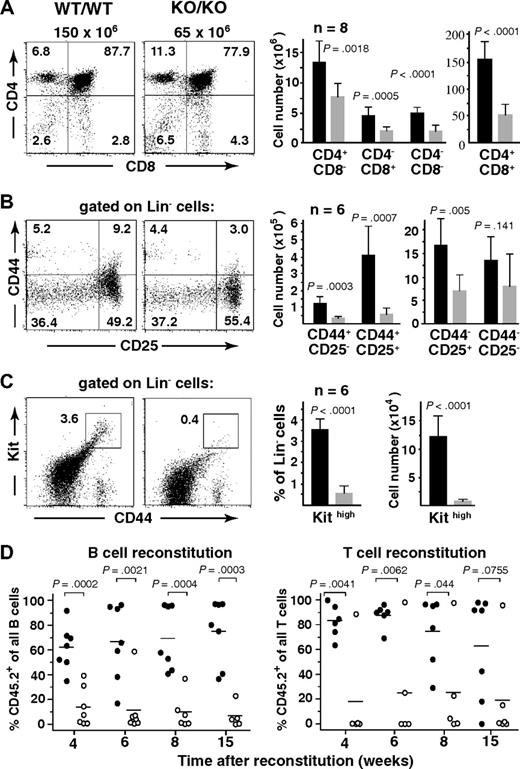
 represents Mll5−/− mice; ■, WT littermates. (D) Competitive BM reconstitution into alymphoid Rag2−/−γc−/− recipients. FACS-sorted Lin-negative BM cells (CD45.2) from Mll5−/− mice (KO) or WT littermates were mixed with an equal number of Lin-negative competitor cells (CD45.1) and injected into sublethally irradiated (400 cGy) recipients. Individual recipients reconstituted with Mll5−/− donor cells (○) or donor cells from WT littermates (●) are shown (data from 2 independent experiments combined).
represents Mll5−/− mice; ■, WT littermates. (D) Competitive BM reconstitution into alymphoid Rag2−/−γc−/− recipients. FACS-sorted Lin-negative BM cells (CD45.2) from Mll5−/− mice (KO) or WT littermates were mixed with an equal number of Lin-negative competitor cells (CD45.1) and injected into sublethally irradiated (400 cGy) recipients. Individual recipients reconstituted with Mll5−/− donor cells (○) or donor cells from WT littermates (●) are shown (data from 2 independent experiments combined).