Abstract
MLL5 is a novel trithorax group gene and a candidate tumor suppressor gene located within a 2.5-Mb interval of chromosome band 7q22 that frequently is deleted in human myeloid malignancy. Here we show that inactivation of the Mll5 gene in mice results in a 30% reduction in the average representation of hematopoietic stem cells and in functional impairment of long-term hematopoietic repopulation potential under competitive conditions. Bone marrow cells from Mll5-deficient mice were defective in spleen colony-forming assays, and the mutant mice showed enhanced susceptibility to 5-fluorouracil–induced myelosuppression. Heterozygous and homozygous Mll5 mutant mice did not spontaneously develop hematologic cancers, and loss of Mll5 did not alter the phenotype of a fatal myeloproliferative disorder induced by oncogenic Kras in vivo. Collectively, the data reveal an important role for Mll5 in HSC homeostasis and provide a basis for further studies to explore its role in leukemogenesis.
Introduction
Cytogenetic abnormalities that arise frequently in cancer cells are starting points for identifying genes that initiate or cooperate in tumorigenesis. In the study of leukemia in particular, major advances have come from the cloning of genes located at translocation breakpoints. Most notably, these genes have included BCR, ABL, MLL, PML, RARA, AML1/ RUNX1, and CBFβ,1 many of which have proven to be essential for normal hematopoiesis in addition to their involvement in cancer
Segmental deletions, such as those involving chromosome bands 5q31, 7q22, and 20q12, are also recurring leukemia-associated genomic abnormalities. In contrast to translocations, however, much less is known about how the deletions influence tumorigenesis. It is hypothesized that the deleted intervals contain tumor suppressor genes (TSGs) and that the deletion of these genes deregulates cell division, survival, or differentiation. It is unclear, however, whether such deregulation occurs solely as a consequence of haploinsufficiency of the putative TSGs2,3 or whether “second-hit” mutations that result in biallelic inactivation are required.
Loss of chromosome 7 (monosomy 7) and deletion of a segment of the long arm [del(7q)] are recurring cytogenetic abnormalities in myeloid malignancies, including myelodysplastic syndrome (MDS), acute myelogenous leukemia (AML), therapy-associated leukemia, and leukemias that arises in the context of inherited predispositions such as Fanconi anemia or neurofibromatosis type 1.4 Many of these myeloid malignancies are associated with resistance to current treatments and a poor prognosis. Cytogenetic analysis of patients with myeloid disorders characterized by a del(7q) has revealed frequent involvement of band q22, and fluorescence in situ hybridization (FISH) studies of cases with proximal or distal breakpoints within 7q22 defined a commonly deleted segment (CDS) spanning 2.5 Mb that is bounded by D7S1503 and D7S18415 . Molecular analysis of the 14 known genes within this interval has not uncovered either pathogenic mutations or epigenetic silencing.6,7
Here, we focus on the function of the mouse homolog of MLL5, a candidate TSG that is located within the 7q22 CDS.8 MLL5 is a member of the mammalian Trithorax Group (trxG) of genes, several of which have been implicated in cancer. Most notably, MLL is a frequent target of translocations in leukemia, with more than 40 different fusion partner genes described to date.9 Inactivation of the mouse homologue of MLL blocks definitive hematopoiesis in mouse embryos10,11 and compromises hematopoietic stem cell (HSC) function.12,13
Little is known about the normal function of MLL5. The gene is active in multiple tissues, with prominent expression in the hematopoietic system.8,14 Overexpression or small interfering RNA-mediated knockdown of MLL5 causes cell-cycle arrest in multiple cell lines,15,16 and an MLL5-GFP fusion protein localizes to foci in the nuclei of transfected cells.15 Consistent with a possible role in cell-cycle regulation, Mll5 was recently identified as a gene associated with myoblast quiescence through a novel gene-trap screen.17 Like other members of the trxG, MLL5 contains a supressor of variegation-enhancer of zeste-trithorax (SET) domain that is expected to influence chromatin accessibility and gene expression through histone methylation.18 Despite its large size (1858 aa) the only other recognizable protein motif in MLL5 is a PHD finger, which is also found in other trxG proteins and may be essential for interactions with modified histones.19 MLL5 is most closely related to SETD5, a protein of unknown function encoded by an unlinked gene in the human genome. Both proteins have SET domains that bear similarity to that of SET3,20 a meiosis-specific repressor of sporulation genes in Saccharomyces cerevisiae,21 and MLL5 is part of a nuclear complex in HeLa cells that is potentially equivalent to the yeast SET3 complex.22
On the basis the importance of other MLL family members in development and cancer, the chromosomal location of MLL5 within the 7q22 CDS, and prominent expression in the hematopoietic system, we generated a loss-of-function mutation in the mouse MLL5 homolog to explore its function in vivo. We find that the inactivation of Mll5 results in a modest reduction in the average size of the HSC compartment, markedly compromises HSC function, and causes dysregulation of Hoxb2 and Hoxb5. Mll5 mutant mice do not spontaneously develop hematologic malignancies. The data reveal MLL5 as a regulator of hematopoiesis and provide a basis for further studies to explore its role in leukemogenesis.
Methods
Animals
An Mll5-null mutation was generated by gene targeting in embryonic stem cells with the use of a construct that deleted DNA from exons 3 and 4 and replaced it with an IRES-GFP element and associated loxP-flanked neomycin resistance gene (Y.Z., J.W., K.M.S., N.K., manuscript in preparation). The neo gene was not removed for the studies presented here. Targeting was confirmed by Southern blot by using probes that mapped on either side of the targeted region and an internal probe. Northern blot and reverse-transcription polymerase chain reaction (RT-PCR) analysis confirmed that the Mll5 gene was inactivated as expected (Y.Z., J.W., K.M.S., N.K., manuscript in preparation). The mutant mice were backcrossed for 6 (or 9) generations to C57BL/6 partners before intercrossing them for use in experiments. The experiments reported here involved N6 mice unless otherwise stated. C57BL/6-CD45.1 congenic mice were purchased from the National Cancer Institute Animal Production Program (Frederick, MD) and were intercrossed or bred with C57BL/6 mice (to generate F1 mice expressing both CD45 allotypes). Mx1-cre, KrasG12D mice were generated, treated with polyI:C, and analyzed as described previously.23,24 All mice were housed in a specific pathogen-free facility at the University of California San Francisco (UCSF), and all animal experiments were conducted under protocols approved by the UCSF Institutional Animal Care and Use Committee.
Flow cytometry
Bone marrow cells were recovered from the femurs and tibias of mice. Lineage staining was performed with a cocktail of biotinylated antibodies specific for CD3(145-2C11), CD4(RM4-5), CD5(53-7.3), CD8(53-6.7), B220(RA3-6B2), CD11b(M1/70), Gr-1(RB6-8C5), and Ter119. These and other antibodies (including anti Sca-1 [D7], -c-kit [ACK2], -CD34 [RAM34], -Flt-3[A2F10], -CD48 [HM48-1], -CD150 [TC15-12F12.2], and -IL-7Rα [A7R34]) were purchased from eBioscience (San Diego, CA), BD Biosciences (San Jose, CA), or Biolegend (San Diego, CA). Annexin-V was purchased from BD Biosciences. Cells were analyzed and sorted with a Becton Dickinson (Franklin Lakes, NJ) LSRII and FACSAria, respectively.
For cell-cycle analysis, bone marrow cells were stained with antibodies to detect LSK cells. They were then fixed with Cytofix buffer (BD Biosciences) for 20 minutes at room temperature in the dark before permeabilization in Perm/Wash buffer (BD Biosciences) at 4°C overnight. The cells were resuspended in phosphate-buffered saline containing bovine serum albumin (0.3% wt/vol), 14 μmol/L 4′,6-diamidino-2-phenylindole (Sigma-Aldrich, St Louis, MO) and 4 μg/mL Pyronin Y (Sigma-Aldrich) before analysis by flow cytometry. For labeling of dividing cells in vivo, mice were treated with 1 mg 5-bromo-2-deoxyuridine (BrdU) given intraperitoneally 2 hours before staining with fluorescein isothiocyanate–conjugated anti-BrdU and 7-amino-actinomycin D according to the manufacturer's instructions (BD Biosciences).
Adoptive transfer, spleen colony-forming units, and 5-FU experiments
CD45.1 congenic C57BL/6 mice were gamma-irradiated (2 × 550 rad) before the intravenous injection of 2 × 106 cells comprising a 1:1 mix of bone-marrow or fetal liver cells from Mll5−/− or littermate control mice.
For short-term transfers, bone marrow cells were labeled with 5 μmol/L carboxyfluorescein diacetate succinimidyl ester (Invitrogen, Carlsbad, CA) in accordance with the manufacturer's instructions. The cells were injected into the tail veins of 8- to 12-week-old C57BL/6 mice that had been lethally irradiated 24 hours previously.
Next, 105 bone marrow cells were injected intravenously into lethally irradiated C57BL/6 mice. Spleens were taken from the recipients on day 8 or day 12 after transplantation, fixed in Bouin's solution (Sigma-Aldrich) for 10 minutes, and then transferred into 10% buffered formalin solution (Fisher Scientific, Santa Clara, CA). Colonies on the surfaces of the spleens were counted 2 days later. Wild-type or Mll5−/− mice were injected weekly with 5-fluorouracil (5-FU; Sigma-Aldrich) intraperitoneally at a dose of 200 mg/kg.
Gene expression analysis
Bone marrow cells were pooled from 8-week-old Mll5−/− mice and sex-matched wild-type littermates (pools of 6 mice; 2 pools per genotype). Total RNA was isolated from flow-sorted LSK cells by using the RNeasy Mini kit (QIAGEN, Valencia, CA), and its quality was assessed by using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). RNA amplification and array hybridizations were performed by the UCSF Shared Microarray Core Facilities. In brief, RNA was amplified and labeled with Cy3-CTP by using the Agilent low RNA input fluorescent linear amplification kit. Labeled cRNA was then hybridized for 14 hours to Agilent whole mouse genome 4 × 44K Ink-jet microarrays. Arrays were washed and then scanned using the Agilent microarray scanner. Raw signal intensities were extracted with Feature Extraction version 9.1 software (Agilent, Santa Clara, CA). The dataset was normalized using the quantile normalization method proposed by Bolstad et al25 with the Bioconductor package limma.26 No background subtraction was performed. Microarray data have been deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE10817).
For real-time RT-PCR analysis of gene expression, total RNA was isolated as described previously and converted into complementary DNA (cDNA) by using Superscript reverse transcriptase (Stratagene, La Jolla, CA). Primers and probes for PCR were designed using Primer3 software27 or selected from the PrimerBank database (Table S2, available on the Blood website; see the Supplemental Materials link at the top of the online article).28 All reactions were performed by using a Chromo4 system (Bio-Rad, Hercules, CA) with AmpliTaq gold DNA polymerase (Applied Biosystems, Foster City, CA) or universal SYBR Greener reagents (Invitrogen).
Statistical analysis
We performed 2-tailed paired or unpaired Student t tests by using Prism software (GraphPad Software, La Jolla, CA). Survival data from 5-FU experiments were analyzed by using a log-rank nonparametric test, and the data were expressed as Kaplan-Meier survival curves. We considered P values less than .05 to be significant in all cases.
Results
Mll5 mutant mice
The Mll5 gene was inactivated in mouse embryonic stem cells by gene targeting (Y.Z., J.W., K.M.S., N.K., manuscript in preparation). The mutant allele lacks DNA spanning exons 3 and 4 and, in place of this, carries a large insertion (of a loxP-flanked neo gene and an IRES-GFP element) that terminates translation and transcription. Mice that are homozygous for this allele displayed incompletely penetrant postnatal lethality; 70% of them failed to develop to weaning age, and most died within the first 24 hours of life. Surviving Mll5−/− mice showed mild growth retardation but were otherwise healthy (Y.Z., J.W., K.M.S., N.K., manuscript in preparation).
Mll5 inactivation perturbs hematopoietic differentiation.
All hematopoietic cells were present in Mll5−/− mice in near-normal proportions in peripheral blood (Table 1). There were, however, reproducible anomalies in the representation of specific cell types that became more obvious as the mice were backcrossed onto the C57BL/6 background. In particular, neutrophil numbers were increased in the peripheral circulation although not in the bone marrow (Table 1, Figure S1A). There were also significantly decreased numbers of natural killer (NK) and NK T cells, especially in the liver (Figure S1B). Monocytes, macrophages, dendritic cells, thymocytes, and peripheral lymphocytes all were present in normal numbers in the mutant animals (Figures S1A, S2).
As in humans, mouse Mll5 exhibits a broad pattern of expression with transcripts detectable in most tissues examined (Figure S3A). Of importance, we detected Mll5 transcripts in the bone marrow (Figure S3A) and also in purified populations of hematopoietic stem and progenitor cells (Figure S3B). Given this broad pattern of expression, we used flow cytometry to test whether the absence of Mll5 might impact the representation of stem cells and/or progenitors in the bone marrow.
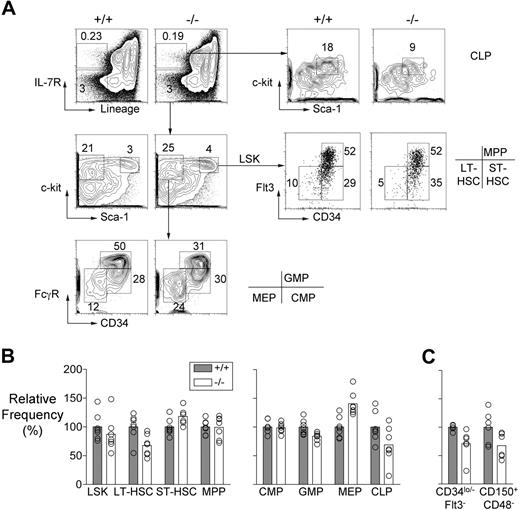
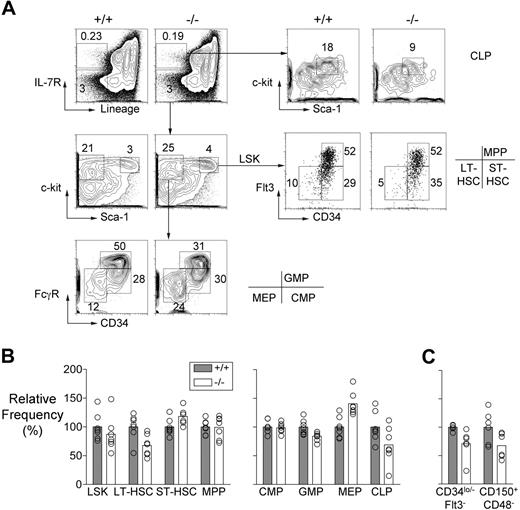
Mll5−/− bone marrow demonstrated a reduction in the frequency of cells with a Lin−, Sca-1+, c-kit+ (LSK), CD34lo/−, and Flt-3lo surface phenotype, which is associated with long-term repopulating HSC (LT-HSC; Figure 1A,B). The 30% reduction in the average number of LT-HSCs was consistently apparent in mice that had been backcrossed either 6 or 9 times to C57BL/6 partners, and it was also evident when LT-HSCs were identified as CD150+CD48−LSK cells29 (Figure 1C). By contrast, the mutant mice had increased numbers of cells expressing surface markers associated with short-term HSC (ST-HSC), that is, LSK cells that are Flt-3lo CD34+.30
HSC and progenitor populations in Mll5−/− bone marrow. (A) Representative FACS data showing LT-HSC (Lin−, Sca-1+, c-kit+, CD34lo/−, Flt3lo), ST-HSC (Lin−, Sca-1+, c-kit+, CD34+, Flt3 lo), MPP (Lin−, Sca-1+, c-kit+, CD34+, Flt3+), CMP (Lin−, Sca-1−, c-kit+, CD34+, FcγRlo), GMP (Lin−, Sca-1−, c-kit+, CD34+, FcγRhi), MEP (Lin−, Sca-1−, c-kit+, CD34−, FcγRlo), and CLP (Lin−, IL-7R+, Sca-1lo, c-kitlo) populations in an Mll5−/− mouse and a wild-type littermate. (B) Graph showing the average relative frequencies of the indicated cell types in wild-type and Mll5−/− mice (n = 7, both groups) determined using the gating strategies shown in panel A. The data were normalized to the wild-type averages and expressed as percentages. Using the Student t test, we discovered that the following populations showed significant differences in frequency between the 2 groups: LT-HSC, ST-HSC, GMP, MEP (all P < .01) and CLP (P < .05). (C) LT-HSC numbers determined in wild-type and Mll5−/− mice (n = 6, both groups) on the basis of CD34 and Flt-3 or CD150 and CD48 expression. The cells for this experiment were obtained from mice that had been backcrossed 9 times to C57BL/6 partners. The differences in LT-HSC numbers were significant in both cases by Student t test (P < .05).
HSC and progenitor populations in Mll5−/− bone marrow. (A) Representative FACS data showing LT-HSC (Lin−, Sca-1+, c-kit+, CD34lo/−, Flt3lo), ST-HSC (Lin−, Sca-1+, c-kit+, CD34+, Flt3 lo), MPP (Lin−, Sca-1+, c-kit+, CD34+, Flt3+), CMP (Lin−, Sca-1−, c-kit+, CD34+, FcγRlo), GMP (Lin−, Sca-1−, c-kit+, CD34+, FcγRhi), MEP (Lin−, Sca-1−, c-kit+, CD34−, FcγRlo), and CLP (Lin−, IL-7R+, Sca-1lo, c-kitlo) populations in an Mll5−/− mouse and a wild-type littermate. (B) Graph showing the average relative frequencies of the indicated cell types in wild-type and Mll5−/− mice (n = 7, both groups) determined using the gating strategies shown in panel A. The data were normalized to the wild-type averages and expressed as percentages. Using the Student t test, we discovered that the following populations showed significant differences in frequency between the 2 groups: LT-HSC, ST-HSC, GMP, MEP (all P < .01) and CLP (P < .05). (C) LT-HSC numbers determined in wild-type and Mll5−/− mice (n = 6, both groups) on the basis of CD34 and Flt-3 or CD150 and CD48 expression. The cells for this experiment were obtained from mice that had been backcrossed 9 times to C57BL/6 partners. The differences in LT-HSC numbers were significant in both cases by Student t test (P < .05).
Multipotent progenitor cells (MPPs) are derived from ST-HSC, express both Flt-3 and CD34, but lack self-renewal ability. These cells were present in normal numbers in the bone marrow of Mll5 mutant mice (Figure 1A,B). Whereas the numbers of common myeloid progenitor cells (CMP; Lin−, Sca-1−, c-kit+, CD34+, FcγRlo) were also normal, there was evidence of defective differentiation of this population. Specifically, granulocyte/macrophage progenitors (GMP; Lin−, Sca-1−, c-kit+, CD34+, FcγRhi) were decreased in number whereas megakaryocyte/erythrocyte progenitors (MEPs; Lin−, Sca-1−, c-kit+, CD34−, FcγRlo) were present in increased numbers (Figure 1A,B). We also found that the mutant mice had significantly reduced numbers of common lymphoid progenitors (CLP; Lin−, IL-7R+, Sca-1lo, c-kitlo; Figure 1A,B).
Collectively, the data just summarized suggest that Mll5 regulates early hematopoiesis at 3 discrete stages: it is required for homeostasis of the HSC compartment because in its absence there was an imbalance in LT- and ST-HSC representation; it is also required for commitment of CMP cells to granulocyte/macrophage versus megakaryocyte/erythrocyte lineages, and for formation of common lymphocyte progenitors. With these data in mind, we performed additional experiments to test for defects in HSC function caused by inactivation of Mll5.
Mll5−/− LT-HSC show impaired repopulating potential
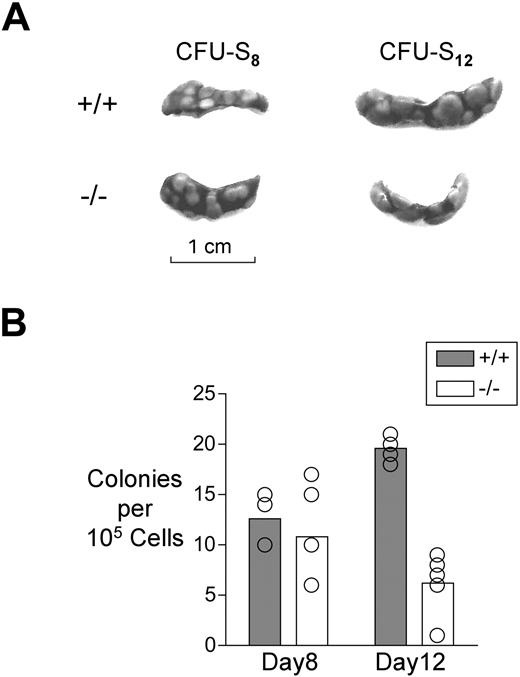
Transplanted bone marrow cells form colonies in the spleens of lethally irradiated recipients in a fashion that is determined by progenitor and HSC numbers and activity.31-33 At day 8 after transplantation, the number of colonies formed reflects the number of CMP and MEP in the donor inoculum, whereas at day 12, half of the colonies are derived from CMP and MEP and half from HSC and MPP.32 Although clearly not a definitive test of HSC activity,34 both LT-HSC and ST-HSC are capable of forming spleen colonies at day 12,32,33 and mutations that impact the HSC compartment can interfere with this.35,36 With this in mind, and with a view to obtaining a preliminary assessment of whether the observed abnormalities in HSC numbers might impact short-term repopulation ability, we transplanted 105Mll5−/− or wild-type bone marrow cells into irradiated recipients and counted colonies on their spleens at day 8 or 12 (CFU-S8 or -S12, respectively).
Loss of Mll5 had no effect on CFU-S8, which is consistent with the immunophenotypic data indicating that CMP and MEP numbers were not reduced in the mutant mice (Figure 2). Colony replating assays, which also are sensitive to progenitor cell number and fitness, similarly revealed no defect in cells from the mutant mice (data not shown). In marked contrast, however, Mll5−/− bone marrow cells were significantly reduced in CFU-S12 (Figure 2). This finding was suggestive of abnormal progenitor/HSC activity in the mutant mice.
Analysis of hematopoiesis using the spleen colony formation assay. (A) Representative spleens of irradiated mice that received 105 bone marrow cells 8 or 12 days before analysis. (B) Graph showing the average number of colonies per spleen at day 8 or day 12 after transplantation. The difference between genotypes in colony number at day 8 was statistically significant by Student t test (P < .01).
Analysis of hematopoiesis using the spleen colony formation assay. (A) Representative spleens of irradiated mice that received 105 bone marrow cells 8 or 12 days before analysis. (B) Graph showing the average number of colonies per spleen at day 8 or day 12 after transplantation. The difference between genotypes in colony number at day 8 was statistically significant by Student t test (P < .01).
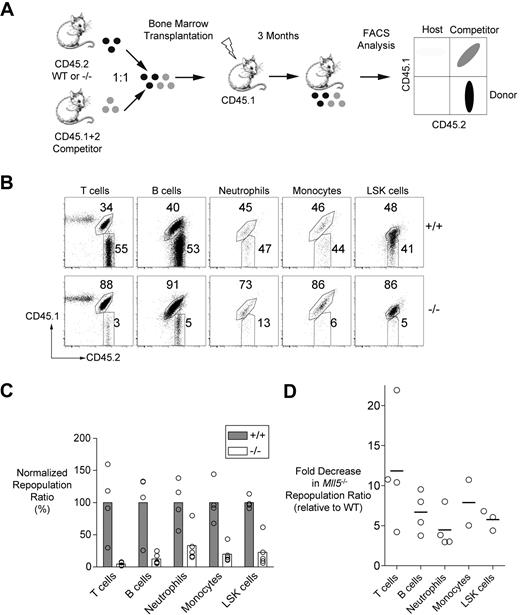
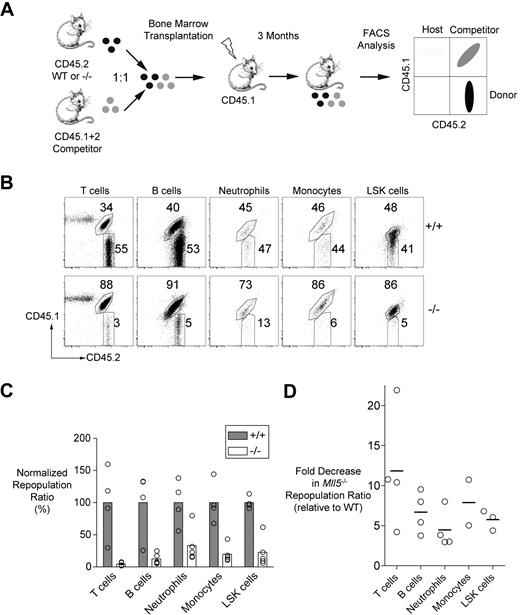
As a more stringent test of LT-HSC fitness, we used competitive repopulation assays, which directly compare the repopulating capacities of mutant and wild-type cells in individual mice. We mixed Mll5−/− (or control littermate) bone marrow cells with equal numbers of wild-type competitor bone marrow cells before transfer into lethally irradiated recipients. The donor, competitor, and residual host cells in the chimeras differed in their expression of CD45 allotypes, and, thus, all 3 cell types could be distinguished by flow cytometry using allotype-specific antibodies (Figure 3A).
Impaired repopulation of irradiated recipients by Mll5−/− bone marrow cells. (A) Experimental strategy for the competitive repopulation assay. (B) Representative FACS data showing the frequencies of T cells (CD3+), B cells (CD19+), neutrophils (Gr-1hi, CD11b+), monocytes/macrophages (Gr-1+, CD11b+), and LSK cells in recipient mice 3 months after transplantation with mixtures of CD45.2+ test cells (wild-type or Mll5−/−) and CD45.1+CD45.2+ competitor cells. (C) The graph shows the averages of the ratios of test versus competitor populations of the indicated cell types in repopulated mice 3 months after transplantation. The data for each cell type are normalized to the average ratios obtained with mice that received wild-type test cells and are expressed as percentages. A value of less than 100% indicates impaired competitiveness of the test population. The differences between genotypes were statistically significant by Student t test (P < .05 for all comparisons except LSK cells, P < .01). The data are representative of 3 independent experiments performed with bone marrow cells and 1 experiment performed with fetal liver cells (Figure S5). (D) The graph shows the fold decrease in the Mll5−/− repopulation ratios in the 4 independent experiments (relative to wild-type in each experiment).
Impaired repopulation of irradiated recipients by Mll5−/− bone marrow cells. (A) Experimental strategy for the competitive repopulation assay. (B) Representative FACS data showing the frequencies of T cells (CD3+), B cells (CD19+), neutrophils (Gr-1hi, CD11b+), monocytes/macrophages (Gr-1+, CD11b+), and LSK cells in recipient mice 3 months after transplantation with mixtures of CD45.2+ test cells (wild-type or Mll5−/−) and CD45.1+CD45.2+ competitor cells. (C) The graph shows the averages of the ratios of test versus competitor populations of the indicated cell types in repopulated mice 3 months after transplantation. The data for each cell type are normalized to the average ratios obtained with mice that received wild-type test cells and are expressed as percentages. A value of less than 100% indicates impaired competitiveness of the test population. The differences between genotypes were statistically significant by Student t test (P < .05 for all comparisons except LSK cells, P < .01). The data are representative of 3 independent experiments performed with bone marrow cells and 1 experiment performed with fetal liver cells (Figure S5). (D) The graph shows the fold decrease in the Mll5−/− repopulation ratios in the 4 independent experiments (relative to wild-type in each experiment).
At 3 months after adoptive transfer, the spleens of recipients that received wild-type control and competitor bone marrow cells were repopulated with hematopoietic cells derived from both sources in roughly equal measure (Figure 3B,C). In striking contrast, Mll5−/− cells were markedly underrepresented in all of the hematopoietic lineages examined (Figure 3B,C). This underrepresentation was present but less striking at 4 weeks after transfer, consistent with the presence of abundant ST-HSC and progenitor cells in the mutant mice (Figure S4A). The magnitude of the decrease in representation at 3 months (3-12 × average decrease in the repopulation ratio) was similar across all lineages within individual mice and was observed in 3 independent experiments (Figure 3D). A fourth experiment involving fetal liver cells revealed similar effects, indicating that Mll5 is necessary for both embryonic and adult hematopoiesis (Figures S5, 3D). Finally, we noted that mutant LT-HSC and ST-HSC were markedly underrepresented in the bone marrow at 3 months to the same extent as the peripheral lineages and that there were similar reductions in multipotent (MPP) or myeloerythroid (CMP, GMP, and MEP) progenitor populations (Figure S4B).
To test whether loss of Mll5 impaired homing to the bone marrow, we performed short-term transfer experiments by using carboxyfluorescein diacetate succinimidyl ester-labeled bone marrow cells (Figure S6A) and found that Mll5−/− and control cells were present in equivalent numbers in the femurs and tibias of recipient mice 12 hours after transfer (Figure S6B). Moreover, Mll5-deficient LSK cells also showed normal expression of CXCR4, VLA-4, and VLA-5, which are integral to homing to the bone marrow (Figure S6C). Collectively, the CFU-S and short-term repopulation data indicate that Mll5 acts subsequent to engraftment to regulate HSC fitness and homeostasis.
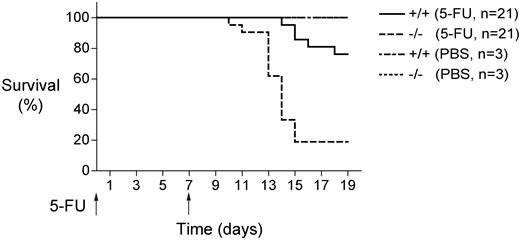
Impaired survival of Mll5−/− mice injected with 5-FU
The nucleotide analog 5-FU is toxic for dividing cells and consequently causes rapid depletion of hematopoietic cells when administered to mice.37 Hematopoietic recovery from 5-FU is dependent, in part, on the regenerating properties of quiescent HSCs that are induced to divide and differentiate in the immediate aftermath of 5-FU treatment.38 Although progenitor or other non-HSC defects can also result in increased sensitivity to 5-FU, defects in HSC representation or function have been associated with increased 5-FU sensitivity in some contexts.39
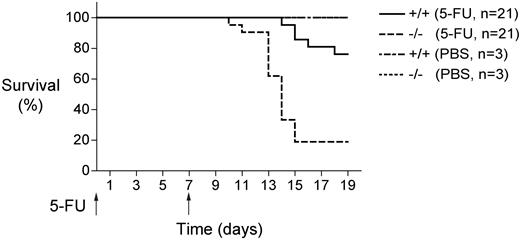
Mll5-deficient mice showed reduced survival after weekly injections of 5-FU (Figure 4). The mutant mice began to die as early as 10 days after initiating 5-FU treatment, and more than 80% of them had died by day 15. By contrast, none of the wild-type mice died until 2 weeks after the initial injection, and nearly 80% of them survived until day 19. These results are consistent with the reduced numbers of LT-HSC in Mll5−/− bone marrow (Figure 1) and with the markedly impaired repopulating potential of these cells under competitive conditions (Figure 3).
Mll5−/− bone marrow cells show increased susceptibility to myelosuppression. Survival of Mll5−/− and wild-type littermate control mice (21 per group) that received weekly intraperitoneal doses of 5-fluorouracil (5-FU; 200 mg/kg). Mll5−/− mice showed significantly increased sensitivity to 5-FU (log-rank nonparametric test; P < .0001).
Mll5−/− bone marrow cells show increased susceptibility to myelosuppression. Survival of Mll5−/− and wild-type littermate control mice (21 per group) that received weekly intraperitoneal doses of 5-fluorouracil (5-FU; 200 mg/kg). Mll5−/− mice showed significantly increased sensitivity to 5-FU (log-rank nonparametric test; P < .0001).
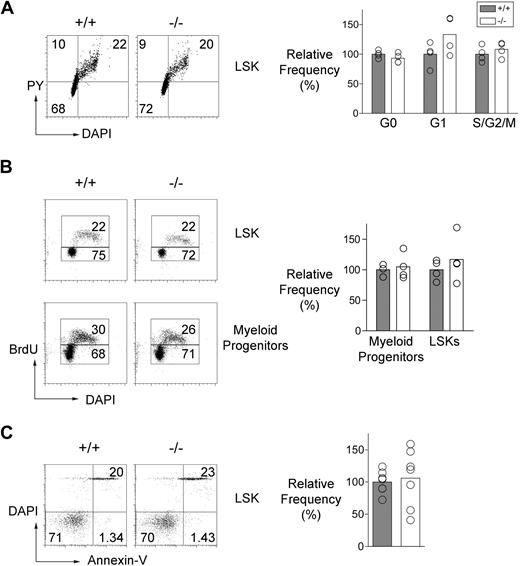
Cell-cycle status and apoptosis of Mll5−/− cells
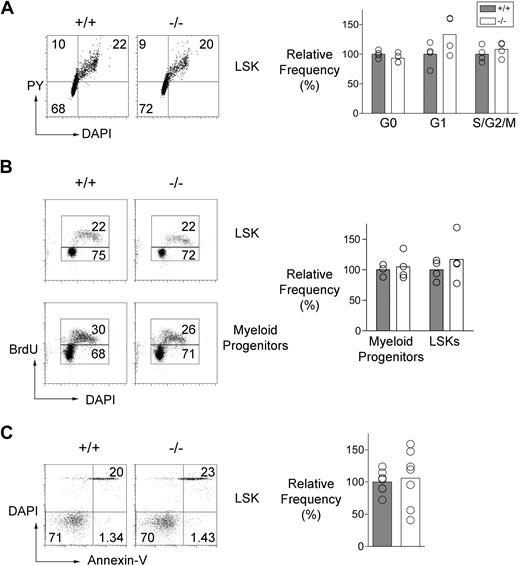
A deficit of LT-HSCs could be a consequence of imperfect regulation of stem cell quiescence, differentiation, or survival.40 With this in mind, we used sensitive flow cytometric assays to analyze the cell-cycle status of the LSK compartment and the frequency of apoptotic cells within it. DNA and RNA content of the LSK cells were measured with DAPI and Pyronin-Y, respectively (Figure 5A), and we also labeled proliferating cells in vivo by injecting mice with BrdU (Figure 5B). Apoptotic cells were detected in freshly isolated preparations of LSK cells by using fluorescent annexin-V (Figure 5C). These assays revealed no detectable effect of the absence of Mll5 on the cell-cycle status or frequency of apoptotic cells in the LSK compartment.
Cell-cycle status and apoptosis of Mll5−/− HSC. (A, left) Representative FACS data showing Pyronin-Y and DAPI staining of LSK cells from an Mll5−/− mouse and a control littermate. The gates used to discriminate cells in different stages of the cell cycle are shown. (Bottom left quadrant) G0; top left quadrant: G1; top right quadrant: S/G2/M. (Right panel) The graph summarizes cell-cycle data obtained from Mll5−/− and control mice (4 mice per group). (B, left) Representative FACS data showing BrdU incorporation in LSK and myeloid progenitor (Lin−, Sca-1−, c-kit+) cells from an Mll5−/− mouse and a control littermate following a single injection of BrdU (1 mg) 2 hours before analysis. (Right panel) The graph shows the relative frequencies of BrdU+ cells in LSK and myeloid progenitor populations from Mll5−/− and control mice (4 mice per group). (C, left) Representative FACS data showing annexin-V and DAPI staining of LSK cells in an Mll5−/− mouse and a control littermate. (Right) The graph shows the relative frequencies of apoptotic (annexin-V+ DAPI−) LSK cells in Mll5−/− and control littermate mice (7 mice per group).
Cell-cycle status and apoptosis of Mll5−/− HSC. (A, left) Representative FACS data showing Pyronin-Y and DAPI staining of LSK cells from an Mll5−/− mouse and a control littermate. The gates used to discriminate cells in different stages of the cell cycle are shown. (Bottom left quadrant) G0; top left quadrant: G1; top right quadrant: S/G2/M. (Right panel) The graph summarizes cell-cycle data obtained from Mll5−/− and control mice (4 mice per group). (B, left) Representative FACS data showing BrdU incorporation in LSK and myeloid progenitor (Lin−, Sca-1−, c-kit+) cells from an Mll5−/− mouse and a control littermate following a single injection of BrdU (1 mg) 2 hours before analysis. (Right panel) The graph shows the relative frequencies of BrdU+ cells in LSK and myeloid progenitor populations from Mll5−/− and control mice (4 mice per group). (C, left) Representative FACS data showing annexin-V and DAPI staining of LSK cells in an Mll5−/− mouse and a control littermate. (Right) The graph shows the relative frequencies of apoptotic (annexin-V+ DAPI−) LSK cells in Mll5−/− and control littermate mice (7 mice per group).
Gene expression in Mll5−/− LSK cells
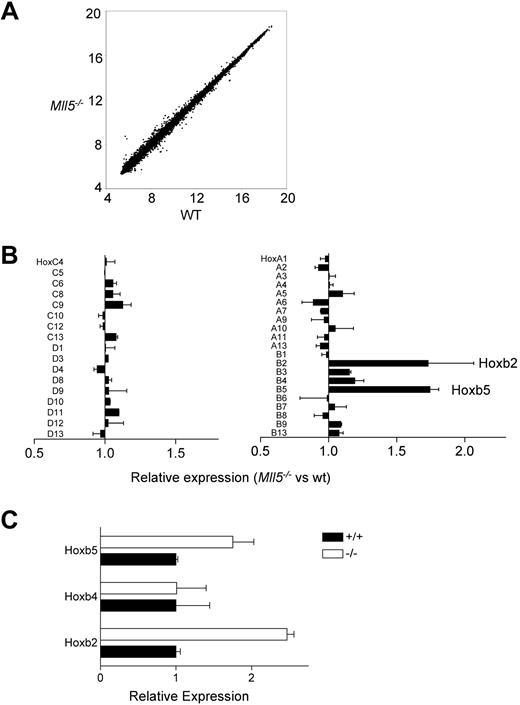
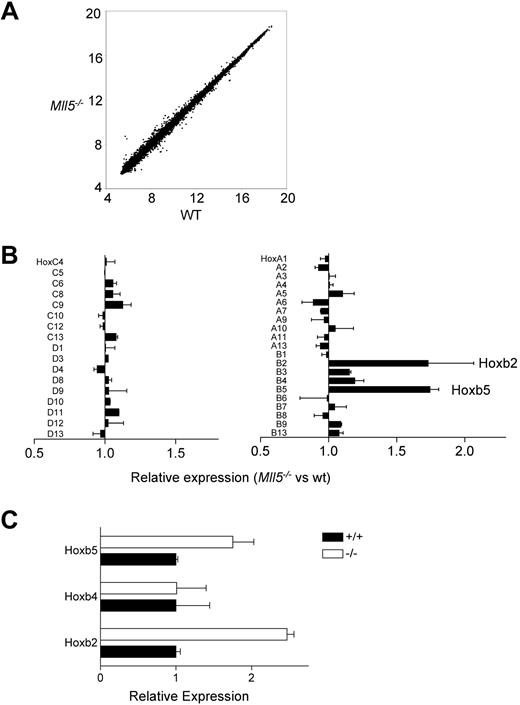
Trithorax group genes regulate gene expression through posttranslational modification of histones. We used 2 approaches to search for evidence of changes in gene expression that correlated with the loss of Mll5 and thus might identify genes that are directly or indirectly regulated by it. One approach was to use quantitative RT-PCR to test for changes in the expression of known regulators of HSC homeostasis by using cDNA prepared from wild-type and mutant LSK cells. None of the genes we tested were changed (Figure S7), suggesting that Mll5 may act independently of them. We also conducted a more general test for changes in gene expression using microarrays. These experiments also did not uncover dramatic changes in gene expression (Figure 6A), but we nonetheless detected 2-fold increases in the expression of Hoxb2 and Hoxb5 (Figure 6B), and these increases were subsequently verified by independent quantitative RT-PCR experiments (Figure 6C). Other Hox genes, including Hoxb4, Hoxa7, and Hoxa9, were unchanged in their expression (Figure 6B).
Gene expression in Mll5−/− HSC. (A) Genome-wide analysis of gene expression in Mll5−/− and wild-type LSK cells. The plot shows normalized (log2) hybridization signals for individual features on the microarrays probed with Mll5−/− or wild-type labeled cRNA. The data were generated by using 4 independent preparations of RNA from flow-sorted LSK cells (2 preparations from Mll5−/− mice and 2 from littermate controls). In each case, the LSK cells were prepared from bone marrow pooled from 6 mice. (B) Hox gene expression in Mll5−/− LSK cells as determined by the microarray analysis shown in panel A. The graph shows means plus or minus SEM. (C) Quantitative RT-PCR analysis of Hoxb2 and Hoxb5 expression in Mll5−/− LSK cells. Levels of expression were determined in triplicate and normalized to that of β-actin. The graph shows means plus or minus SD for samples from 1 pair of mice. The data are representative of 3 independent experiments.
Gene expression in Mll5−/− HSC. (A) Genome-wide analysis of gene expression in Mll5−/− and wild-type LSK cells. The plot shows normalized (log2) hybridization signals for individual features on the microarrays probed with Mll5−/− or wild-type labeled cRNA. The data were generated by using 4 independent preparations of RNA from flow-sorted LSK cells (2 preparations from Mll5−/− mice and 2 from littermate controls). In each case, the LSK cells were prepared from bone marrow pooled from 6 mice. (B) Hox gene expression in Mll5−/− LSK cells as determined by the microarray analysis shown in panel A. The graph shows means plus or minus SEM. (C) Quantitative RT-PCR analysis of Hoxb2 and Hoxb5 expression in Mll5−/− LSK cells. Levels of expression were determined in triplicate and normalized to that of β-actin. The graph shows means plus or minus SD for samples from 1 pair of mice. The data are representative of 3 independent experiments.
Effects of Mll5 deficiency in Mx1-Cre, LSL-KrasG12D mice
The MLL5 gene is located within 7q22 and is therefore frequently rendered haploid in myeloid malignancies. However, inactivating mutations or epigenetic silencing of the retained MLL5 allele have not been detected in 7q22 leukemia cells.8 Mll5+/− and Mll5−/− mice did not spontaneously develop malignant hematologic disease when they were observed for more than a year (data not shown). Thus, decreased expression of Mll5 alone was insufficient to cause leukemia in mice. Because chromosome 7 deletions are associated with mutations that cause hyperactive Ras signaling in some human leukemias,4,41 we reasoned that deregulated Ras signaling might be an important collaborating event for the involvement of Mll5 in leukemia. To investigate this, we crossed the Mll5 mutant mice to mice carrying a (Cre-recombinase–dependent) conditionally oncogenic Ras allele (LSL-KrasG12D) and the Mx1-Cre allele.42,43 These mice manifest an aggressive myeloproliferative disorder (MPD) after a single injection of polyI:C at weaning to induce Cre recombination in the bone marrow.23 Loss of one or both Mll5 alleles neither accelerated nor retarded MPD development in these mice (Figure S8A). Mx1-cre, KrasG12D mice of all 3 Mll5 genotypes showed a progressive increase in blood leukocyte counts and died between 73 and 81 days of age (Figure S8A). Whereas most mice died of MPD, such as is typical of this model (Figure S8B-E),23 4 of 10 Mll5+/+, 4 of 17 Mll5+/−, and 1 of 5 Mll5−/− mice developed T-lineage malignances that presented as large thymic masses. The T-cell tumors of Mll5+/−, Mx1-Cre, KrasG12D retained the wild-type Mll5 allele (data not shown). Thus, partial or full loss of Mll5 does not significantly alter the spectrum or aggressiveness of the malignant hematologic diseases observed in Mx1-Cre, LSL-KrasG12D mice.
Discussion
Mutations in genes for multiple SET domain-containing proteins impair mouse development, consistent with the expected significance of these proteins in global regulation of organogenic gene expression programs.20,44,45 Despite the broad pattern of Mll5 expression, the homozygosity of an Mll5 null mutation did not detectably impair embryogenesis. Thus, in contrast to other members of the trxG, such as Mll and Mll2,20,44 Mll5 is largely dispensable for organogenesis, with the one clear exception to this being the hematopoietic system.
Inactivation of Mll5 resulted in a 30% reduction in the average number of LT-HSC in the bone marrow of the mutant mice. Despite this result, Mll5−/− mice showed near-normal steady-state hematopoiesis, indicating that their HSCs retained the capacity to differentiate along all lineages. Strikingly, however, we found that loss of Mll5 severely compromised the capacity of HSCs to repopulate irradiated recipients under competitive conditions. This cell autonomous fitness effect was evident in both adult and embryonic HSCs, and it could not be readily explained by evidence of a failure of transplanted cells to relocate into the recipient bone marrow. Moreover, its magnitude (a 7- to 8-fold average decrease in the repopulation ratio across multiple lineages in 4 independent experiments) far exceeded the observed decrease in LT-HSC numbers in the donor mice. More precise quantitation of the repopulation defect will depend on additional repopulation assays featuring varied ratios of donor and competitor cells (in contrast to the uniform 50:50 mix we used here) and/or serial transplantation assays.
Mutations in multiple genes have been reported to cause decreases in LT-HSC numbers and impaired repopulation capacity.46-52 In several cases, these changes have been linked to abnormalities in cell-cycle status suggestive of a loss of HSC quiescence and thus implicating the mutated genes in HSC self-renewal.46,48,52 Despite evidence that Mll5 overexpression or underexpression can cause cell-cycle arrest,15,16 and that it is part of a nuclear complex (www.nursa.org/10.1621/datasets.01002) implicated in mitotic regulation,22 we did not detect cell-cycle abnormalities in HSCs in the Mll5−/− mice. The reason could be because the sensitivity of the assays used was insufficient to detect modest effects commensurate with the 30% reduction in LT-HSCs. Alternatively, Mll5 may affect HSC population size independently of cell-cycle regulation. We note that deficiencies of Smad4 or Foxo3a also have small or no effect on HSC numbers but nonetheless compromise repopulation capacity and only in the latter has a defect in cell-cycle regulation been described.53,54
There are similarities between the hematopoietic phenotype exhibited by Mll5−/− mice and that caused by inactivation of the related Trithorax group gene Mll.12,13 In both cases, the deficiencies reduced the size of the LT-HSC compartment and impaired the capacity to reconstitute hematopoiesis in irradiated recipients. Mll deficiency caused a severe deficit in HSC fitness, resulting in death caused by bone marrow failure within 3 weeks of somatic inactivation.12 By contrast, Mll5−/− mice could survive well into adulthood without overt hematopoietic defects. Inactivation of Mll resulted in rapid attrition of quiescent LSK cells and an increase in the frequency of cycling cells,12 unlike our findings on Mll5−/− mice. Enhanced apoptosis was not evident in either case, but the detection of apoptotic cells in fresh ex vivo samples is complicated by their rapid clearance in vivo.55 It is possible, therefore, that selective apoptosis of quiescent LSK cells might be at least partially responsible for the repopulation defects exhibited by cells from both strains of mice.
In addition to compromising HSC fitness, Mll5 deficiency altered the representation of certain fully differentiated leukocytes (NK cells, NK T cells, and neutrophils) as well as specific progenitor populations in the bone marrow. We found that the mutant mice had increased numbers of MEP and reduced numbers of GMP, suggestive of an effect of the mutation on the commitment of CMP. CLP also were reduced in number either because of a direct effect at this stage or because of impaired differentiation from the MPP or HSC stage. Interestingly, the absence of Mll also impacted progenitor populations without detectably affecting mature leukocytes (ie, in the context of late-stage inactivation of a conditional null Mll allele).12 In this respect, therefore, as with regulation of HSC fitness, Mll and Mll5 appear to have similar spheres of influence.
As a member of the Trithorax group of genes, Mll5 likely regulates chromatin accessibility through SET domain-dependent modification of histones.18 This putative role has not been tested experimentally and, therefore, it is unknown whether Mll5 imparts principally activating or silencing effects on chromatin. We addressed this issue indirectly through a comparative analysis of gene expression in Mll5−/− and wild-type LSK cells using microarrays. A major conclusion from these experiments was that the deficiency had little detectable impact on gene expression in these cells. It is important to note, however, that LT-HSC represent only 10% of the LSK compartment,30 so the sensitivity of this experiment was not optimal and additional experiments are required to explore the issue more carefully. Despite these limitations, we verified an approximately 2-fold increase in the expression of both Hoxb2 and Hoxb5. Unlike Hoxb4, neither of these genes has yet been associated with roles in the hematopoietic compartment. Other members of the Trithorax group, such as Mll, have recognized roles in the regulation of Hox genes, including Hoxa7, Hoxa9, Hoxc8, and Hoxc9.44,56,57 It remains to be determined whether the observed changes in Hoxb2 and Hoxb5 represent direct or indirect effects of Mll5 or whether they are at least partially explained by differences in the proportions of cells within the LSK compartment.
Our interest in generating Mll5 mutant mice was based, in part, on the fact that MLL5 was identified as a candidate 7q22 myeloid tumor suppressor. A role of Mll5 in regulating stem cell fitness and/or self-renewal is consistent with it being mutated in leukemia, although not perhaps obviously reconcilable with tumor suppression. However, AML1/RUNX1, which is essential for normal hematopoietic development and is mutated in myeloid malignancies,58,59 is an interesting precedent. Whereas AML1-ETO fusions encode dominant oncogenic proteins, germ line and somatic loss-of-function AML1 mutations also contribute to myeloid leukemogenesis by haploinsufficiency.60-65 Indeed, our analysis of Mll5−/− mice suggests that the absence of “second-hit” MLL5 mutations in human myeloid malignancies may reflect the fact that homozygous inactivation would be deleterious to a leukemia-initiating cell. Although Mll5 haploinsufficiency has no overt consequences in mice, heterozygous MLL5 inactivation may cooperate with other genes rendered haploid by 7q22 deletion in leukemogenesis. We conclude that (1) investigating the consequences of Mll5 haploinsufficiency in the context of other known leukemia-associated mutations by using Mll5−/− mice as a sensitized background for retroviral insertional mutagenesis and (2) functionally characterizing other 7q22 candidate TSGs are complementary strategies for fully defining the role of MLL5 mutations in human myeloid malignancies.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the members of the Killeen laboratory for helpful discussions. We thank Ayako Kuroda, Vinh Nguyen, Helen Lu, and Gavina Bentivez for expert technical assistance and Ernesto Diaz-Flores for advice on HSC staining and analysis. We thank Emmanuelle Passegué and members of her laboratory for helpful comments and advice. We gratefully acknowledge the assistance of the UCSF Functional Genomics and Shared Microarray Core Facility and the UCSF Comprehensive Cancer Center Transgenic and Targeted Mutagenesis Core Facility. We thank David Tuveson and Tyler Jacks for LSL-KrasG12D mice and Michelle Le Beau and Ben Braun for insightful comments.
This study was supported by grants from the National Institutes of Health (CACA84221, K.S. and N.K.; CA40046, K.S.), by Scholar (N.K.) and Fellow (J.W.) awards from the Leukemia & Lymphoma Society, and by a postdoctoral fellowship from the Natural Sciences and Engineering Research Council of Canada (M.K.).
National Institutes of Health
Authorship
Contribution: Y.Z., J.W., M.K., and M.T.T. performed research and analyzed data; N.K. and K.S. designed research and analyzed data; and Y.Z., N.K., and K.S. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nigel Killeen, Department of Microbiology and Immunology, 513 Parnassus Avenue, Room HSE 1001, University of California, San Francisco, CA 94143-0414; e-mail: nigel.killeen@ucsf.edu.