Abstract
The BCR-ABL1 fusion gene defines the subgroup of acute lymphoblastic leukemia (ALL) with the worst clinical prognosis. To identify oncogenic lesions that combine with BCR-ABL1 to cause ALL, we used Affymetrix Genome-Wide Human SNP arrays (250K NspI and SNP 6.0), fluorescence in situ hybridization, and genomic polymerase chain reaction to study 106 cases of adult BCR-ABL1–positive ALL. The most frequent somatic copy number alteration was a focal deletion on 7p12 of IKZF1, which encodes the transcription factor Ikaros and was identified in 80 (75%) of 106 patients. Different patterns of deletions occurred, but the most frequent were those characterized by a loss of exons 4 through 7 (Δ4-7) and by removal of exons 2 through 7 (Δ2-7). A variable number of nucleotides (patient specific) were inserted at the conjunction and maintained with fidelity at the time of relapse. The extent of the Δ4-7 deletion correlated with the expression of a dominant-negative isoform with cytoplasmic localization and oncogenic activity, whereas the Δ2-7 deletion resulted in a transcript lacking the translation start site. The IKZF1 deletion also was identified in the progression of chronic myeloid leukemia to lymphoid blast crisis (66%) but never in myeloid blast crisis or chronic-phase chronic myeloid leukemia or in patients with acute myeloid leukemia. Known DNA sequences and structural features were mapped along the breakpoint cluster regions, including heptamer recombination signal sequences recognized by RAG enzymes during V(D)J recombination, suggesting that IKZF1 deletions could arise from aberrant RAG-mediated recombination.
Introduction
Acute lymphoblastic leukemia (ALL) is a heterogeneous disease characterized by multiple subtypes.1 The Philadelphia chromosome (Ph) results from a reciprocal translocation fusing the abelson (ABL1) proto-oncogene from chromosome 9 with the breakpoint cluster region (BCR) sequences on chromosome 22, creating the Bcr-Abl fusion protein, a constitutively activated form of the Abl tyrosine kinase.2 The Ph chromosome is considered the hallmark of chronic myeloid leukemia (CML), but it is also the most frequent cytogenetic aberration associated with ALL, found in 20% to 40% of patients with ALL and in more than 50% of patients ages 50 years or older.3 The presence of the BCR-ABL1 rearrangement worsens the prognosis of ALL and represents the most significant adverse prognostic marker that influences disease outcome.4 Ph-positive (Ph+) ALL is a more aggressive disease than CML, indicating that factors other than BCR-ABL1 are involved in its development and progression.4-6 By using high-resolution single nucleotide polymorphism (SNP) arrays, Mullighan et al7 recently described a deletion on 7p12 of IKZF1, which encodes the transcription factor Ikaros, in 83.7% of BCR–ABL1 ALL cases but not in chronic-phase CML, suggesting that loss of Ikaros8-11 is an important step in the progression of BCR-ABL1–positive ALL. Ikaros is the prototypical member of the Kruppel-like zinc finger (ZnF) transcription factor subfamily, which is required for normal hematopoietic differentiation and proliferation, particularly in lymphoid lineages.8,12,13 The Ikaros gene is transcribed in several isoforms as the result of alternative splicing, essentially altering the expression of exons 3 through 5 that encode the N-terminal DNA-binding domain. The long isoforms (Ik1 through Ik3), which have at least 3 Znfs, can bind efficiently to DNA, unlike the shorter isoforms (Ik4 through Ik8), which lack Znfs and instead behave as dominant-negative isoforms upon homodimerization as well as heterodimerization.14 The most frequent alteration involving the IKZF1 gene was a deletion of an internal subset of exons from 4 through 7 (Δ4-7; Ikaros genomic organization and exon designation is according to Kaufman et al15 ). Because heptamer recombination signal sequences (RSSs) recognized by RAG enzymes during V(D)J recombination were located immediately internal to the deletion breakpoints,16 the authors suggested that the IKZF1 Δ4-7 deletion was the result of aberrant RAG-mediated recombination.
Here, we conducted high-resolution interrogation of genomic copy number alterations (Affymetrix GeneChip Human Mapping 250K NspI and Genome-Wide Human SNP 6.0 array GeneChip microarrays), fluorescence in situ hybridization (FISH), genomic polymerase chain reaction (PCR) analysis, cloning, and direct sequencing of the genomic profile of adult chronic and acute leukemias, including 106 de novo BCR-ABL1–positive ALL adult patients. Our aim was to identify oncogenic lesions that escape standard cytogenetic observations and combine with BCR-ABL1 to induce ALL. The focal deletions on 7p12 in the IKZF1 gene were the most frequent somatic copy number alterations in BCR-ABL1–positive ALL (75%), were predominantly monoallelic, and gave rise in the majority of cases to an Ikaros isoform with cytoplasmic localization and oncogenic activity or a transcript that lacked the translation start site. Furthermore, by mapping the genomic breakpoints, we found that 2 major deletions primarily accounted for BCR-ABL1–positive ALL: Δ4-7, characterized by loss of exons 4 through 7 (55% of IKZF1 deleted patients or 42% considering all patients), and Δ2-7, removing exons 2 through 7 (24% of IKZF1 deleted patients or 18% considering all patients). In a small proportion of cases we also found deletions involving the IKZF1 promoter region (4%). Finally, heptamer RSSs were mapped along the breakpoint cluster regions, confirming that IKZF1 deletions could arise from aberrant RAG-mediated recombination.
Methods
Patients
One hundred six BCR-ABL1–positive adult ALL patients were analyzed by Affymetrix Genome-wide Human SNP arrays (78 patients by 250K NspI and 28 patients by SNP 6.0; 2 patients by both). The median age was 53 years (range, 18-76 years), and the median blast percentage at diagnosis was 90% (range, 18%-99%). The characteristics of the patients are shown in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). The diagnosis of all ALL cases was made on the basis of morphologic, biochemical, and immunologic features of the leukemic cells. In addition, the human lymphoblastoid SD-1 and the human B-cell precursor leukemia BV-173 cell lines also were included in the analysis. Human cell lines were obtained from DMSZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH) and maintained in culture following the DMSZ recommendations.
SNP microarray analysis
Genomic DNA was extracted by the use of the DNA Blood Mini Kit (QIAGEN) from mononuclear cells isolated from peripheral blood or bone marrow aspirates samples by Ficoll gradient centrifugation. DNA was quantified by use of the NanoDrop spectrophotometer, and quality was assessed by the NanoDrop and by agarose gel electrophoresis.
Samples were genotyped with GeneChip Human Mapping 250K NspI and Genome-Wide Human SNP 6.0 array microarrays (Affymetrix) and according to the manufacturer's instructions. For all samples, 250 ng of DNA was digested with NspI (New England Biolabs) and 250 ng with Sty enzymes for cases analyzed by SNP 6.0 array GeneChip. Digested DNA was adaptor-ligated and PCR-amplified by use of the Clontech Titanium TAQ DNA polymerase (CELBIO) in 4 100-μl PCRs for NspI and 3 for StyI. PCR products from each set of reactions were pooled, purified, and fragmented. Fragmented PCR products were then labeled, denatured, and hybridized to the arrays. Arrays were then washed by the use of Affymetrix fluidics stations and scanned with the Gene Chip Scanner 3000. Array image data were analyzed with Affymetrix GCOS 1.4 operating software and Genotyping Console 3.0 to derive .cel data files, which were also exported to the Partek Genomic Suite (Partek Inc) for further data visualization and analysis. Copy number aberrations were scored with the Hidden Markov Model (HMM) and the segmentation approach available within the Partek software package as well as by visual inspection. For the analysis by the HMM, we set the following parameters: max probability (specify the max probability of retaining the same state between neighboring observations) equal to 0.9; genomic decay (the decay parameter describes how quickly the HMM retention of state will decay toward the initial probability) equal to 0, and sigma (specify the Gaussian bandwidth of the distribution from which observation are drawn) equal to 0.8. All aberrations were calculated with respect to a set of 270 HapMap-normal persons and a set of samples obtained from acute leukemia cases in remission to reduce the noise of raw copy number data. When available, comparisons with paired constitutional DNA and with paired remission DNA were performed to exclude inherited copy number variants. Copy number aberrations involving IKZF1 gene were also analyzed and eventually confirmed with the Genotyping Console 3.0 (Affymetrix). SNP array files are available for free download at http://gebbama-prod.cineca.it/MicroarrayRepository/MicroarrayRepository.htm by use of the following username and password: public public. Files are visualized in the Project management section, go to http://gebbama-prod.cineca.it/alfresco/webdav/MICCROARRAY%20LABORATORY/PUBLIC/DOWNLOAD/public and use “public public.”
IKZF1 genomic qPCR
IKZF1 quantitative PCR (qPCR) of all IKZF1 coding exons was performed as described by Mullighan et al7 by the use of a 7900 Real-Time PCR system and 7900 System Software (Applied Biosystems).
IKZF1 reverse transcription PCR
Mononuclear cells were separated by Ficoll-Hypaque density gradient centrifugation, and samples were stored at −190°C in RPMI 1640 with 20% fetal bovine serum and 10% dimethylsulfoxide or in guanidine thiocyanate at −80°C, as needed. Total cellular RNA was extracted from cells by use of the RNeasy total RNA isolation kit (QIAGEN) according to the instructions of the manufacturer, and 1 μg of the total RNA sample was used for cDNA synthesis with Moloney murine leukemia virus reverse transcriptase (Invitrogen) in the presence of dNTPs. Reverse transcription PCR with primers specific for exon 1 and exon 8 (A3 and A2, respectively, in supplemental Table 2) of IKZF1 and nucleotide sequencing was performed to identify the specific Ikaros isoforms as previously described.17 RNA integrity was confirmed by PCR amplification of the GAPDH mRNA, which is expressed ubiquitously in human hematopoietic cells.
IKZF1 gene expression analysis was performed by the use of the Syber Green PCR master Mix (Applied Biosystems) and primers B1 and B2 in supplemental Table 2. Taqman Syber Green assays were performed by the use of a 7900 Real-Time PCR system and 7900 System Software (Applied Biosystems). The quantitative PCR thermal protocol consisted of 50°C for 2 minutes, followed by 95°C for 10 minutes, then 40 cycles of 95°C for 1 minute and 60°C for 1 minute. GAPDH was used as control gene, using primers E3 and E4 in supplemental Table 2.
FISH
FISH analysis was performed as previously described.18 In brief, chromosome preparations from bone marrow cells were hybridized in situ with 1 μg of probe labeled by nick translation. Hybridization was performed at 37°C in 2X saline sodium citrate, 50% (vol/vol) formamide, 10% (wt/vol) dextran sulfate, 5 μg of COT1 DNA (Bethesda Research Laboratories), and 3 μg of sonicated salmon sperm DNA in a volume of 10 μL. Posthybridization washing was performed at 60°C in 0.1× saline sodium citrate (3 times). In cohybridization experiments, the probes were directly labeled with fluorescein and Cy3. Chromosomes were identified by DAPI staining. Digital images were obtained by the use of a Leica DMRXA epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments). Cy3 (red; New England Nuclear), fluorescein (green; Fermentas Life Sciences), and DAPI (blue) fluorescence signals, which were detected by the use of specific filters, were recorded separately as gray-scale images. Pseudocoloring and merging of images were performed with Adobe Photoshop software.
Probes
The whole chromosome paints used for chromosomes 7, derived from flow-sorted chromosomes, was a gift of the Sanger Center (Dr Nigel Carter).
Fosmid probes specific for the IKZF1 gene (G248P800745C8 [chr7:50 381,496-50 422,338] and G248P87926C7 [chr7:50 418,455-50 458,507]), as well as a bacterial artificial chromosome probe-specific for BCR (RP11-164N13 [chr22:21 897,904-22 091,572]), were properly selected accordingly to the latest release (March 2006) of the UC Santa Cruz (UCSC) Human Genome Browser (http://genome.ucsc.edu/).
Cloning of deletion junctions
The information derived from the SNP-array experiments were used to design appropriate forward and reverse primers mapping in the distal and proximal breakpoint regions and used in long-range PCR experiments (supplemental Table 2), aimed at sequencing the deletion junction regions. In case of positive results, nested PCR experiments were performed. The long-range PCR analyses were performed by the use of TaKaRa LA Taq (Cambrex BioScience), and the nested PCR products, containing the junction fragments, were sequenced by use of the Big Dye Terminator Sequencing kits (Applied Biosystems). In the cases of breakpoints occurred in intron 1 (Δ2-7 deletion), the primers C1/C2 and C5 were used in long PCR experiments, and then we chose the primers C3/C4 (sense) and C5 (anti-sense) to obtain a smaller amplified. PCR was performed with 100 ng of genomic DNA in a 25-μL reaction with 2U of FastStart Taq DNA Polymerase (Roche Diagnostics). The long-range PCR cycling parameters were as follows: 95°C for 5 minutes (complete denaturation), followed by 40 cycles at 95°C for 30 seconds, 62°C for 30 seconds, and extension at 72°C for 90 seconds, with an additional extension at the end for 7 minutes. This protocol was used to easily screen for IKZF1 deletions all patients and confirm SNP results. In the case of Δ4-7 deletion, we used the primers D1 and D2 for genomic PCR experiments and to perform an extensive screening. The PCR protocol was the same before reported. Reference genome sequence data were obtained from the University of Santa Cruz browser (http://genome.ucsc.edu/cgi-bin/hgGateway?db = hg18; March 2006 release), and sequence comparison was performed with the BLAST software tool (www.ncbi.nlm.nih.gov/BLAST/).
Western blotting
Cells were lysed with sample buffer (2% sodium dodecyl sulfate in 125 mmol/L Tris HCL, pH 6.8). Cell lysates were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis on 12% gels and then transferred to nitrocellulose membranes (Amersham Biosciences). The blots were incubated for 60 minutes in Odyssey blocking buffer before incubation overnight (4°C) with polyclonal anti-Ikaros antibody (Santa Cruz Biotechnology). Blotted proteins were detected and quantified by use of the Odyssey infrared imaging system LI-COR.
Subcellular localization studies using confocal laser scanning microscopy
The subcellular localization of Ikaros protein(s) was examined by immunofluorescence and confocal laser scanning microscopy as previously described.17
Bioinformatic analysis
Known motifs were searched within the IKZF1 breakpoint regions (intron1_2 and intron7_8), using the following algorithms:
EMBOSS/fuzznuc (http://emboss.sourceforge.net/apps/release/5.0/emboss/apps/fuzznuc.html) to search for the regular expression pattern match of the motifs listed in supplemental Table 3;
EMBOSS/marscan (http://emboss.sourceforge.net/apps/release/5.0/emboss/apps/marscan.html) to find out Matrix/Scaffold Attachment Regions (MAR/SAR) sites;
MAR-Wiz (http://www.futuresoft.org/MAR-Wiz/) to disclose the occurrence of DNA patterns that have been known to occur in the neighborhood of MARs;
RepeatMasker (http://www.repeatmasker.org/) to detect the presence of known human interspersed repeated elements.
Results
SNP microarray analysis detected frequent and recurrent deletions in the IKZF1 gene
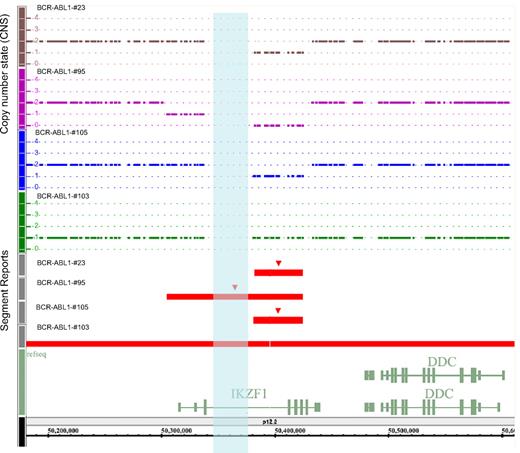
To identify oncogenic lesions that combine with BCR-ABL1 to induce ALL, we profiled the genomes of 106 BCR-ABL1–positive ALL patients by using high-resolution SNP array. We identified regions of high-level amplification and homozygous deletion in all patients, with a mean of 10 somatic copy number alterations per case (range, 22-2), with 2 gains (range, 1-10) and 6.5 losses (range, 2-20). Deletions outnumbered amplifications by almost 3:1. Lesions varied from loss or gain of complete chromosome arms (trisomy 4; monosomy 7; loss of 9p, 10q, 14q, 16q; and gain of 1q and 17q) to microdeletions and microduplications targeting genomic intervals. Deletions of known leukemia-associated genes occurred with a high frequency, as well as deletions in genes involved in the G1/S cell cycle checkpoint and B-cell lineage differentiation (eg, CDKN2A in 42%, BTG1 in 19%, PAX5 in 29%, and EBF1 in 5%). A detailed list of the alterations is shown in supplemental Table 4. The most frequent somatic copy number alteration was a deletion on 7p12 involving the IKZF1 gene (80 [75%] of 106 adult BCR-ABL1 ALL cases) encoding the transcription factor Ikaros, which is required for the earliest stages of lymphoid lineage commitment and acts as a tumor suppressor in mice.8 Deletions involving the IKZF1 gene were monoallelic in 85% of cases and were limited to the gene in all patients, except for 1 patient (BCR-ABL1-#47) and patients with monosomy of chromosome 7 (Figure 1; supplemental Table 5).
Representation from Genotyping Console 3.0 (Affymetrix) of IKZF1 deletions or monosomy 7 in 4 BCR-ABL1–positive ALL patients. Patients #23, #105, and #103 have a monoallelic deletion (DNA copy number state = 1) of different regions of IKZF1. Patient #95 shows a biallelic deletion. Normal diploid DNA content is shown as a continuous line of copy number state = 2. Red segment reports indicate the genomic extent of the IKZF1 deletions. The opalescent vertical box indicates the gap (chr7:50338126-50378125) inside the IKZF1 gene.
Representation from Genotyping Console 3.0 (Affymetrix) of IKZF1 deletions or monosomy 7 in 4 BCR-ABL1–positive ALL patients. Patients #23, #105, and #103 have a monoallelic deletion (DNA copy number state = 1) of different regions of IKZF1. Patient #95 shows a biallelic deletion. Normal diploid DNA content is shown as a continuous line of copy number state = 2. Red segment reports indicate the genomic extent of the IKZF1 deletions. The opalescent vertical box indicates the gap (chr7:50338126-50378125) inside the IKZF1 gene.
IKZF1 deletions were confirmed by FISH analysis and Q-PCR
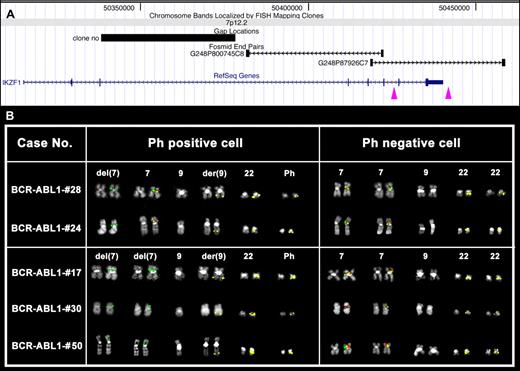
SNP array data were confirmed by the use of the overlapping fosmid clones G248P800745C8 (chr7:50 381,496-50 422,338), G248P82319E12 (chr7:50 400,870-50 443,154), and G248P87926C7 (chr7:50 418,455-50 458,507) in cohybridization with a probe encompassing the BCR gene (bacterial artificial chromosome clone RP11-164N13, chr22:21 897,904-22 091,572), to specifically identify Ph+ cells.
Patients BCR-ABL1-#24 and BCR-ABL1-#28 showed a heterozygous deletion for the fosmid G248P800745C8 in Ph+ cells only (Figure 2A). Patients BCR-ABL1-#17, BCR-ABL1-#30, and BCR-ABL1-#50 showed homozygous IKZF1 deletions (Figure 2B). Interestingly, patient BCR-ABL1-#17 showed 2 deletions with a different proximal breakpoint (Figure 2A-B) in Ph+ cells; Ph− cells were normal. FISH analysis not only confirmed the SNP array results but also demonstrated that BCR-ABL1 rearrangements and IKZF1 deletions were in the same clones and, further, that all Ph+ metaphases also had the IKZF1 deletion.
FISH analysis of IKZF1 deletions. (A) Map of the deletion proximal breakpoints within the IKZF1 gene is shown. Purple arrowheads indicate the breakpoint regions mapped by FISH experiments. Particularly, the left arrow corresponds to both Δ4-7 and Δ2-7 deletion proximal breakpoint regions, whereas the right arrow indicates a novel proximal breakpoint region identified in patient #17 (see panel B). (B) FISH results obtained in some of the cases under study showing homozygous (cases #17, #30, and #50) or heterozygous (cases #24 and #28) deletion of the fosmid clone G248P800745C8 (red) only in Ph+ (middle column), as shown by the FISH pattern of the RP11-164N13 (BCR) probe (in yellow). Clone G248P87926C7 (green) is always retained on deleted chromosomes 7. Notably, in patient #17, 1 of the 2 green signals is fainter than the other, revealing the occurrence of a partial deletion of G248P87926C7 in 1 of the 2 chromosome 7 homologs.
FISH analysis of IKZF1 deletions. (A) Map of the deletion proximal breakpoints within the IKZF1 gene is shown. Purple arrowheads indicate the breakpoint regions mapped by FISH experiments. Particularly, the left arrow corresponds to both Δ4-7 and Δ2-7 deletion proximal breakpoint regions, whereas the right arrow indicates a novel proximal breakpoint region identified in patient #17 (see panel B). (B) FISH results obtained in some of the cases under study showing homozygous (cases #17, #30, and #50) or heterozygous (cases #24 and #28) deletion of the fosmid clone G248P800745C8 (red) only in Ph+ (middle column), as shown by the FISH pattern of the RP11-164N13 (BCR) probe (in yellow). Clone G248P87926C7 (green) is always retained on deleted chromosomes 7. Notably, in patient #17, 1 of the 2 green signals is fainter than the other, revealing the occurrence of a partial deletion of G248P87926C7 in 1 of the 2 chromosome 7 homologs.
Real-time Q-PCR of promoter and all IKZF1 exons was performed to confirm SNP results and to characterize the focal deletions. A big concordance between the extension of deletions previously identified by SNP array and the Q-PCR results was found. Furthermore, Q-PCR allowed us to determine the extension of homozygous deletions, suggesting that in some cases different proximal and distal breakpoints can occur on the 2 alleles (supplemental Table 6). Q-PCR on Ikaros mRNA demonstrated that the deletion of IKZF1 correlated with a down-modulation at the transcript level of the Ikaros DNA-binding isoforms (supplemental Figure 1).
Two types of deletions in the IKZF1 gene account for most cases of BCR-ABL1–positive ALL
The information derived from the SNP array, genomic Q-PCR, and FISH experiments was used to design appropriate forward and reverse primers mapping in the distal and proximal breakpoint regions, which were used in genomic PCR and sequencing experiments aimed at defining the deletion junction regions and at investigating the mechanisms responsible for IKZF1 deletions. This combined approach (genomic PCR and direct sequencing) has been demonstrated to be a power tool to identify IKZF1 deletions in those cases in which SNP array analysis has failed.
The Δ4-7 deletion was identified in 42% of patients with BCR-ABL1–positive ALL
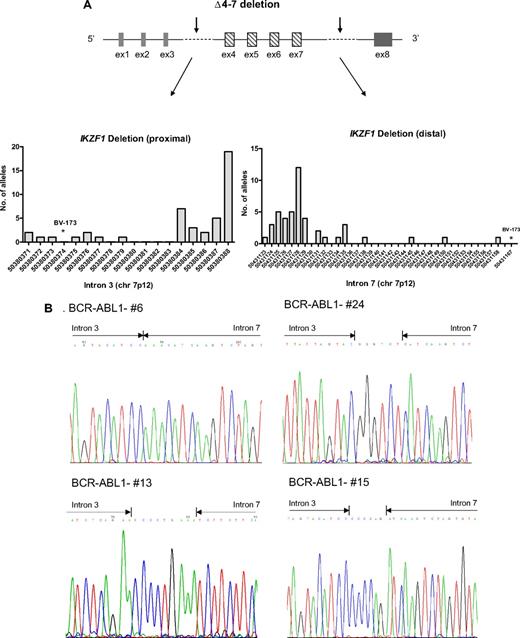
In 44 (42%) 106 patients, the deletion of IKZF1 was limited to exons 4-7 (Δ4-7), with breakpoints occurring in the region ranging from 50380371 to 50380388 and from 50431123 to 50431167 in introns 3 and 7 (supplemental Figure 2), respectively, on chromosome 7p12. A variable number of patient-specific nucleotides were inserted at the conjunction, and cloning analysis showed that the breakpoints were the same in each BCR-ABL1–positive cell (supplemental Table 7; Figure 3A-B). As previously shown,7 the extent of the deletion correlated with the expression of a dominant-negative isoform, Ik6, with cytoplasmic localization and oncogenic activity (supplemental Figure 3). The Ik6 transcript and protein, which lack exons 4-7 and all ZnF motifs, respectively, were exclusively observed in cases harboring the IKZF1 Δ4-7 deletion. This finding validates the concept that the expression of non–DNA-binding Ikaros isoforms is caused by IKZF1 genomic abnormalities and not aberrant splicing after transcription induced by BCR-ABL1.19
IKZF1 Δ4-7 breakpoints. (A) Schematic representation of Δ4-7 deletion with arrows indicating the region in which the breakpoints occur. In the graphs are shown the chromosome positions of breakpoints in the proximal (intron 3) and distal (intron 7) regions. The most frequent breakpoints occur at 50380384 and 50380388 in the proximal region and at 50431125 and 50431128 in the distal region. Primers used were D1 and D2 (supplemental Table 2); products were then directly sequenced to characterize the sequence flanking deletion breakpoints. (B) Pherograms of sequencing of IKZF1 Δ4-7 breakpoints. Regions matching the reference genomic IKZF1 sequence are shown by arrows, separated by additional nucleotides not matching the consensus sequence.
IKZF1 Δ4-7 breakpoints. (A) Schematic representation of Δ4-7 deletion with arrows indicating the region in which the breakpoints occur. In the graphs are shown the chromosome positions of breakpoints in the proximal (intron 3) and distal (intron 7) regions. The most frequent breakpoints occur at 50380384 and 50380388 in the proximal region and at 50431125 and 50431128 in the distal region. Primers used were D1 and D2 (supplemental Table 2); products were then directly sequenced to characterize the sequence flanking deletion breakpoints. (B) Pherograms of sequencing of IKZF1 Δ4-7 breakpoints. Regions matching the reference genomic IKZF1 sequence are shown by arrows, separated by additional nucleotides not matching the consensus sequence.
The Δ2-7 deletion (type B) was identified in 18% of patients with BCR-ABL1–positive ALL
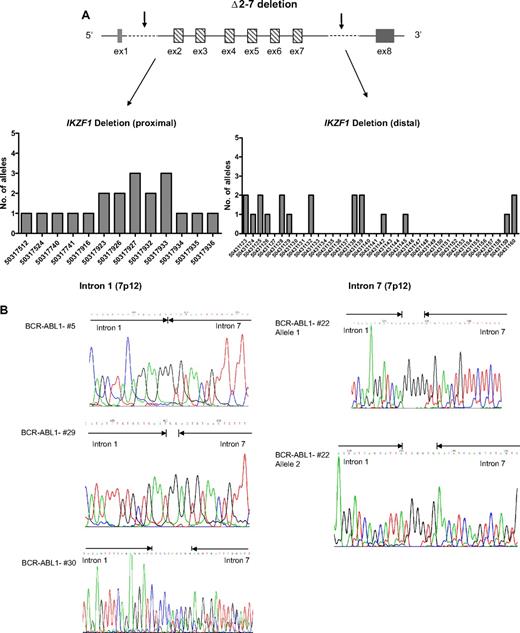
In 19 (18%) of 106 patients, the IKZF1 deletion involved exons 2-7 (Δ2-7), with a variable pattern of breakpoints in intron 1 (from 50317112 to 50317936) not previously reported (supplemental Table 8; Figure 4A-B), whereas the breakpoints in intron 7 were in the same region of those of the Δ4-7 deletion. There was a correlation between the extension of this deletion and the expression of an aberrant transcript containing only exons 1 and 8, which lacked the translation start (supplemental Figure 4).
IKZF1 Δ2-7 breakpoints. (A) Schematic representation of Δ2-7 deletion with arrows indicating the region in which the breakpoints occur. In the graphs are shown the chromosome positions of breakpoints in the proximal (intron 1) and distal (intron 7) regions. The most frequent breakpoints occur at 50317927 and 50317933 in the proximal region and at 50431128 in the distal region. Primers used were C3/C4 and C5 (supplemental Table 2); products were then directly sequenced to characterize the sequence flanking deletion breakpoints. (B) Pherograms of sequencing of IKZF1 Δ2-7 breakpoints. Regions matching the reference genomic IKZF1 sequence are shown by arrows, separated by additional nucleotides not matching the consensus sequence.
IKZF1 Δ2-7 breakpoints. (A) Schematic representation of Δ2-7 deletion with arrows indicating the region in which the breakpoints occur. In the graphs are shown the chromosome positions of breakpoints in the proximal (intron 1) and distal (intron 7) regions. The most frequent breakpoints occur at 50317927 and 50317933 in the proximal region and at 50431128 in the distal region. Primers used were C3/C4 and C5 (supplemental Table 2); products were then directly sequenced to characterize the sequence flanking deletion breakpoints. (B) Pherograms of sequencing of IKZF1 Δ2-7 breakpoints. Regions matching the reference genomic IKZF1 sequence are shown by arrows, separated by additional nucleotides not matching the consensus sequence.
Other deletions may occur in the IKZF1 gene at low frequency
Other deletions in the IKZF1 gene were observed at low frequency. Interestingly, 3 patients had a homozygous IKZF1 deletion with 2 different proximal and distal breakpoints, giving rise to Δ4-7 and Δ4-8 deletions (BCR-ABL1-#17) and Δ2-7 and Δ4-7 (BCR-ABL1-#50 and BCR-ABL-#95; supplemental Tables 9-10). In 1 patient (BCR-ABL-#36), we identified a deletion of the entire IKZF1 gene and part of GRB10, and in 2 patients (BCR-ABL1-#47 and BCR-ABL1-#71), we identified a smaller deletion spanning the whole promoter and first exon of IKZF1. Another uncommon deletion was that spanning from promoter to exon 7 (supplemental Tables 5-6). Overall, deletions involving the IKZF1 gene were identified in 68 (64%) of 106 BCR-ABL1-positive ALL patients. It is important to consider that in 13 patients, we had a monosomy 7; in these cases, therefore, there was a deletion of the entire IKZF1 gene on one allele. In only one case, we also found the Δ4-7 deletion on the other allele. Because monosomy 7 results in Ikaros haploinsufficiency, like many focal deletions, if we include these cases in our analysis, the rate of patients with IKZF1 loss and/or haploinsufficiency increases to 75% (80/106).
IKZF1 deletion is not detected in other types of leukemia except for lymphoid blast crisis CML
To investigate the possibility that the IKZF1 alteration is a frequent event across different types of leukemia, we used high-resolution SNP arrays and genomic PCR to examine 30 CML chronic-phase patients, 10 CML in blast crisis patients (3 lymphoid and 7 myeloid), 28 adult AML patients with different French-American-British classification, and 2 hypereosinophilic syndrome patients (supplemental Table 11). No focal deletions were identified in these leukemia types except for 2 patients with CML in lymphoid blast crisis. In particular, we found the Δ4-7 deletion in one case (CML-ly-BC#1) and the Δ2-7 deletion in another case (CML-ly-BC#3), suggesting a role for IKZF1 genomic alterations in the progression from chronic phase to blast crisis in CML, given that the deletion was not detected at diagnosis (supplemental Table 10).
RSS-flanked IKZF1 genomic breakpoints
To investigate the mechanisms responsible for the generation of the IKZF1 Δ4-7 and Δ2-7 deletions in BCR-ABL1–positive leukemia, we searched within the IKZF1 breakpoint regions for regular expression pattern matches of the motifs listed in supplemental Table 3, such as activation-induced cytidine deaminase (AID) motif (DGYW/WRCH), V(D)J RSS, topoisomerase II binding site, translin binding site, Chi-like sequence, PurI binding site, Eukaryotes replication origin sequence, ARS-S cerevisiae, putative triple helices, pyrimidine trait, Human minisatellite core sequence, and Satellite III core. As mentioned previously,7 RSSs recognized by RAG enzymes during V(D)J recombination were located immediately internal to the deletion breakpoints, and a variable number of additional nucleotides (patient-specific) were present between the consensus intron 3 and 7 sequences, which is suggestive of the action of terminal deoxynucleotidyl transferase
Interestingly, the breakpoints in the IKZF1 gene and the additional nucleotides inserted at the junction were maintained with fidelity at the time of relapse and could be exploited for monitoring minimal residual disease. We also noted that AID consensus sequences (DGYW/WRCH corresponding to AGTA and TGTT or TGTA in our cases) flanked the genomic breakpoints in both intron 3 and intron 7 (supplemental Table 6). Furthermore, AID consensus sequences were found very close to the breakpoints in intron 1 in the case of the Δ2-7 deletion (supplemental Figure 5). Because there was no evidence of nucleotide changes to suggest AID activity and because the AID consensus sequence is highly degenerate and may likely occur very frequently by chance, we hypothesized that aberrant RAG activity could be responsible for the IKZF1 deletions.
Discussion
The Ph chromosome encodes the oncogenic Bcr-Abl kinase and defines a subgroup of ALL with a particularly unfavorable prognosis.1 The reasons for the aggressive nature of BCR-ABL1–positive ALL are still under investigation and have not yet been elucidated. To identify additional oncogenic lesions involved in the generation of BCR-ABL1–positive ALL and responsible for its poor outcome and biologic difference from CML, we performed a high-resolution genomic study of copy number alterations in a large cohort of adult patients with acute and chronic leukemia, including 106 adult patients with BCR-ABL1–positive leukemia. We found that homozygous or heterozygous deletions in the IKZF1 gene frequently occur only in BCR-ABL1–positive ALL (75%) patients or lymphoid blast crisis CML (66%), as previously demonstrated by Mullighan et al,7 in 21 pediatric and 22 adult BCR–ABL1 ALL patients.
IKZF1 encodes a ZnF protein required for lymphoid lineage differentiation, proliferation, and function.19,20 Ikaros contains 2 separate regions with ZnF domains: 4 DNA-binding ZnFs near the N-terminus and 2 ZnFs for protein–protein interactions near the C terminus. Alternative splicing can generate multiple functionally different Ikaros isoforms that lack variable numbers of internal exons. Isoforms that lack the N-terminal ZnFs are unable to bind transcriptional targets normally but retain the C-terminal ZnFs and the ability to dimerise and act as dominant negative inhibitors of Ikaros function. Ikaros transgenic and mutant mouse models have clearly demonstrated the important role of Ikaros in both normal hematopoiesis and tumor suppression. Mutant Ikaros −/− mice have severe lymphoid cell defects,21,22 but heterozygous Ikaros DN +/− mice invariably develop T-cell malignancies.
During the last decade, several groups11,23-27 have reported the expression of aberrant Ikaros isoforms caused by alternative splicing in different acute leukemias. In 2006, Klein et al19 suggested that the expression of aberrant Ikaros isoforms occurred at a posttranscriptional level as a result of the action of the Bcr-Abl fusion protein. This hypothesis was reversed by the findings of Mullighan et al7 and by our study, which strongly demonstrated that intragenic deletions in the IKZF1 gene were responsible for the generation of different aberrant isoforms. Here, we characterized and mapped all breakpoints, demonstrating that 2 frequent deletions occur in BCR-ABL1–positive lymphoid leukemias: Δ4-7 (42%), removing exons 4 through 7, and Δ2-7 (18%), resulting in a transcript with only exons 1 and 8.
An elevated frequency of genomic aberrations could be directly caused by an abnormally high incidence of DNA double-strand breaks. In normal cells, DNA lesions are detected and repaired by sophisticated physiologic machinery and a system of cell cycle checkpoints, preventing cells that have sustained DNA damage from proliferating further. In this study, we performed a bionformatic analysis to determine the mechanisms responsible for the generation of IKZF1 deletions. Known motifs associated with deletions were searched within the IKZF1 breakpoint regions by the use of different algorithms, and known DNA sequence and structural features were mapped along the breakpoint cluster regions, including heptamer RSSs, suggesting that IKZF1 deletions could arise from aberrant RAG-mediated recombination.
In conclusion, our results confirmed, as previously showed by Mullighan et al,7 that IKZF1 deletion is the most frequent somatic copy number alteration in Ph+ ALL, and they provided new details on other common breakpoints and on the mechanisms involved in the rearrangement. It is likely that Ikaros loss combines with BCR-ABL1 to induce lymphoblastic leukemia, arresting B-lymphoid maturation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Serena Formica (University of Bologna), Anna Ferrari (University of Bologna), Monica Messina (University of Rome) for help in performing SNP arrays, and Silvia Giuliani and Elda Rossi (CINECA) for help in file repository (CINECA, Bologna).
This work was supported by AIL, European LeukaemiaNet, AIRC, Fondazione Del Monte di Bologna e Ravenna, FIRB 2006, Ateneo 60% grants, and Gimema Onlus Working Party ALL and CML, and Biopharmanet project.
Authorship
Contributions: I.I. designed research, performed SNP array analysis, and drafted the manuscript; C.T.S., L.I., A.L., and C.B. performed molecular and conventional cytogenetic analysis; P.D. performed bioinformatic analysis; E.O. and A.A. contributed to SNP array analysis; S.S., A.L., C.P., S.C., P.P.P., S.P., D.R., F.P., G.S., and M.B. contributed in the development of the study; S.C., A.V., M.V, and R.F. contributed in data collection and data analysis; D.C. and F.M. contributed in proteomic analysis and data analysis; and G.M. designed research and gave final approval to the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A list of the GIMEMA AL WP participants is available in the supplemental Appendix.
Correspondence: Prof Giovanni Martinelli, Molecular Biology Unit, Department of Hematology/Oncology Seràgnoli, University of Bologna, Via Massarenti, 9-40138 Bologna, Italy; e-mail: giovanni.martinelli2@unibo.it.
References
Supplemental data
Additional supplemental tables and supplemental figures can be seen here.