Minimally differentiated acute myeloid leukemia (AML-M0) is defined by immature morphology and expression of early hematologic markers. By gene expression profiling (GEP) and subsequent unsupervised analysis of 35 AML-M0 samples and 253 previously reported AML cases, we demonstrate that AML-M0 cases express a unique signature that is largely separated from other molecular subtypes. Hematologic transcription regulators such as CEBPA, CEBPD, and ETV6, and the differentiation associated gene MPO appeared strongly down-regulated, in line with the primitive state of this leukemia. AML-M0 frequently carries loss-of-function RUNX1 mutation. Unsupervised analyses revealed a subdivision between AML-M0 cases with and without RUNX1 mutations. RUNX1 mutant AML-M0 samples showed a distinct up-regulation of B cell–related genes such as members of the B-cell receptor complex, transcription regulators RUNX3, ETS2, IRF8, or PRDM1, and major histocompatibility complex class II genes. Importantly, prediction with high accuracy of the AML-M0 subtype and prediction of patients carrying RUNX1 mutation within this subtype were possible based on the expression level of only a few transcripts. We propose that RUNX1 mutations in this AML subgroup cause lineage infidelity, leading to aberrant coexpression of myeloid and B-lymphoid genes. Furthermore, our results imply that AML-M0, although originally determined by morphology, constitutes a leukemia subgroup.
Introduction
Minimally differentiated acute myeloid leukemia, known in the French-American-British (FAB) classification as AML-M0, represents approximately 5% of all AML cases, frequently occurs in elderly patients, and has a remarkably poor prognosis.1,,–4 It is molecularly characterized by low expression of MPO and expression of at least 1 myeloid antigen (CD13, CD33, CD15).1 In some cases, coexpression of lymphoid-associated antigens occurs.5 The immature status of this leukemia is emphasized immunologically by the frequent expression of CD34, TdT, HLA-DR, and CD117.2,–4 Morphologically, AML-M0 blasts are large and agranular, sometimes resembling lymphoblasts. Contrary to other AML subtypes, there are no characteristic chromosomal translocations related to AML-M0. Nevertheless, the incidence of abnormal karyotypes, complex karyotypes, and unbalanced chromosome changes is more frequent in this leukemia (71%-81%) than in other subtypes.3,4,6,7 Thus, although these leukemias share common features, morphologic, immunologic, and cytogenetic analysis reveals variability among cases. These observations raise the question whether AML-M0 leukemias represent 1 or multiple subtypes.
The most characteristic molecular alterations found in AML-M0 are RUNX1 (alias AML1) mutations. Biallelic or dominant-negative mutations in RUNX1 have been reported in 15% to 35% of this leukemia subtype.8 RUNX1 is a transcription factor that binds DNA through its Runt domain.9 Runx1 protein has been demonstrated to be essential for definitive hematopoiesis in various mouse models, and in Runx1 conditional knockouts it has been shown that the encoded transcription factor is required for the development of both myeloid and lymphoid lineages.9,10 RUNX1 is frequently involved in chromosomal aberrations in various hematologic malignancies.11 Although less frequent, FLT3, RAS, PTPN11, and ETV6 mutations have also been reported in AML-M0.12,13
Gene expression profiling (GEP) has been successfully applied to identify molecularly well-characterized AML subtypes, such as cases that carry translocations t(8;21), t(15;17), or inv(16).14 Moreover, by applying GEP several novel AML subtypes have been uncovered, of which some appeared to carry signatures associated with specific molecular lesions, for example, mutations in the genes encoding for CEBPA or NPM1.15 Besides the fact that genome-wide approaches offer the potential to improve AML diagnostics, clinical outcome prediction, and discovery of new subtypes, it may also contribute to the finding of new therapeutic targets by increasing the knowledge on deregulated pathways in leukemia.
Here we applied gene expression profiling on a cohort of 35 AML-M0 samples. We demonstrate that AML-M0 has a distinct gene expression signature, which can be further divided into 2 unique AML subtypes. Moreover, we show that one of these AML-M0 subgroups is fully associated with RUNX1 mutations. Interestingly, the RUNX1 mutant subgroup carries a signature of genes, of which many are related to early B-cell development. Importantly, these AML-M0 subtypes can be predicted with high accuracy by GEP using a minimal set of transcripts, providing the possibility to develop a diagnostic assay to identify patients with these subtypes.
Methods
Patient material
After informed consent was obtained from patients in accordance with the Declaration of Helsinki, bone marrow aspirates or peripheral blood samples of AML patients with a diagnosis of primary AML, classified morphologically and immunophenotypically as AML-M0, were collected from the medical centers of the University of Berlin, Leiden University, University of Groningen, Erasmus University, Rotterdam, and University of Vienna. Blasts and mononuclear cells were purified by Ficoll-Hypaque (Nygaard) centrifugation and cryopreserved. The 35 patients used in this study were a subgroup of a cohort of 52 patients for whom sufficient mRNA was available (patients 1-3, 5-9, 18-28, 30-35, 39, 41, 42, 45, 52, 55-58, 60).16 These AML-M0 cases were morphologically, immunophenotypically, cytogenetically, and genetically characterized,16 and relevant information is provided in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). The CEL files of the AML-M0 expression data discussed in this paper are available in the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) accession number GSE17061.17 Clinical, cytogenetic, and molecular information as well as the gene expression profiles of all 253 reference primary AML cases and 3 normal CD34+ sorted bone marrow cell samples18 were obtained from the GEO (accession number GSE1159). Approval was obtained from the Erasmus University Medical Center Institutional Review Board for these studies.
RNA extraction, labeling, and hybridization
mRNA for gene expression analysis was isolated using QIAamp RNA Blood Mini Kit following the manufacturer's instructions (QIAGEN). RNA concentration, quality, and purity were examined using the RNA 6000 Nano assay on the Agilent 2100 Bioanalyzer (Agilent). None of the RNA samples showed degradation (28S/18S rRNA ratio ≥ 2) or contamination by DNA.
RNA labeling and hybridization were performed as previously described with few modifications.18 In short, 2 μg total RNA of each sample was used to prepare antisense biotinylated RNA. Single- and double-stranded cDNA was synthesized according to the manufacturer's protocol (Invitrogen Life Technologies). cDNA was transcribed into biotin-labeled cRNA using a MEGAScript T7 labeling kit (Ambion). After cleanup and fragmentation, the labeled cRNA was hybridized to an Affymetrix GeneChip Human Genome U133 Plus 2.0 array (containing targets for more than 54 000 probe sets) for 16 hours at 45°C. Arrays were washed, stained, and finally scanned in a GeneChip Scanner 3000 station (Affymetrix).
Data analysis
The measured intensity values were analyzed using GeneChip Operating Software (Affymetrix). The percentage of present calls (40.2 ± 3.6), background (59.2 ± 8.6), and 3′ to 5′ ratio of ACTIN (1.65 ± 0.36) and GAPDH (1.30 ± 0.24) indicated a high quality of samples and an overall comparability.
Scanned array images were normalized by Robust Multichip Analysis. After normalization, the data were back-transformed to normal intensity values. For each probe set, the geometric mean of the hybridization intensities of all samples from the patients was calculated. The level of expression of each probe set in every sample was determined relative to this geometric mean and log2 transformed to ascribe equal weight to gene expression levels with similar relative distances to the geometric mean.
Unsupervised cluster analysis was performed in Omniviz (Version 5.1.1; BioWisdom Inc) using Pearson correlation in the Correlation View.18
Supervised analysis was based on Significance Analysis of Microarrays (SAM; Stanford University).19 Cutoff values for significantly expressed genes were chosen based on the false discovery rate (FDR) and the fold change (FC). We compared AML-M0 versus all other AML subtypes (SAM: FDR = 0, FC > 2) and AML-M0 group A versus group B (see “Expression of genes involved in B-cell development is upregulated in AML-M0 with RUNX1 mutations”; SAM: FDR = 0.05, FC ≥ 1.5). All genes significantly differentially expressed are available in supplemental Tables 2 and 6 (annotation based on Affymetrix file HG-U133_Plus_2.na23.annot.txt). Functional analysis of the results was done using Ingenuity Pathway Analysis (Ingenuity Systems, http://www.ingenuity.com) and DAVID.20,21
All supervised class-prediction analyses were performed by applying Prediction Analysis for Microarray (PAM; Stanford University) software (Version 1.28) in R (Version 2.1.0).22 We used the method of the nearest shrunken centroids to identify a minimal set of genes that best characterizes a predefined class as defined by the smallest number of false-negative predictions. The prediction error was calculated by 10-fold cross validation within the training set (two-thirds of the samples) followed by the use of a validation set (one-third of the samples) for AML-M0 prediction. For prediction of RUNX1 mutation in AML-M0, the prediction error was calculated by a 10-fold cross validation without splitting between training and validation set due to the reduced number of samples. For this analysis, only 25 samples for which the RUNX1 mutation status was clear were used.
Results
AML-M0 leukemias represent a distinct group based on gene expression profiling
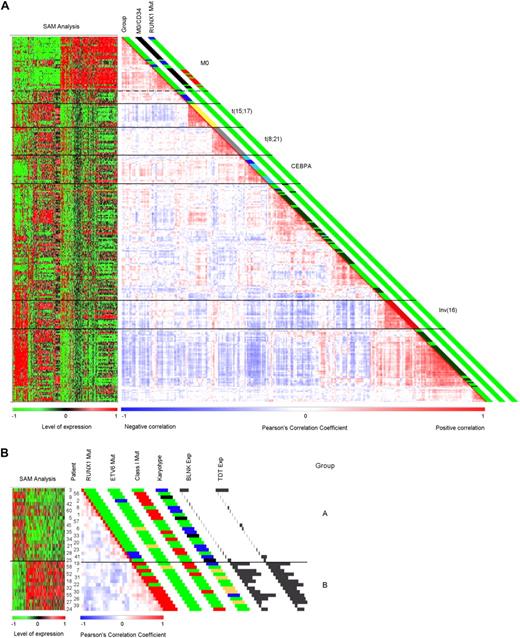
To investigate whether the AML-M0 patient samples carry a specific gene expression signature that distinguishes them from other primary AML cases, we performed mRNA hybridizations for 35 cases using Affymetrix GeneChip Human Genome U133 Plus 2.0 arrays, and compared them with a previously published representative cohort of 253 AMLs.18 Unsupervised clustering using Pearson correlation showed a distinct grouping of the AML-M0 samples (Figure 1A). The clustering of AML-M0 was stable using different numbers of genes in the unsupervised analysis (cutoffs ranging from 8- to 12-fold differential expression to the geometric mean; 3472 to 1794 probe sets). We selected a cutoff of 8-fold as the most optimal setting (Figure 1A), because at this level the previously published clusters characterized by the AML subtypes carrying an inv(16), t(8;21), t(15;17), CEBPA, and NPM1 mutants were best recognized.18
Pairwise correlation plots and SAM analyses heat maps. Samples were clustered based on pairwise correlation of gene expression profiles and are displayed based on a color gradient ranging from deep blue for a negative correlation to vivid red for a positive correlation. A negative correlation between 2 samples indicates that the genes used in the analysis have opposite expression tendencies (high/low). To the left of each correlation plot, a heat map showing the results from the SAM analysis is presented. Results are shown as an increasing green to red gradient representing the expression level. (A) Unsupervised clustering of AML-M0 with 253 AML samples of other subtypes and 3 CD34 sorted normal samples and corresponding SAM analysis (cutoff: 8-fold; 3472 probe sets). A dotted line delimits the AML-M0 samples within the overall AML-M0 cluster. The first column to the right of the correlation plot refers to the major mutation group the samples belong to and patients belonging to cluster no. 1018 (dark blue: cluster no. 10; yellow: t(15;17); gray: t(8;21); light blue: CEBPA mutation; dark red: inv(16); dark green: NPM1 mutations). The second column indicates the AML-M0 patients (in black) and CD34+ samples (in blue). The last column refers to the mutation status of RUNX1 in the AML-M0 samples. Mutations expected to lead to complete loss of function of RUNX1 are red and heterozygous mutations outside the runt domain are blue. In all 3 columns, green represents either a negative result for the variable(s) or no information available. (B) Unsupervised cluster analyses of all 35 AML-M0 samples and SAM analysis comparing groups A and B (cutoff: 9-fold; 1327 probe sets). Unsupervised clustering shows a strong correlation between samples with RUNX1 mutation, a tendency also evident in panel A. The first column to the left of correlation plot shows the status of the RUNX1 mutation as in panel A. The second column shows the mutation status for ETV6, blue representing a translocation involving this gene, whereas orange shows point mutations or insertions.13 In the third column, different detected mutations leading to a proliferative advantage (class I Mut) are represented in red (these include mutation of RAS, PTPN11, FLT3, and JAK2). The fourth column provides cytogenetic information. Trisomy 13, represented in yellow, is correlated to RUNX1 mutation.23,24 Blue stands for a normal karyotype; black, for a complex karyotype (more than 5 abnormalities). The last 2 columns represent expression levels for BLNK and TDT in all samples in which the histograms are proportional to the level of expression (probe set BLNK: 207655_s_at and TDT: 210487_at). In all columns, green represents either a negative result for the variable(s) or no information available.
Pairwise correlation plots and SAM analyses heat maps. Samples were clustered based on pairwise correlation of gene expression profiles and are displayed based on a color gradient ranging from deep blue for a negative correlation to vivid red for a positive correlation. A negative correlation between 2 samples indicates that the genes used in the analysis have opposite expression tendencies (high/low). To the left of each correlation plot, a heat map showing the results from the SAM analysis is presented. Results are shown as an increasing green to red gradient representing the expression level. (A) Unsupervised clustering of AML-M0 with 253 AML samples of other subtypes and 3 CD34 sorted normal samples and corresponding SAM analysis (cutoff: 8-fold; 3472 probe sets). A dotted line delimits the AML-M0 samples within the overall AML-M0 cluster. The first column to the right of the correlation plot refers to the major mutation group the samples belong to and patients belonging to cluster no. 1018 (dark blue: cluster no. 10; yellow: t(15;17); gray: t(8;21); light blue: CEBPA mutation; dark red: inv(16); dark green: NPM1 mutations). The second column indicates the AML-M0 patients (in black) and CD34+ samples (in blue). The last column refers to the mutation status of RUNX1 in the AML-M0 samples. Mutations expected to lead to complete loss of function of RUNX1 are red and heterozygous mutations outside the runt domain are blue. In all 3 columns, green represents either a negative result for the variable(s) or no information available. (B) Unsupervised cluster analyses of all 35 AML-M0 samples and SAM analysis comparing groups A and B (cutoff: 9-fold; 1327 probe sets). Unsupervised clustering shows a strong correlation between samples with RUNX1 mutation, a tendency also evident in panel A. The first column to the left of correlation plot shows the status of the RUNX1 mutation as in panel A. The second column shows the mutation status for ETV6, blue representing a translocation involving this gene, whereas orange shows point mutations or insertions.13 In the third column, different detected mutations leading to a proliferative advantage (class I Mut) are represented in red (these include mutation of RAS, PTPN11, FLT3, and JAK2). The fourth column provides cytogenetic information. Trisomy 13, represented in yellow, is correlated to RUNX1 mutation.23,24 Blue stands for a normal karyotype; black, for a complex karyotype (more than 5 abnormalities). The last 2 columns represent expression levels for BLNK and TDT in all samples in which the histograms are proportional to the level of expression (probe set BLNK: 207655_s_at and TDT: 210487_at). In all columns, green represents either a negative result for the variable(s) or no information available.
Thirty-three of the 35 M0 and 7 of 253 AML patient samples were present in the distinct AML-M0 cluster (Figure 1A). These 7 patients were classified as FAB-M1 or -M5. Moreover, 10 other patients who clustered immediately adjacent to the newly defined FAB-M0 cluster were also classified as FAB-M1 or -M5. Nine of these 17 patients were previously shown to cluster in a subgroup with adverse prognosis (cluster no. 10).18 The 2 AML-M0 patients who did not cluster in the newly defined M0-cluster carried atypical translocations involving chromosome 11 but did not associate with any distinct molecular abnormality or one of the previously defined gene expression clusters.18 Interestingly, profiles of 3 normal CD34+ sorted progenitor bone marrow cell samples that were analyzed also grouped within the AML-M0 cluster (Figure 1A), which may relate to the early differentiation stage of hematopoiesis of AML-M0 cases based on molecular analysis.
Genes involved in normal hematopoiesis are specifically deregulated in AML-M0
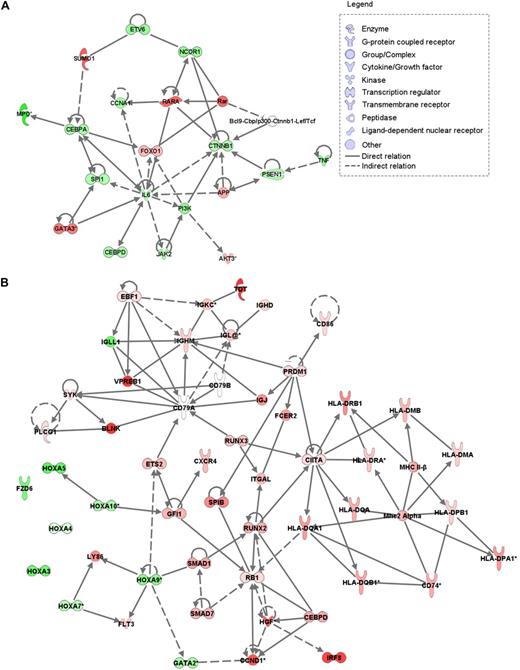
To identify the genes important in AML-M0 pathogenesis, we compared the expression profiles of the 35 AML-M0 patients with the 253 AML samples of other subtypes (Figure 1A). The comparison yielded 1635 significantly differentially expressed probe sets corresponding to 1064 genes and 348 probe sets without annotation (supplemental Table 2). In line with the AML-M0 immunophenotype, the top down-regulated gene was MPO (Figure 2A). Genes known to be particularly involved in myeloid development, for example, CEBPA, CEBPD, SPI1 (PU.1), ETV6, and JAK2, appeared to be significantly down-regulated, whereas RARA, AKT3, and GATA3 were up-regulated in AML-M0 (Figure 2A). Further functional annotation of the differentially expressed genes showed an enrichment of mitochondria-related genes and ribosomal genes (supplemental Tables 3-4). Genes related to the phosphorylative oxidation complexes (I, II, and IV) and several ATP synthethases encoding genes were found up-regulated, whereas the pathways involved in NAD+ degradation (the first electron acceptor in the oxidative phosphorylation) were down-regulated (supplemental Table 3 and supplemental Figures 1-2).
Overview of biologic networks of differently expressed genes. Relation between different genes is based on Ingenuity knowledge database and was generated using Ingenuity Pathways Analysis (Ingenuity Systems). (A) SAM analysis of the AML-M0 samples versus the primary AML samples showed down-regulation of several transcription factors depicted here in an integrated network. Genes up- and down-regulated in AML-M0 are represented in red and green, respectively. (B) SAM analysis of AML-M0s groups A and B showed up-regulation of many B cell–related genes in group B (associated with RUNX1 mutation), several of which are here represented together with other genes. Red represents up-regulation in group B; green, down-regulation; and white, no difference in expression.
Overview of biologic networks of differently expressed genes. Relation between different genes is based on Ingenuity knowledge database and was generated using Ingenuity Pathways Analysis (Ingenuity Systems). (A) SAM analysis of the AML-M0 samples versus the primary AML samples showed down-regulation of several transcription factors depicted here in an integrated network. Genes up- and down-regulated in AML-M0 are represented in red and green, respectively. (B) SAM analysis of AML-M0s groups A and B showed up-regulation of many B cell–related genes in group B (associated with RUNX1 mutation), several of which are here represented together with other genes. Red represents up-regulation in group B; green, down-regulation; and white, no difference in expression.
AML-M0 cases with RUNX1 mutations have a distinct gene expression signature
Unsupervised class discovery analyses of the 35 AML-M0 samples independent of other AML subtypes showed the existence of 2 subgroups (Figure 1B), which were already apparent in the overall clustering analysis (Figure 1A). These AML-M0 subgroups (A and B) were stable at different cutoffs (ranging from 6- to 9-fold corresponding to 2692 to 1427 probe sets). Importantly, RUNX1 mutations were strongly associated with AML-M0 subgroup B (Fisher exact test, P < .001). Notably, of the patients with a RUNX1 mutation, only those with a biallelic mutation or with a dominant-negative mutation clustered within group B (see supplemental Table 5 for the RUNX1 mutations). Patients 28 and 41, with heterozygous mutations outside the runt domain did not cluster within this group (Figure 1B). Patients 22, 52, and 58, for whom no RUNX1 mutations were detected, also clustered in group B. However, whole-genome genotyping of patient 52 showed copy number neutral loss of heterozygosity of chromosome 21, indicative of the existence of a biallelic RUNX1 mutation, and patient 58 showed a chromosome 21 alteration, which could point to the presence of a dominant-negative RUNX1 mutation.16 In the reference group of 253 AML patients, who were analyzed for RUNX1 mutations in the runt domain only, only 3 patients with RUNX1 mutations were detected. One of these, with a homozygous mutation, clustered within AML-M0 group B (Figure 1A). Sequencing RUNX1 in the remaining 6 cases of the reference group that segregated with AML-M0 (Figure 1A) revealed 2 additional AML cases with a heterozygous mutation outside the runt domain (data not shown). Other mutations present in the AML-M0 samples, such as mutations of ETV6, RAS, FLT3, JAK2, PTPN11, and chromosomal alterations (supplemental Table 1), were not significantly associated with either group A or B (Figure 1B). An association between trisomy 13 and RUNX1 mutation is apparent, as was reported before.23,24
Expression of genes involved in B-cell development is up-regulated in AML-M0 with RUNX1 mutations
We compared AML-M0 groups A and B (RUNX1 mutation) using SAM to define a gene expression signature for each subtype (Figures 1B and 2B). The difference in gene expression between the 2 groups should reflect, at least to some extent, the impact of RUNX1 loss/mutation on transcription and modulation of hematopoiesis in AML-M0. Filtering of the results using a FDR of 0.05 and a 1.5-fold change cutoff yielded 1326 differentially expressed probe sets corresponding to 837 genes and 168 unknown probe sets (supplemental Table 6).
We detected in group B (RUNX1 mutation) a pronounced gene expression signature related to B-cell development and migration (Figures 1B,2B and supplemental Table 6). Among the genes up-regulated were TDT (alias DNTT), several members of the B-cell receptor (BCR) complex, such as VPREB1, IGHM, IGL, and IGKC, and some downstream effectors of this complex, for instance BLNK, SYK, and PLCG1.25,–27 CXCR4, a chemokine receptor involved in the migration of blood cells, several major histocompatibility complex class II genes (included in HLA loci of DR, DQ, DP, DM, and DO), and many transcription and cell cycle regulators, such as SPIB, RUNX3, ETS2, IRF8, PRDM1, CIITA, and EBF1, represent other B cell–related genes found up-regulated in group B.26,,,,,–32 In addition, FLT3, RB1, and CCND1 also showed higher expression in this group. Conversely, several HOX genes (HOXA3, 4, 5, 7, 9, and 10), GATA2, and DZF6, usually linked to stem cell development,33,34 were expressed at significantly lower levels in group B.
Class prediction of AML-M0 and RUNX1 mutation
We performed PAM analysis with the purpose to determine the minimal number of genes necessary to predict AML-M0 in the full cohort of AMLs and a second analysis to predict RUNX1 mutations in a selected AML-M0 cohort.
The 35 AML-M0 samples and the 253 AML samples of other subtypes were mixed and randomly divided into a training set (n = 191) and a validation set (n = 97), which contained equal proportions of AML-M0 samples. We derived an 8 probe set classifier for AML-M0 (Table 1). All genes were also identified as significantly up-regulated in AML-M0 by SAM analysis (supplemental Table 2). In cross validation using the training set, all M0 cases were correctly predicted, and only 3 other AMLs were misclassified as AML-M0 (sensitivity, 100%; specificity, 98%). In the validation set of 97 cases, 1 M0 case was misclassified (sensitivity, 92%), whereas all other AMLs were correctly classified (specificity, 100%).
For class prediction of RUNX1 mutation status in the 35 AML-M0 samples, we excluded 10 cases for which the exact mutation status was not clear. These were samples without detectable RUNX1 mutation that aggregated into group B and samples with hemizygous loss of the RUNX1 locus (supplemental Table 5). We also restricted the analysis to samples with a runt domain mutation (DNA-binding domain of RUNX1) as this was the only area completely screened for mutations in all AML-M0 patients.16 Remarkably, expression of a single gene, BLNK (207655_s_at), was sufficient to obtain optimal class separation in 10-fold cross validation (sensitivity and specificity, 100%). Interestingly, among the 4 top probe sets that could be included in this classifier without affecting the prediction accuracy were 3 probe sets for TDT (210487_at, 1566363_at, and 1566362_at).
Discussion
Unsupervised cluster analyses of gene expression profiles is commonly used for class discovery in AML.15 Here, unsupervised clustering of 35 AML-M0 samples concurrently with 253 primary AMLs of other subtypes revealed AML-M0 as a distinct entity based on genome-wide gene expression patterns (Figure 1A). Thirty-three of the 35 AML-M0 samples clustered in an independent subgroup with a few non-M0 samples. The molecular lesions of these 33 patients were diverse (Figure 1B and supplemental Table 1), implying that the lesions must target a common pathway leading to a similar phenotype. The immature phenotype of AML-M0 was emphasized by the clustering of the CD34+ sorted samples, consisting of hematopoietic stem cells and committed early progenitors, within the AML-M0 group. Some of the non-M0 samples in the AML-M0 cluster were previously shown to have gene expression profiles characteristic of patients with an adverse prognosis (cluster no. 10).18 Because AML-M0 is also associated with a poor prognosis,2,–4 this result may be indicative of the existence of a common gene expression signature for this unfavorable outcome.
Many differentially expressed genes between AML-M0 and the reference samples were identified. One of the top down-regulated genes was MPO (Figure 2A). This result corroborates our analysis, as MPO negativity is a hallmark of AML-M0.1 Other genes found down-regulated were SPI1, CEBPA, CEBPD, and ETV6 (Figure 2A). All of these genes are important transcriptions factors involved in different stages of hematopoiesis and their down-regulation is in agreement with the undifferentiated state of AML-M0.35,36
Another difference between the AML-M0 samples and the remaining AMLs was the significant (up to 3-fold and higher) up-regulation of mitochondrial and ribosomal genes. This could signify an overall higher cellular activity in AML-M0. In particular, up-regulation of genes necessary for oxidative phosphorylation could be related to increased production of ATP, which would confer a significant advantage to cell survival. A number of strategies to target mitochondria in cancer therapy are known or being developed.37
Two distinct subgroups within the AML-M0 cluster (Figure 1A-B) could be identified by unsupervised clustering. RUNX1 mutations were significantly associated with group B. An immediately apparent difference between group A and group B was the expression of a large number of genes involved in B-cell development in AML-M0 with RUNX1 mutations. These genes included most members of the BCR complex, transcriptional regulators (such as RUNX3, EBF1, IRF8, CIITA), major histocompatibility complex class II genes, downstream effectors of the BCR complex (such as BLNK), and lymphoid homing chemokines (such as CXRC4). Furthermore, expression of VPREB1 is seen only at early stages of B-cell development, indicating that the gene expression in these samples reflects these stages of development.38 Two other genes up-regulated in AML-M0 with a RUNX1 mutation were FLT3 and TDT (also an early lymphoid marker).
We and others reported an increase in FLT3 expression in RUNX1 mutated samples in conjunction with and independently of trisomy 13 (chromosome 13 harbors FLT3).23,24 Expression of FLT3 is an early regulatory event in pre-B-cell development.27 The increase in FLT3 expression in RUNX1 mutated AML-M0 could be related to the B-cell characteristics found in these cells. Furthermore, this would be in line with the association between RUNX1 mutations and trisomy 13, which is indicative of a cooperative activity of these mutations with increased FLT3 expression.23,24
Intensity levels measured by 8 probe sets were sufficient to correctly predict all AML-M0 cases in cross validation, and 11 of 12 M0 cases in a validation set. Of note, the 2 patients who did not cluster in the AML-M0 group in the unsupervised clustering (Figure 1A) were correctly classified in this analysis. PAM also showed that one gene, BLNK, was sufficient to discriminate between AML-M0 samples with and without RUNX1 mutation in a 10-fold cross validation. However, in other subtypes, BLNK expression did not correlate to RUNX1 mutation (data not shown). Interestingly, the PAM results also indicate that TDT could be used instead of BLNK in discriminating between AML-M0 samples with and without RUNX1 mutation. In fact the gene expression of these 2 genes is very similar (Figure 1B). Furthermore, we detected a strong association between TdT protein expression and RUNX1 mutation.16 Because TdT expression is measured by immunophenotyping at diagnosis, it could be easily applied in the prediction of RUNX1 mutations in AML-M0, after validation.
In conclusion, we show that, at the gene expression level, AML-M0 is a distinct primary AML subtype. We also show that there are 2 distinct subgroups of AML-M0 characterized by their RUNX1 mutation status. The expression of lymphocyte-related genes is a long-reported occurrence in AML-M0.5 Our results link the expression of these genes to RUNX1 mutation. Expression of lymphoid and myeloid genes in AML-M0 patients with RUNX1 mutation indicates a role for RUNX1 in the regulation of both lineages at early stages in humans in resemblance to mice.9,10 The expression of B cell–related genes can have a critical role in AML-M0 and deserves further consideration.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Inês de Almeida, Monique van Velzen, Sacha Lind, Margriet Ouwens, and Roel Verhaak for microarray analysis and experimental support.
This work was supported by grants from the Calouste Gulbenkian Foundation and the Foundation for Science and Technology (F.P.G.S.).
Authorship
Contribution: F.P.G.S. contributed to conception and design, acquisition, analysis, and interpretation of data, and drafted the article; S.M.A.S., C.E.-V., and B.J.W. performed acquisition and analysis of data; R.D. performed critical paper revision; H.V., P.v.d.S. and P.J.M.V. contributed to interpretation of data and critical paper revision; M.G.-G. initiated, coordinated, and supervised this study and contributed to interpretation of data and critical paper revision; and all authors revised the article critically for important intellectual content and approved the final version to be published.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Micheline Giphart-Gassler, Department of Toxicogenetics, Leiden University Medical Center, PO Box 9600, Postzone S4-P, 2300 RC Leiden, The Netherlands; e-mail: m.giphart-gassler@lumc.nl.