Abstract
The lack of natural killer (NK) cell–specific markers, as well as the overlap among several common surface antigens and functional properties, has obscured the delineation between NK cells and dendritic cells. Here, novel subsets of peripheral blood CD3/14/19neg NK cells and monocyte/dendritic cell (DC)–like cells were identified on the basis of CD7 and CD4 expression. Coexpression of CD7 and CD56 differentiates NK cells from CD56+ monocyte/DC-like cells, which lack CD7. In contrast to CD7+CD56+ NK cells, CD7negCD56+ cells lack expression of NK cell–associated markers, but share commonalities in their expression of various monocyte/DC-associated markers. Using CD7, we observed approximately 60% of CD4+CD56+ cells were CD7neg cells, indicating the actual frequency of activated CD4+ NK cells is much lower in the blood than previously recognized. Functionally, only CD7+ NK cells secrete gamma interferon (IFNγ) and degranulate after interleukin-12 (IL-12) plus IL-18 or K562 target cell stimulation. Furthermore, using CD7 to separate CD56+ NK cells and CD56+ myeloid cells, we demonstrate that unlike resting CD7+CD56+ NK cells, the CD7negCD56+ myeloid cells stimulate a potent allogeneic response. Our data indicate that CD7 and CD56 coexpression discriminates NK cells from CD7negCD56+ monocyte/DC-like cells, thereby improving our ability to study the intricacies of NK-cell subset phenotypes and functions in vivo.
Introduction
Natural killer (NK) cells are a subset of lymphocytes comprising approximately 10% of peripheral blood mononuclear cells in humans.1 NK cells serve an important role in the defense against viral infections, as well as in tumor surveillance,2-4 and are also involved in shaping adaptive immune responses through their production of cytokines.5,6 Recently, mouse NK cells have been observed to demonstrate immune memory, similar to B and T immune cells, after viral infection.7 This finding is particularly intriguing given the interest in developing strategies to apply NK cells as therapeutic agents against a broad range of malignancies8,9 and augmenting NK-cell function during chronic viral infection (eg, HIV-1, hepatitis C virus).
Despite major advances achieved in NK-cell biology, the lack of any specific, NK cell–restricted markers has limited our understanding of their biologic function. NK cells are generally identified by the expression, or lack thereof, of different surface receptors depending on the species being studied. In mice, NK cells are traditionally identified as CD3neg, CD49b+, asialo-GM1+, NK1.1+ lymphocytes and more recently by the expression of NKp46.10 In humans, NK cells are identified as CD3/14/19neg lymphocytes expressing CD56 (neural cell adhesion molecule) and more recently NKp46,10 although not all human NK cells express NKp46.11 However, in both species, there are examples where a particular NK-cell receptor is not expressed on all NK cells and where other immune cell subsets express the same receptor. For example, the NK1.1 epitope in mice is expressed only in some strains of mice,12 and some activated T cells can express NK1.1. In mouse strains lacking NK1.1, NK cells are commonly identified using the DX5 monoclonal antibody that recognizes CD49b,13 an integrin subunit also expressed on some activated T cells, platelets, and basophils.14,15 In humans, CD56 is also present on a subset of CD3+ T cells,16 as well as a subset of monocyte-derived dendritic cells.17 In addition to NK cells, NKp46 is expressed on a subset of γδ T cells,10 as well as on recently identified cells of unknown lineage in the gut.18 The different phenotypic markers used to identify NK cells in mice and humans, as well as the lack of a NK cell–specific marker, increase the difficulty in studying this population.
In addition to the lack of specific markers, human NK cells exhibit developmental and functional properties common to other immune cell subsets, particularly dendritic cells (DCs). NK-cell development is predominantly through a common lymphoid progenitor,19 similar to certain subsets of DCs.19,20 Beyond their ability to kill target cells, NK cells contribute to overall immune function in other capacities. For example, NK cells express many costimulatory molecules typically found on DCs (reviewed in Spits and Lanier21 ) that can influence activation of other immune cells. In contrast to freshly isolated, resting human NK cells, activated human NK cells can express human leukocyte antigen DR (HLA-DR), OX40 ligand, CD80, and CD86 and have been shown previously to have antigen-presenting capabilities similar to certain DC subsets.22-24 Activated NK cells can also express CD4 on their surface,25,26 a molecule typically found on a subset of CD3+ T cells and monocytes/macrophages; however, CD4 has also been observed on many activated cell types including CD8+ T cells,27 B cells,28 monocytes,29 eosinophils,30 and neutrophils.31 Given the small percentage of circulating CD4+ NK cells, previous studies have used in vitro stimulation protocols to expand the number of NK cells expressing CD425,26 ; however, these activated CD4+ NK cells may not be representative of CD4+ NK cells in vivo.
The overlap of potential NK-cell markers and functions with other immune cell subsets indicates that to study rare subsets of NK cells, a marker is required to distinguish this population from other potentially contaminating immune cells, particularly monocytic and dendritic cells. In this study, we compared the ex vivo phenotypic and functional characteristics of peripheral blood cells typically identified as CD3/14/19neg CD56+ NK cells, and determined that a significant percentage of these cells are not NK cells, but rather resemble cells from the monocyte/DC lineage. This study provides a novel means to discriminate NK cells from monocyte/DC-like cells, providing a more precise identification of human NK cells that significantly increases our ability to understand the phenotypic and functional intricacies of NK cells in vivo.
Methods
Human subjects
Peripheral blood mononuclear cells (PBMCs) were obtained from leukocyte concentrates of healthy volunteers (Stanford University Blood Center) after density gradient centrifugation over Ficoll-Paque (GE Healthcare). All persons gave informed consent to participate in this study, in accordance with the Declaration of Helsinki, and the University of California, San Francisco (UCSF) Committee on Human Research approved this study.
Fluorochrome-labeled antibodies and phenotypic characterization of cell subsets
Phenotypic analysis of PBMCs was performed on cryopreserved PBMCs as previously described.32 All antibodies were purchased from BD Pharmingen unless otherwise noted. The following fluorochrome-labeled antibodies were used in all phenotypic panels: fluorescein isothiocyanate (FITC)–conjugated anti-CD7 (Beckman Coulter), phycoerythrin (PE)–conjugated anti-CD7 (Beckman Coulter), phycoerythrin–Texas Red (ECD)–conjugated anti-CD3 (Beckman Coulter), Alexa700-conjugated anti-CD4, phycoerythrin-Cy7 (PE-Cy7)–conjugated anti-CD56, Pacific Blue–conjugated anti-CD16, and allophycocyanin-Cy7 (APC-Cy7)–conjugated anti-CD14 and anti-CD19. Amine Aqua (Invitrogen) was included in each stain to exclude dead cells. Phenotypic characterization of cells was performed with the following antibodies: PE-conjugated anti-KIR3DL1/DS1 (clone Z27; Beckman Coulter), PE-conjugated anti-KIR2DL1 (clone 143211; R&D Systems), APC-conjugated anti-KIR2DL3 (clone 180701; R&D Systems), APC-conjugated anti-CD161, PE-conjugated anti-NKp30 (Beckman Coulter), PE-conjugated anti-NKp46 (Beckman Coulter), APC-conjugated anti-NKG2A (R&D Systems), APC-conjugated anti-NKG2D (R&D Systems), APC-conjugated anti-CD13, FITC-conjugated anti-CD123 (Miltenyi Biotec), PE-conjugated anti–granzyme B, FITC-conjugated antiperforin, and PE-conjugated anti–HLA-DR. All cells were fixed with 2% paraformaldehyde in phosphate-buffered saline (PBS) and analyzed by flow cytometry with a 4-laser LSR-II instrument (BD Biosciences). Anti–mouse immunoglobulin G–coated beads were stained with each fluorochrome-conjugated mouse antibody separately and used for software-based compensation. Final analysis was carried out using FlowJo flow cytometric analysis software (TreeStar).
NK-cell stimulation and intracellular cytokine staining
PBMCs were added at 500 000 per well to 96-well U-bottom plates. PBMCs were cultured in media alone (unstimulated) or stimulated with 50 000 K562 target cells or 50 ng/mL each of interleukin-12 (IL-12) and IL-18 (PeproTech) in the presence of FITC-conjugated anti-CD107a for 1 hour, followed by 5 hours in Brefeldin A and monensin for intracellular cytokine staining. Cells were washed twice with fluorescence-activated cell sorting (FACS) buffer (PBS with 2 mM of ethylenediaminetetraacetic acid and 0.5% bovine serum albumin) before being surfaced stained. Cells were then fixed in 2% paraformaldehyde in PBS and permeabilized with FACS Permeabilizing Solution 2 (Becton Dickinson). Permeabilized cells were stained for intracellular gamma interferon (IFNγ; Becton Dickinson).
Flow cytometric sorting of NK cells and myeloid cells
CD56+ cell subsets were enriched by depleting T cells using anti-CD3 magnetic beads (StemCell Technologies). CD56-enriched PBMCs were stained with FITC-conjugated anti-CD7, ECD-conjugated anti-CD3, Alexa700-conjugated anti-CD4, APC-Cy7–conjugated anti-CD14 and anti-CD19, PE-Cy7–conjugated anti-CD56, and Amine Aqua (live/dead cell marker). Three subpopulations of CD56+ cells were collected: CD7+CD56+CD4neg, CD7negCD56+CD4+, and CD7+CD56+CD4+ on an ARIA sorter (BD Biosciences). However, only CD7+CD56+CD4neg and CD7negCD56+CD4+ cells were collected at sufficient purity (> 90%) to be used in further experiments. Sorted cell subsets were used immediately in enzyme-linked immunosorbent spot (ELISPOT) assays.
ELISPOT analysis of sorted CD56+ cell subsets
Flow cytometry–purified cells were added at a concentration of 1000 cells/well to anti-IFNγ antibody precoated wells. Cells were incubated overnight in medium alone (unstimulated control) or incubated in the presence of K562 target cells or 50 ng/mL each of IL-12 and IL-18. ELISPOT plates were developed as previously described.33
Assessment of antigen presentation capacity by CD56+ cell subsets
The ability of CD56+ cell subsets to serve as antigen-presenting cells was assessed using a mixed leukocyte reaction (MLR). Stimulator cells were isolated first by depleting T cells using anti-CD3 magnetic beads (StemCell Technologies). CD3-depleted cells were then stained with one or more of the following antibodies: PE-conjugated anti-CD7, ECD-conjugated anti-CD3, APC-Cy7–conjugated anti-CD14 and anti-CD19, or PE-Cy7–conjugated anti-CD56. Three populations of CD56+ cells were purified on a FACSAria cell sorter (BD Biosciences) to greater than 95% purity: NK cells defined as CD3negCD56+; NK cells defined as CD3/14/19negCD7+CD56+; and a subset of cells defined as CD3/14/19/7negCD56+. As a positive control in the MLR, a mixed population of CD14+ monocytes and CD19+ B cells was also purified by flow cytometry. Flow cytometry–purified stimulator cells were γ-irradiated before use in the MLR. Responder T cells derived by negative selection from an allogeneic donor using a CD3 Enrichment Kit (StemCell Technologies) were carboxyfluorescein succinimidyl ester (CFSE)–labeled to track proliferation. The MLR was performed by incubating 105 stimulator cells with 105 responder T cells in a 96-well U-bottom plate in the presence or absence of 250 IU/mL of IL-2. As a positive control, some wells were incubated with 1 μg/mL of phytohemagglutinin (PHA; Sigma-Aldrich). On day 7, cells were harvested and analyzed on an LSR-II (BD Biosciences) for the dilution of CFSE dye. Final analysis was carried out using FlowJo flow cytometric analysis software (TreeStar).
Statistical analysis
Statistical analysis was performed with GraphPad Prism statistical software (GraphPad Software). The nonparametric Mann-Whitney U test was used to compare between-group distributions. The statistical significance threshold was set at P value less than .05.
Results
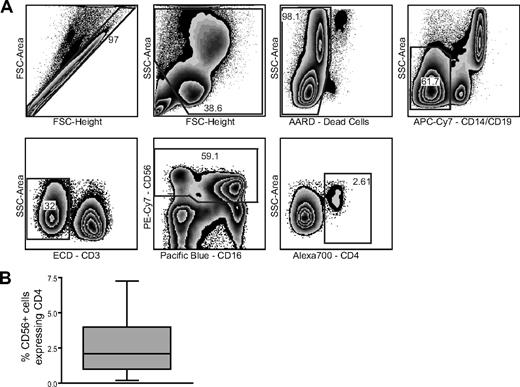
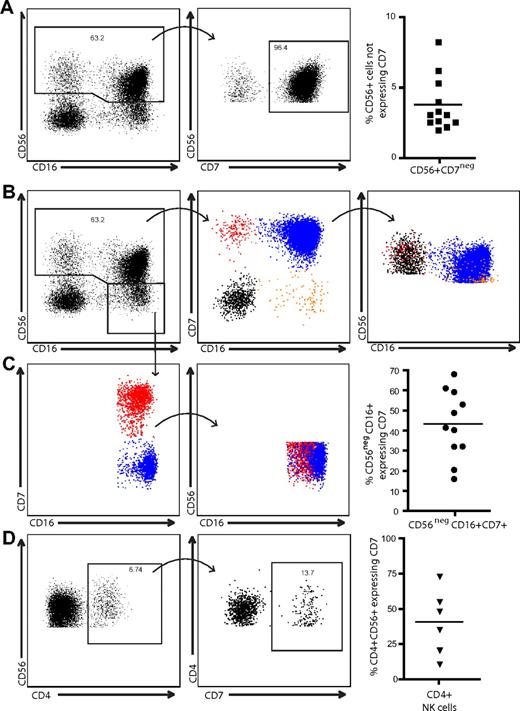
CD7 identifies 2 subsets of CD4+CD56+ cells
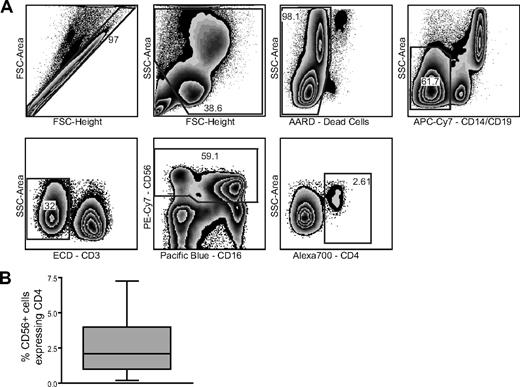
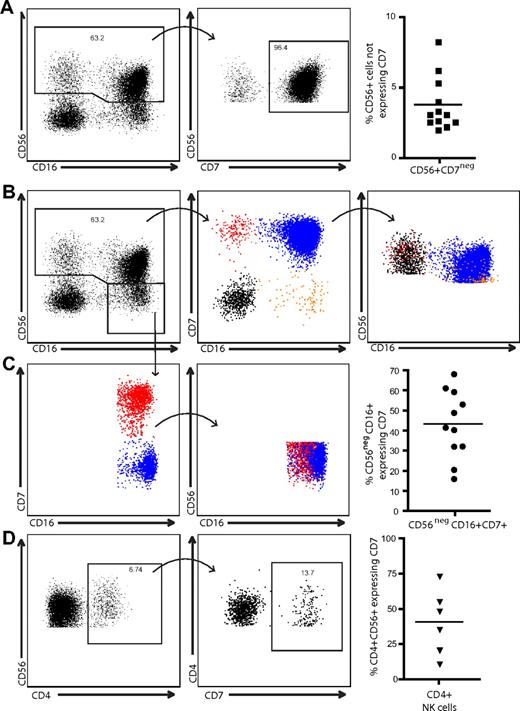
The expression of CD56, classically thought to be limited to NK cells, has also been observed on a subset of CD3+ T cells16 and more recently on monocyte-derived dendritic cells generated in the presence of IFNα and granulocyte-macrophage colony-stimulating factor.17 In agreement with a previously published report,25 we observed a distinct population of peripheral blood CD56+CD3/14/19neg cells expressing CD4 (Figure 1A) that comprised 0.2% to 7.5% of the peripheral blood CD56+ cell population (Figure 1B) within the lymphocyte gate. To distinguish between CD4+CD56+ NK cells and other potentially contaminating cell types, such as monocytic or dendritic cells, we used CD7 as an additional NK cell–specific marker. CD7 is expressed on T cells, NK cells, and pre-B cells, but is absent on mature myeloid cells.34,35 Expression of CD7 was observed on 90% to 98% of CD56+CD3/14/19neg cells; however, 2% to 10% of CD56+CD3/14/19neg cells are CD7neg (Figure 2A). After gating on CD56+CD3/14/19neg cells, 4 populations of cells were identified based on CD7 and CD16 expression (Figure 2B). Overlaying these 4 populations in a graph of CD56 and CD16, we determined that CD7+CD56+CD16neg cells comprise the CD56bright population of NK cells and the CD7+CD56+CD16+ cells are equivalent to the CD56dimCD16+ NK cells. In contrast, the majority of the CD56dimCD16neg fraction of cells is composed of CD7negCD56+ cells (Figure 2B). Moreover, when we gated on the CD56dim/negCD16+ cells, which are elevated in certain diseases such HIV-1 infection,36 approximately 43% (range, 16%–69%) were CD7+ (Figure 2C). The ability of CD7 to distinguish NK cells from other CD56+ cells revealed that only approximately 40% (range, 11%–74%) of the CD4+CD56+ cells expressed CD7 (Figure 2D). Direct ex vivo assessment of CD56+ cells, which are typically defined as NK cells, indicates that among healthy donors a significant proportion of this subset (up to 10% of CD56+ cells) is CD7negCD56+.
Identification of a distinct population of CD4+CD56+ cells. (A) Representative gating strategy used to identify a population of CD56+CD4+ cells. (B) Percentage of CD4+ cells within the CD56+ cell population (n = 10). Box plot indicates the median, upper and lower quartiles. Whiskers represent the minimum and maximum data values observed.
Identification of a distinct population of CD4+CD56+ cells. (A) Representative gating strategy used to identify a population of CD56+CD4+ cells. (B) Percentage of CD4+ cells within the CD56+ cell population (n = 10). Box plot indicates the median, upper and lower quartiles. Whiskers represent the minimum and maximum data values observed.
CD7 identifies 2 distinct populations of CD56+ cells. (A) CD3/14/19neg cells were gated for CD56 expression and CD56+ cells were plotted against CD7 to determine the percentage of CD7+ cells within the CD56 gate. In peripheral blood, as many as 10% of CD56+ CD3/14/19neg cells are CD7neg. (B) Classically defined NK cells (CD3negCD56+CD16+/neg) were gated and plotted against CD7 versus CD16 to identify 4 populations of cells. Each population was then overlaid back to a CD56 versus CD16 plot to observe where the CD7neg non–NK-cell subsets were distributed among the CD7+ NK cells. (C) Similarly, CD56dim/negCD16+ cells were gated and plotted against CD7 versus CD16. Although approximately 43% of the CD56dim/negCD16+ population was CD7+ NK cells, there was extensive overlap with the CD7neg cell fraction. (D) CD4+CD56+ cells were assessed for CD7 expression. On average, less than 50% of CD4+CD56+ cells coexpress CD7 (n = 6).
CD7 identifies 2 distinct populations of CD56+ cells. (A) CD3/14/19neg cells were gated for CD56 expression and CD56+ cells were plotted against CD7 to determine the percentage of CD7+ cells within the CD56 gate. In peripheral blood, as many as 10% of CD56+ CD3/14/19neg cells are CD7neg. (B) Classically defined NK cells (CD3negCD56+CD16+/neg) were gated and plotted against CD7 versus CD16 to identify 4 populations of cells. Each population was then overlaid back to a CD56 versus CD16 plot to observe where the CD7neg non–NK-cell subsets were distributed among the CD7+ NK cells. (C) Similarly, CD56dim/negCD16+ cells were gated and plotted against CD7 versus CD16. Although approximately 43% of the CD56dim/negCD16+ population was CD7+ NK cells, there was extensive overlap with the CD7neg cell fraction. (D) CD4+CD56+ cells were assessed for CD7 expression. On average, less than 50% of CD4+CD56+ cells coexpress CD7 (n = 6).
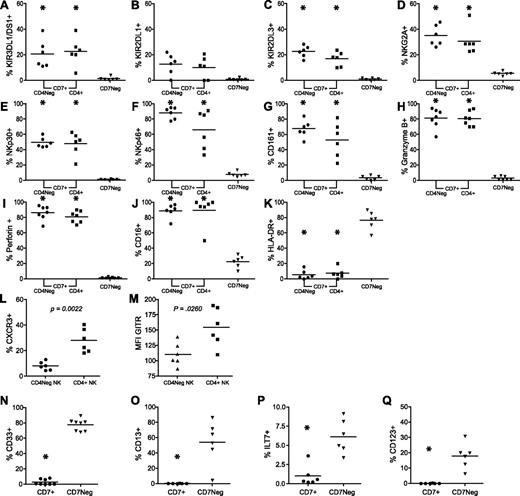
Only CD7+CD56+ cells express NK cell–associated receptors
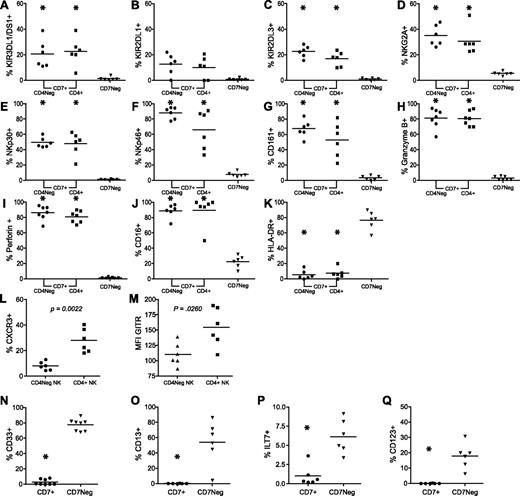
The initial goal of this study was to determine whether CD4+ NK cells were phenotypically or functionally distinct from CD4neg NK cells. Therefore, further characterization of the phenotypic differences between CD4+CD7+CD56+ NK cells and CD4negCD7+CD56+ NK cells, as well as between CD7+CD56+ NK cells and CD7negCD56+ cells, was undertaken. The expression of typical NK-cell markers such as CD16, killer cell immunoglobulin-like receptor (KIRs; KIR3DL1/DS1, KIR2DL1, and KIR2DL3), the C-type lectinlike receptor NKG2A, the inhibitory receptor CD161, activating natural cytotoxicity receptors (NKp30 and NKp46), and effector molecules (perforin, granzyme B), as well as the monocyte/DC markers CD13, ILT7, CD33, and CD123 were assessed (Figure 3). Careful assessment of the NK cell–associated markers, as well as the NK cell–associated effector molecules on the CD4+ and CD4neg CD7+ NK cells, revealed no significant differences between these subsets (Figure 3A-I). In addition, the majority of both the CD7+CD4neg and the CD7+CD4+ NK cells expressed CD16 (Figure 3J). The only significant difference observed in this study pertained to the activation status of the CD7+CD4+ NK cells. In agreement with a previous report proposing that CD4+ NK cells are activated and capable of producing cytokines,25 we observed that a significantly greater percentage of CD7+CD4+ NK cells express the T helper 1 chemokine receptor CXC chemokine receptor 3 (CXCR3) and possess significantly higher levels of glucocorticoid-induced tumor necrosis factor (TNF) receptor (GITR; Figure 3L-M). We also observed that approximately 30% and 18% of the IFNγ+ NK cells express CD4 after IL-12 plus IL-18 and K562 stimulation, respectively (data not shown). In contrast, CD7negCD56+ cells did not express any of the NK cell–associated receptors examined (Figure 3A-G), nor did they express either of the effector molecules assessed (Figure 3H-I). An assessment of CD16 expression revealed that approximately 90% of CD7+CD56+ NK cells were CD16+ as opposed to approximately 25% of CD7negCD56+ cells (Figure 3J). Because the CD7negCD56+ fraction lacked expression of typical NK-cell markers, we examined several monocyte/DC-associated markers including CD13, CD33, ILT7, and CD123 (Figure 3N-Q). CD33 was expressed on 70% to 80% of CD7negCD56+ cells, but less than 1% of CD7+CD56+ NK cells (Figure 3N). Further characterization using CD13, ILT7, and CD123 indicated that CD7+CD56+ NK cells lacked expression of any of these markers (Figure 3O-Q); however, a significant percentage of CD7negCD56+ cells demonstrated CD13, ILT7, or CD123 staining (Figure 3O-Q). Furthermore, many of the CD7negCD56+ cells coexpressed CD13 and CD33, and a subset of CD13+ and CD33+ cells also coexpressed CD123 (data not shown). In addition, more than 75% of CD7negCD56+ cells expressed HLA-DR compared with only 5% of the CD7+CD56+ NK cells (Figure 3K). To determine whether CD56 was expressed on a significant proportion of dendritic cells, we gated on 2 populations of dendritic cells (CD123+ and ILT7+) and a population of monocytes (CD13+) defined by the phenotype CD3/14/19negHLA-DR+CD7neg. Importantly, we observed approximately 10% of plasmacytoid dendritic cells (pDCs) and monocytes expressed CD56. Depending on the marker used to identify the pDC populations, 2.6% to 15.3% (mean = 9.8% ± 5.47%) of CD123+ pDCs and 0% to 25.3% (mean = 10.8% ± 8.5%) of ILT7+ pDCs expressed CD56. Furthermore, 6.9% to 11.6% (mean = 9.57% ± 1.87%) of CD13+ monocytes expressed CD56 (data not shown). These data indicate that CD7negCD56+ cells are not NK cells, but rather are composed of a mix of monocytes and DCs and may be readily distinguished from CD56+ NK cells by the expression of CD7 on NK cells.
Only CD7+CD56+ cells express typical NK-cell markers, whereas CD7negCD56+ cells express myeloid-associated receptors. (A-K) CD3negCD14negCD19neg cells were gated for CD56 expression and CD56+ cells were plotted against CD7 to identify CD7+ NK cells and CD7neg monocyte/DC-like cells. CD7+CD56+ NK cells were further subset into CD4+ NK cells and CD4neg NK cells. The percentage of cells expressing each receptor was determined on each subset of cells. (L-M) Percentage of cells expressing CXCR3 (L) and mean fluorescent intensity (MFI; M) of GITR were assessed on CD4neg and CD4+CD7+ NK cells. (N-Q) Assessment of DC and monocytic markers on CD7+CD56+ cells and CD7negCD56+ cells. *P < .05. P values in panels A through K were determined between CD4negCD7+ NK cells and CD7neg monocyte/DC-like cells and between CD4+CD7+ NK cells and CD7neg monocyte/DC-like cells (n = 6).
Only CD7+CD56+ cells express typical NK-cell markers, whereas CD7negCD56+ cells express myeloid-associated receptors. (A-K) CD3negCD14negCD19neg cells were gated for CD56 expression and CD56+ cells were plotted against CD7 to identify CD7+ NK cells and CD7neg monocyte/DC-like cells. CD7+CD56+ NK cells were further subset into CD4+ NK cells and CD4neg NK cells. The percentage of cells expressing each receptor was determined on each subset of cells. (L-M) Percentage of cells expressing CXCR3 (L) and mean fluorescent intensity (MFI; M) of GITR were assessed on CD4neg and CD4+CD7+ NK cells. (N-Q) Assessment of DC and monocytic markers on CD7+CD56+ cells and CD7negCD56+ cells. *P < .05. P values in panels A through K were determined between CD4negCD7+ NK cells and CD7neg monocyte/DC-like cells and between CD4+CD7+ NK cells and CD7neg monocyte/DC-like cells (n = 6).
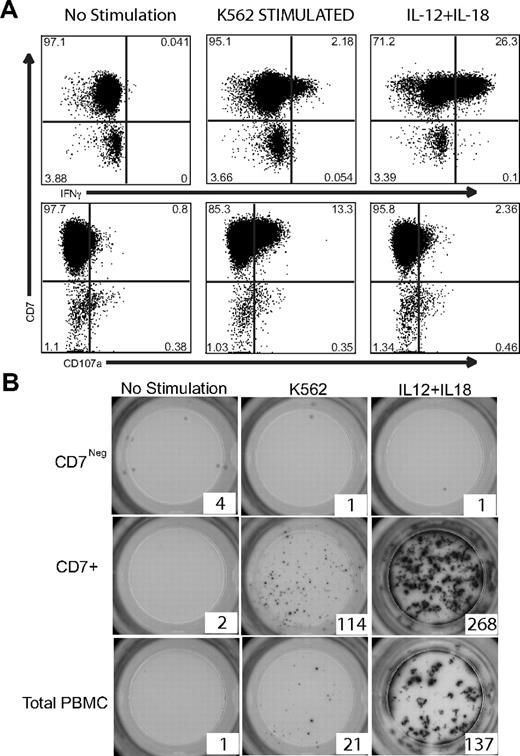
CD7+CD56+ NK cells respond to K562 and IL12 plus IL18 stimulation
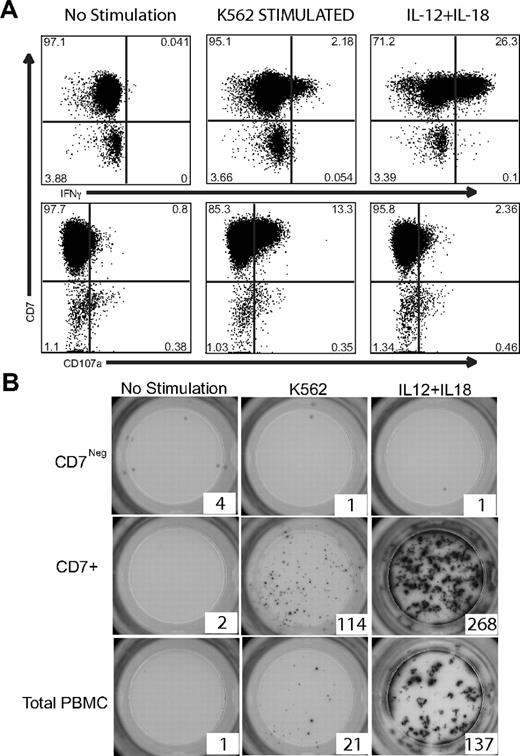
To determine whether effector molecules segregated with different populations of NK cells, we assessed perforin and granzyme B expression within the CD7 versus CD16 populations of NK cells. We observed that CD7+CD16neg NK cells are similar to the CD56bright subset of NK cells and as such did not express or expressed very little perforin and granzyme B, whereas CD7+CD16+ NK cells are similar to CD56dimCD16+ NK cells and expressed high amounts of perforin and granzyme B (data not shown). To ascertain whether a functional difference exists between CD7+CD56+ NK cells and CD7negCD56+ monocytic cells, total PBMCs were stimulated with either K562 target cells or IL-12 plus IL-18 and then assayed for IFNγ expression or CD107a as a marker of degranulation by flow cytometry. K562 target cell stimulation, as well as IL-12 plus IL-18 exposure, resulted in IFNγ detection and CD107a degranulation only in CD7+CD56+ NK cells (Figure 4A). CD7negCD56+ cells neither produced IFNγ, nor degranulated (CD107aneg), in response to these prototypic NK-cell stimuli. To further investigate these populations, CD56+CD3/14/19neg cells were purified by sorting into CD7+CD56+ NK-cell or CD7negCD56+ monocytic fractions, stimulated, and evaluated for the frequency of IFNγ-producing cells using an ELISPOT assay. Similar to the intracellular cytokine staining, CD7negCD56+ cells failed to produce IFNγ in response to either K562 target cells or IL-12 plus IL-18; however, CD7+CD56+ NK cells demonstrated a strong IFNγ response to either stimuli (Figure 4B). Taken together, these data demonstrate that CD7+CD56+ cells are NK cells that are phenotypically and functionally distinct from CD7negCD56+ monocyte/DC-like cells.
CD7+CD56+ NK cells, but not CD7negCD56+ cells, produce IFNγ in response to K562 target cells or IL-12 + IL-18 stimulation. (A) Intracellular cytokine flow cytometry was performed on total PBMCs after no stimulation or stimulation with K562 target cells or IL-12 + IL-18. CD3/14/19neg cells were gated for CD56 expression. These CD56+ cells were analyzed for IFNγ and CD107a expression in the CD7+ and CD7neg fractions. Only CD7+CD56+ NK cells expressed IFNγ and CD107a after stimulation. One representative example of 6 are shown for IFNγ and CD107a expression. (B) CD7+CD56+ NK cells and CD7negCD56+ monocyte/DC-like cells were sorted by flow cytometry and 1000 of each cell type were plated per well of a precoated anti-IFNγ ELISPOT plate. Cells were cultured either in media alone (not stimulated) or were stimulated overnight with K562 target cells or IL-12 + IL-18. One representative example of 3 experiments is shown.
CD7+CD56+ NK cells, but not CD7negCD56+ cells, produce IFNγ in response to K562 target cells or IL-12 + IL-18 stimulation. (A) Intracellular cytokine flow cytometry was performed on total PBMCs after no stimulation or stimulation with K562 target cells or IL-12 + IL-18. CD3/14/19neg cells were gated for CD56 expression. These CD56+ cells were analyzed for IFNγ and CD107a expression in the CD7+ and CD7neg fractions. Only CD7+CD56+ NK cells expressed IFNγ and CD107a after stimulation. One representative example of 6 are shown for IFNγ and CD107a expression. (B) CD7+CD56+ NK cells and CD7negCD56+ monocyte/DC-like cells were sorted by flow cytometry and 1000 of each cell type were plated per well of a precoated anti-IFNγ ELISPOT plate. Cells were cultured either in media alone (not stimulated) or were stimulated overnight with K562 target cells or IL-12 + IL-18. One representative example of 3 experiments is shown.
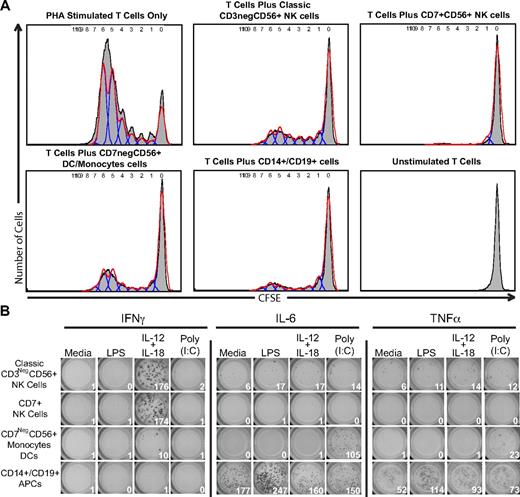
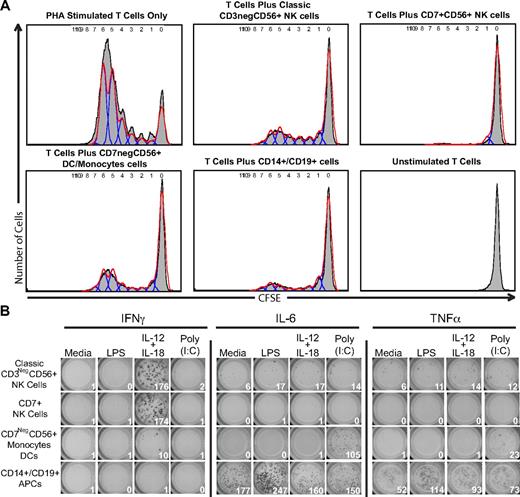
CD7+CD56+ NK cells do not stimulate an allogeneic response
To determine whether there is a functional consequence to separating CD7negCD56+ DCs/monocytes from CD7+ NK cells, PBMCs were separated into classical antigen-presenting cells (CD14+ monocytes and CD19+ B cells), CD7negCD56+ DCs/monocytes, and CD7+CD56+ NK cells. To test the ability of the CD7negCD56+ DCs/monocytes and CD7+CD56+ NK-cell subsets to induce proliferation, a mixed leukocyte reaction (MLR) was performed with 3 purified stimulator cell populations: classical antigen-presenting cells (CD14+ monocytes and CD19+ B cells), CD7negCD56+ DCs/monocytes, and CD7+CD56+ NK cells. After incubating the irradiated stimulator cells with allogeneic T cells for 7 days, the proliferation of the responder T cells was assessed for the dilution of CFSE by flow cytometry (Figure 5A). Compared with PHA-stimulated T cells in which more than 80% of the T cells divided, classically defined NK cells (CD3negCD56+) were capable of supporting a lower level of T-cell proliferation as evidenced by the dilution of CFSE in approximately 40% of T cells. Interestingly, the percentage of proliferating T cells stimulated with either classically defined NK cells (CD3negCD56+) or purified CD7negCD56+ DC/monocyte population was similar (∼ 40% of T cells proliferated). However, purified CD7+CD56+ NK cells did not induce a significant proliferation of the responder T cells as approximately 5% of T cells proliferated. Another function attributed to DCs and monocytes, as well as NK cells, is their ability to directly respond to Toll-like receptor agonists. To determine whether our purified populations of CD7+CD56+ NK cells and CD7negCD56+ DCs/monocytes could respond to Toll-like receptor (TLR) agonists, IFNγ, TNFα, and IL-6 ELISPOTs were performed on lipopolysaccharide (LPS)– and polyinosinic:polycytidylic acid [poly(I:C)]–stimulated cells. IFNγ was detected only in classical CD3negCD56+ NK cells and CD7+CD56+ NK cells after stimulation with IL-12 plus IL-18, but not after LPS or poly(I:C) stimulation (Figure 5B). Furthermore, CD7+CD56+ NK cells did not produce any detectable IL-6 or TNFα responses after any of the stimulation conditions. In contrast, CD7negCD56+ DCs/monocytes did not produce IFNγ after any of the stimulation conditions, but did produce significant IL-6 and a low-level TNFα after poly(I:C) stimulation (Figure 5B). The CD7negCD56+ DCs/monocytes did not produce IL-6 or TNFα after LPS stimulation; however, the mixed population of B cells and monocytes were capable of producing IL-6 and TNFα after LPS stimulation. Taken together, these data indicate that resting CD7+CD56+ NK cells do not induce a significant allogeneic MLR response, nor are they capable of directly responding to LPS or poly(I:C).
CD7+ NK cells do not induce allogeneic T-cell proliferation in a mixed leukocyte reaction. PBMCs were purified by flow cytometry into 4 populations: classical NK cells (CD3negCD56+), CD7+CD56+ NK cells, CD7negCD56+ DCs/monocytes, and CD14+ and CD19+ antigen-presenting cells. (A) Flow cytometry–purified cells were γ-irradiated and incubated in a 1:1 ratio with CFSE-labeled allogeneic T cells. After incubating for 7 days, cells were analyzed for dilution of CFSE as a marker of proliferation. PHA-stimulated T cells were used as a positive control. One representative donor (n = 3) is shown. Similar results were also obtained when the reaction was performed in the presence or absence of 250 IU/mL of IL-2. (B) Flow cytometry–purified cells (1000 per well) were added to wells containing media alone, lipopolysaccharide (LPS), IL-12 + IL-18, or poly(I:C) for 18 hours. One representative experiment of 3 is shown.
CD7+ NK cells do not induce allogeneic T-cell proliferation in a mixed leukocyte reaction. PBMCs were purified by flow cytometry into 4 populations: classical NK cells (CD3negCD56+), CD7+CD56+ NK cells, CD7negCD56+ DCs/monocytes, and CD14+ and CD19+ antigen-presenting cells. (A) Flow cytometry–purified cells were γ-irradiated and incubated in a 1:1 ratio with CFSE-labeled allogeneic T cells. After incubating for 7 days, cells were analyzed for dilution of CFSE as a marker of proliferation. PHA-stimulated T cells were used as a positive control. One representative donor (n = 3) is shown. Similar results were also obtained when the reaction was performed in the presence or absence of 250 IU/mL of IL-2. (B) Flow cytometry–purified cells (1000 per well) were added to wells containing media alone, lipopolysaccharide (LPS), IL-12 + IL-18, or poly(I:C) for 18 hours. One representative experiment of 3 is shown.
Discussion
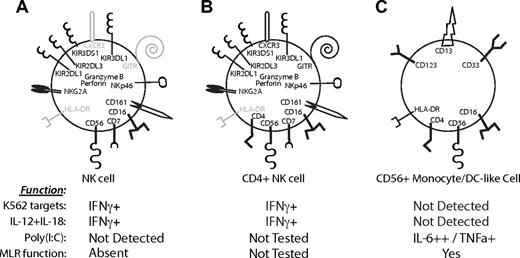
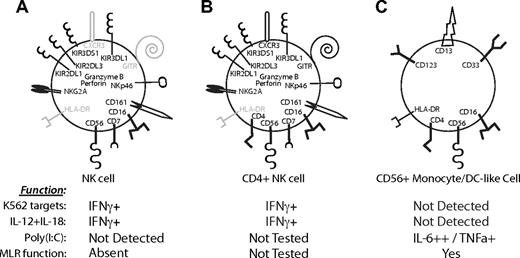
The lack of a NK cell–specific marker, as well as the potential overlap of phenotypic and functional properties of NK cells with other immune cell subsets, creates difficulties in distinguishing NK-cell functions from myeloid cell functions. Indeed, several reports in murine models, as well as in humans, have described dendritic cells with natural killer cell activity,37-41 possibly resulting from the limited markers available to distinguish NK cells from contaminating DC populations. Herein, we demonstrate that CD7 expression on NK cells clearly distinguishes this cell subset from CD7negCD56+ monocyte/DC-like cells. Interestingly, these CD7neg monocyte/DC-like cells display forward and side light scatter characteristics typical for lymphocytes. Unlike NKp46, which is not readily detected on all human NK cells11,42 and can be down-regulated under some physiologic conditions,42-44 CD7 is expressed homogeneously by all NK cells and does not appear to be significantly down-regulated after activation. Although our results confirm that CD4+CD56+ NK cells are indeed activated NK cells,25,26 the use of CD7 in our study provides for a precise determination of the percentage of activated NK cells present in the blood of healthy donors and allows for analysis of their functional behavior. Because the majority of CD4+CD56+ cells are CD7neg monocyte/DC-like cells, we now demonstrate that the frequency of activated NK cells in blood is much lower (1.44% ± 0.99% of CD56+ cells) than previously reported. To better visualize the differences between CD7+CD4neg NK cells, CD7+CD4+ NK cells, and CD7neg monocyte/DC-like cells, their phenotypic and functional characteristics are summarized in Figure 6. NK cells, either expressing or lacking CD4 expression, contained equal frequencies of cells expressing typical NK-cell markers (Figure 6A-B). The most significant differences in expression between CD4+ and CD4neg NK cells were the significantly greater percentage of CXCR3+ cells and significantly increased surface expression (higher MFI) of GITR within the CD4+ NK cells. Although the function of CD4 on these non–T-cell subsets is not completely understood, these data are in agreement with previous studies,25 which indicate that CD4+ NK cells are activated and perhaps able to migrate quickly to sites of inflammation in response to CXCR3 ligands (eg, CXC ligand 10 [CXCL10], CXCL9). Further studies are needed to determine whether the frequency of activated CD4+ NK cells is increased during acute (eg, influenza infection) or chronic (eg, HIV-1) infections or in autoimmune diseases such as rheumatoid arthritis.
Significant differences in receptor expression between CD56+ immune cell subsets. Model depicting the distribution of receptors on (A) CD4neg NK cells, (B) CD4+ NK cells, and (C) CD56+ monocyte/DC-like cells. Conclusions are based on results in Figures 3 and 5. Receptors expressed on the majority of cells of that subset are represented in black. No expression or < 15% of cells expressing a receptor is indicated in gray. Receptor expression in panel C is representative of a heterogeneous population of monocytes and DC-like subsets. For example, CD16 is not expressed on CD123+ cells, but is expressed on many of the CD13+ cells. The expression of cytokines after stimulation with K562 target cells, IL-12 + IL-18, and poly(I:C) for the particular cell subset is indicated below. Only CD7+ NK cells produced IFNγ after stimulation with NK cell–specific stimuli, whereas a strong IL-6 and TNFα response was observed in the CD7negCD56+ cells after poly(I:C) stimulation. The ability of CD7+ NK cells and CD7negCD56+ DCs/monocytes to induce proliferation in a mixed leukocyte reaction (MLR function) is also indicated; only CD7negCD56+ cells are inducers of proliferation in accordance with their phenotype as DCs/monocytes.
Significant differences in receptor expression between CD56+ immune cell subsets. Model depicting the distribution of receptors on (A) CD4neg NK cells, (B) CD4+ NK cells, and (C) CD56+ monocyte/DC-like cells. Conclusions are based on results in Figures 3 and 5. Receptors expressed on the majority of cells of that subset are represented in black. No expression or < 15% of cells expressing a receptor is indicated in gray. Receptor expression in panel C is representative of a heterogeneous population of monocytes and DC-like subsets. For example, CD16 is not expressed on CD123+ cells, but is expressed on many of the CD13+ cells. The expression of cytokines after stimulation with K562 target cells, IL-12 + IL-18, and poly(I:C) for the particular cell subset is indicated below. Only CD7+ NK cells produced IFNγ after stimulation with NK cell–specific stimuli, whereas a strong IL-6 and TNFα response was observed in the CD7negCD56+ cells after poly(I:C) stimulation. The ability of CD7+ NK cells and CD7negCD56+ DCs/monocytes to induce proliferation in a mixed leukocyte reaction (MLR function) is also indicated; only CD7negCD56+ cells are inducers of proliferation in accordance with their phenotype as DCs/monocytes.
In contrast to CD7+CD56+ NK cells, the CD7negCD56+ monocyte/DC-like cells clearly had a different surface phenotype that lacked NK cell–associated markers, but was enriched for markers associated with monocytes and dendritic cells (Figure 6A,C). In our MLR assay, classically defined NK cells (CD3negCD56+) were capable of supporting a low level of T-cell proliferation; however, purified CD7+CD56+ NK cells did not induce any significant proliferation of the responder T cells. Furthermore, in agreement with our phenotypic analysis of CD7negCD56+ cells, the purified CD7negCD56+ DC/monocyte population was an efficient inducer of allogeneic T-cell proliferation. Based on our results, the weak antigen-presenting capabilities of freshly isolated, resting NK cells in a classical preparation appears to be the result of the contaminating CD7negCD56+ DCs/monocytes. Another function of NK cells we investigated was their ability to respond directly to the TLR3 agonist poly(I:C). In contrast to a previous study,45 we did not observe IFNγ responses from poly(I:C)-stimulated classical NK cells or CD7+ NK cells. We did, however, observe a low frequency of IL-6 and TNFα ELISPOT-forming cells in the classical preparation of CD3negCD56+ NK cells, again indicating the presence of a contaminating DC/monocyte population. In the CD7+ NK-cell preparation, IL-6 and TNFα spot-forming cells were completely absent, indicating that contaminating DCs/monocytes had been efficiently removed. The detection of IL-6 and TNFα spot-forming cells in CD7negCD56+ DCs/monocytes indicates these cells may have been the cells previously described as NK cells that were responding to poly(I:C).45 In classic NK-cell preparations, which contain a low level of contaminating CD56+ DCs/monocytes, these DCs/monocytes are likely producing cytokines that stimulate NK cells to produce IFNγ in a paracrine manner. The absence of IL-6 and TNFα responses to LPS from the CD7negCD56+ DCs/monocytes is not surprising as these cells lack CD14 expression. A more thorough analysis of the TLR repertoire expressed by NK cells is now possible using CD7 to gate out contaminating DCs/monocytes.
As our data indicate, the combination of CD7 and CD56 greatly improves our ability to distinguish NK cells from contaminating cell subsets. Interestingly, the function of both of these markers, CD7 and CD56, on NK cells is not well understood. CD56, or neural cell adhesion molecule, is a glycoprotein that binds fibroblast growth factor 1, which is constitutively expressed by fibroblasts.46 The expression of CD56 on NK cells may be important in the differentiation of NK cells through contact with fibroblasts.47 The functional purpose of CD56 expression of the monocyte/DC populations we observed here also remains unknown; however, it may also play a role in differentiation of these cell subsets. Because CD56 is often used as an exclusion marker in DC studies, this subset of DCs remains largely unstudied. Our observation that CD56 may be expressed by as many as 10% of DC subsets suggests that instead of using CD56 as an exclusion marker, CD7 should be used to eliminate NK cells from DC preparations. The function of CD7 on NK cells is likewise relatively unstudied. CD7 is a member of the immunoglobulin superfamily that binds to K12 (secreted and transmembrane 1), as well as galectin-1.35,48,49 Plate-bound anti-CD7 induces IFNγ production and proliferation of NK cells35 ; however, the effect of K12 or galectin-1 binding to CD7 on NK cells remains unknown. In T cells, galectin-1 has an anti-inflammatory effect by inducing apoptosis50 ; however it is unknown whether galectin-1 interaction with CD7 on NK cells induces a similar effect. Thus, although their function on NK cells is not completely understood, CD7 and CD56 coexpression by NK cells allows for improved isolation of this important cell subset.
NK cells have classically been defined as cells capable of killing transformed, stressed, and virally infected target cells without prior sensitization. In contrast, monocytes and dendritic cells are traditionally thought to take up foreign antigens, process them, and present the antigens to T cells. However, several reports have documented antigen presentation by activated human NK cells22-24 and possibly cytolytic capabilities by DCs.37-41 Furthermore, a recent report has found immunologic memory mediated by virus-specific mouse NK cells.7 These overlapping phenotypic and functional characteristics between NK cells, monocyte/DC cells, and B cells and T cells demonstrate the evolving complexity of studying NK cells. Using a combination of markers that includes CD56 and CD7 will greatly increase our ability to investigate the phenotype and function of NK cells, which may be obscured by contaminating DC/monocytic cells. Only when we can study these intricate details of NK-cell biology can we begin to design effective vaccine and therapeutic strategies to suppress or augment NK-cell function during disease.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Matthew Collin for critical comments on the manuscript and Dr J. P. Houchins for generously providing reagents.
This research was supported, in part, by the Department of Health and Human Services funding under National Institutes of Health (NIH) grant number 5T32HL007185 (J.M.M.). Funding was also provided by NIH grant P01-AI64520 (D.F.N. and L.L.L.). L.L.L. is an American Cancer Society Research Professor. J.M. was supported by a grant from the Swedish Research Council.
National Institutes of Health
Authorship
Contribution: J.M.M. designed and performed experiments, analyzed results, made figures, and wrote the manuscript; B.R.L. designed experiments, participated in discussion of the results, and reviewed the paper; J.E.S.-C. designed experiments and reviewed the paper; A.J.C. performed experiments and reviewed the paper; V.A.Y. performed experiments; L.C.N., J.M., and D.F.N. participated in discussion of the results and reviewed the paper; and L.L.L. participated in discussion of the results and edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jeffrey M. Milush, University of California San Francisco, 1001 Potrero Ave, San Francisco, CA 94110; e-mail: jeffrey.milush@ucsf.edu.