Abstract
Type 1 von Willebrand disease (VWD) is the most common inherited human bleeding disorder. However, diagnosis is complicated by incomplete penetrance and variable expressivity, as well as wide variation in von Willebrand factor (VWF) levels among the normal population. Previous work has exploited the highly variable plasma VWF levels among inbred strains of mice to identify 2 major regulators, Mvwf1 and Mvwf2 (modifier of VWF). Mvwf1 is a glycosyltransferase and Mvwf2 is a natural variant in Vwf that alters biosynthesis. We report the identification of an additional alteration at the Vwf locus (Mvwf5), as well as 2 loci unlinked to Vwf (Mvwf6-7) using a backcross approach with the inbred mouse strains WSB/EiJ and C57BL/6J. Through positional cloning, we show that Mvwf5 is a cis-regulatory variant that alters Vwf mRNA expression. A similar mechanism could potentially explain a significant percentage of human VWD cases, especially those with no detectable mutation in the VWF coding sequence. Mvwf6 displays conservation of synteny with potential VWF modifier loci identified in human pedigrees, suggesting that its ortholog may modify VWF in human populations.
Introduction
von Willebrand factor (VWF) is a central component of hemostasis, serving as the adhesive link between platelets and the injured blood vessel wall, as well as the carrier for factor VIII. Deficiencies in VWF result in von Willebrand disease (VWD), the most common inherited bleeding disorder in humans. Elevated VWF levels may also be an important risk factor for thrombosis, both through a direct role in platelet adhesion,1 as well as indirectly by causing elevated levels of factor VIII.2-4 Diagnosis of VWD is elusive in many cases because of its variable expressivity and incomplete penetrance5 and the nonspecific nature of bleeding symptoms.6 VWF plasma protein levels also display a broad distribution in the normal human population. Thus, it is often difficult to determine whether a person has VWD and is at risk for pathologic hemorrhage or simply has VWF levels in the low range of normal.
Levels of plasma VWF have been shown to be largely determined by genetic factors, with estimates of heritability in humans ranging from 25% to 32% by pedigree analysis,7,8 to 66% to 75% in twin studies.9,10 ABO blood group is responsible for one-third of the genetic variability in VWF plasma levels.11 However, the loci responsible for the remaining two-thirds of this genetic component are unknown. Recent evidence from European and Canadian cooperative studies on type 1 VWD have found that disease diagnosis does not segregate with VWF genotype in approximately 50% of families, supporting the existence of additional genetic factors.12-16
Laboratory mice display wide variation in VWF levels with 65% heritability in a cross between the strains A/J and CASA/RkJ,17 strikingly similar to the estimates for humans derived from twin studies.9,10 This variability among inbred mouse strains has been used to identify genetic loci modifying VWF levels, including Mvwf1 (modifier of Vwf), a mouse glycosyltransferase (B4galnt2) that alters clearance of VWF.18 A similar mechanism probably explains the modification of human VWF levels by ABO blood group and some cases of type 1 VWD.19,20 A natural variant of the murine Vwf gene has also been identified (Mvwf2).17
We now report a backcross between 2 additional inbred mouse strains, WSB/EiJ (WSB) and C57BL/6J (B6), with relatively high and low levels of plasma VWF, respectively. Genetic analysis identified 3 significant loci regulating VWF levels. The first is a novel cis-regulatory allele of Vwf, whereas the others map to novel loci on chromosomes 5 and 10.
Methods
Mouse strains and bleeding
Mice were purchased from The Jackson Laboratory. Each individual mouse was bled on 3 separate occasions, at least 1 week apart. Bleeds were performed after isoflurane-induced anesthesia by retro-orbital technique on alternating eyes from week to week, removing approximately 75 μL whole blood with each bleed into heparinized capillary tubes (Thermo Fisher Scientific). For the strain survey, 3 females of each strain were bled between 4 and 8 weeks of age. Further analysis of the C57BL/6J (B6) and WSB/EiJ (WSB) strains were as follows: 2 each of WSB males and females, 3 males and 4 females of B6, and 4 males and 5 females of (B6 × WSB) F1 mice were bled between 3 and 8 weeks of age. For the backcross study, WSB males were crossed to B6 females to generate F1 progeny. Both male and female F1s were backcrossed to B6 to produce the N2 generation. Two hundred seven N2 mice were also bled in the same manner described above, with the first bleed at weaning, the second between 3.5 and 6.5 weeks, and the third from 5 to 8.5 weeks. The mice were then exsanguinated by cardiac puncture after pentobarbital-induced anesthesia, and blood was collected into 0.5M EDTA pH 8.0 at a dilution of 1:40. Organs were harvested for genomic DNA preparation. Mice were housed in microisolator cages, and all procedures were approved by and performed according to the University of Michigan's Committee on Use and Care of Animals guidelines.
VWF plasma protein quantitation and analysis
Platelet-poor plasma was isolated from whole blood by centrifugation at 2000g and stored at −70°C before analysis. VWF levels were quantitated by enzyme-linked immunosorbent assay (ELISA) essentially as previously described.17 A pool of male and female adult B6 plasma (age, 6-8 weeks) was used to generate a standard curve, and a mean VWF level was calculated for each mouse from the 3 retro-orbital bleeds. Because of age and sex differences between the standard pool and experimental groups, the B6 parental strain levels are slightly higher than the value of 10 that was arbitrarily assigned to the standard. Analysis of variance was performed on natural log-transformed VWF values to assess the ELISA assay variance (variation in replicate measurements on the same plasma sample), environmental variance (variation among replicate measurements taken on the same animal), genetic variance (variation among animals), and total backcross population variance. Heritability was estimated by comparing genetic variance with total backcross population variance after correcting for assay variance. Assay variance was determined to be approximately 2% of total variance.
Genotyping
Genomic DNA was isolated from liver by digestion in buffer containing 0.1 mg/mL proteinase K, 0.1M Tris (pH 8.0), 0.2M NaCl, 0.2% SDS, and 5mM EDTA at 55°C overnight, followed by phenol/chloroform extraction, isopropanol precipitation, washing with 70% ethanol, and resuspension in 10mM Tris-Cl, pH 8.0. Genotyping on DNA from 200 mice was performed by the Mammalian Genotyping Service of the National Heart, Lung, and Blood Institute. Seven mice with VWF levels in the middle of the population were removed from the 207 N2 progeny, and 157 simple sequence length polymorphisms (SSLPs) were examined across the genome. At the University of Michigan standard genotyping was performed by polymerase chain reaction (PCR) and agarose gel electrophoresis as described17 to type an additional 28 SSLPs and 4 single nucleotide polymorphisms (SNPs) on all 207 N2 mice, 33 SSLPs selectively applied to mice with the 5% highest and lowest VWF levels, and 7 SSLPs on the entire N2 population excluding the 5% highest and lowest VWF groups. All map positions are noted in millions of base pairs (Mbp), obtained from NCBI (National Center for Biotechnology Information) Build 37 of the mouse genome, unless otherwise noted.
Genome scan analysis and QTL identification
A genome scan was performed by standard interval mapping21 with the natural log of the mean VWF level (averaged across the 3 plasma samples) as a quantitative trait and sex as an additive covariate. LOD (logarithm of odds) scores were obtained as the log-10 likelihood ratio comparing a model with a single QTL (quantitative trait locus) to a model with no QTLs. Because the effect size of the QTL on chromosome 6 was so strong, 2 additional genome scan analyses were also performed with the marker D6Mit366 included as either an additive or interactive covariate, keeping the sex adjustment in both cases. The choice of D6Mit366 was arbitrary because D6Mit329 and D6Mit366 are both at the peak of the chromosome 6 QTL and only 1 Mbp apart. Significance thresholds were obtained by permutation testing, whereby the genome scan analysis was repeated 50 000 times on the autosomes and 1 000 000 times on the X chromosome,22 randomly shuffling phenotypes while keeping genotypes fixed.23 The LOD scores for genomewide significance at levels α equal 0.01 (highly significant), 0.05 (significant), and 0.25 (suggestive) were taken as quantiles of the corresponding permutation distributions, partitioning the type I error rate across the autosomes and the X chromosome, according to the method of Broman et al.22 Note that sample sizes in analyses of the X chromosome were reduced from 207 to 78 individuals, because analysis was restricted to individuals from crosses for which the X chromosome was segregating.22 Finally, 95% confidence intervals were obtained as 96.5% Bayes credible intervals.24,25 Data management and genome scan analyses were performed with the use of R26 and the R/qtl package.27
Allele-specific primer extension analysis
Three male and 3 female 9-week-old (B6 × WSB) F1 mice were anesthetized with isoflurane and pentobarbital and humanely killed. Lungs were dissected and immediately frozen in liquid nitrogen, and total mRNA was isolated with Trizol (Invitrogen). mRNA was treated with DNAse I (Invitrogen) and subjected to reverse transcription (RT)–PCR with SuperScript One-Step (Invitrogen) with the use of Vwf internal exon 7 primers 5′-GGGAGCAATGCCAGCTACT-3′ and 5′-GGCACTGTGGTCAGTCCAG-3′, at 50°C for 30 minutes, 94°C for 2 minutes, 35 cycles of 94°C for 30 seconds, 56°C for 30 seconds, 72°C for 1 minute, concluding with 72°C for 10 minutes. Relative allele-specific mRNA accumulation was determined using a primer extension assay with fluorescently labeled primers as previously described,17,28 using a standard curve composed of B6 and WSB genomic DNA quantitated with PicoGreen (Invitrogen) and mixed in various ratios. The RT-PCR product was treated with ExoSAP-IT (US Biochemical Corp) to remove free nucleotides and primers. Primer extension was carried out with Thermo Sequenase (US Biochemical Corp), a 6-FAM–labeled primer (5′-TGGATCCCGAGTCCTTTGTGGCTC-3′) and a mixture of nucleotides containing ddCTP and run at 94°C for 2 minutes, 30 cycles of 94°C for 30 seconds, 65°C for 30 seconds, 72°C for 1 minute, resulting in differentially sized products because of a T/C SNP +758 bp from the A of the initiation methionine of Vwf. Products were diluted 1:13 in Hi-Di formamide (Applied Biosystems), separated on an Applied Biosystems 3730XL DNA Analyzer at the University of Michigan Sequencing Core, and quantitated with GeneMarker 1.51 (SoftGenetics LLC). The standards were fit to a line resulting in an R2 value of 0.996, calculated by linear regression with Microsoft Excel 2003 (Microsoft Corporation).
Calculation of Vwf locus contribution to plasma VWF levels in the N2 population
The following calculation was used to assess the relative contribution of the Vwf locus to the level of plasma VWF. The N2 population was subdivided into 2 groups based on genotype at a G/T SNP in exon 5 of Vwf (+365 base pairs from the A of the initiation methionine) that produces an XhoI restriction fragment polymorphism that cuts WSB, but not B6 (amplified with the primers 5′-GGCAAGAGAATGAGCCTGTC-3′ and 5′-TGAATCACAGAATCAATGGACTA-3′), and the average VWF level was calculated for the resulting B6:B6 and B6:WSB groups. Any minor modifier loci would be expected to be evenly divided among these 2 groups by Mendelian inheritance; therefore, their effects on plasma VWF levels should be evenly distributed.
Results
VWF plasma levels in WSB/EiJ mice are 3.5-fold higher than C57BL/6J
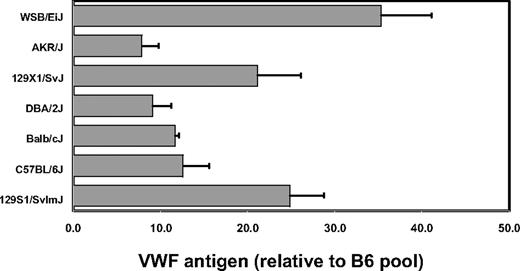
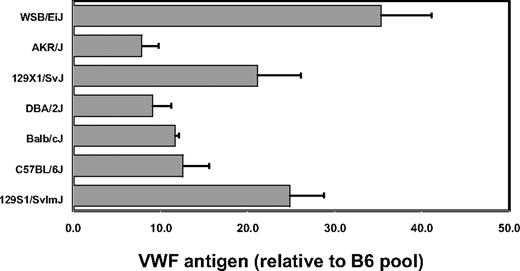
VWF levels were quantitated by ELISA on plasma obtained from 6 laboratory inbred strains as well as the wild-derived inbred strain WSB. VWF plasma protein levels were determined as the average of data from 3 separate retro-orbital bleeds, done at least 1 week apart. WSB exhibited the highest plasma VWF, with levels 3.5-fold greater than most of the common inbred strains, including B6 (Figure 1). Two 129/Sv substrains tested each displayed intermediate levels of VWF, which were 2-fold greater than B6.
Inbred mouse strain survey for VWF plasma protein levels. Individual mice were bled, and the results were averaged and normalized to an adult B6 plasma pool arbitrarily assigned a value of 10. Error bars represent SD.
Inbred mouse strain survey for VWF plasma protein levels. Individual mice were bled, and the results were averaged and normalized to an adult B6 plasma pool arbitrarily assigned a value of 10. Error bars represent SD.
(B6 × WSB) N2 progeny display a wide range of VWF levels
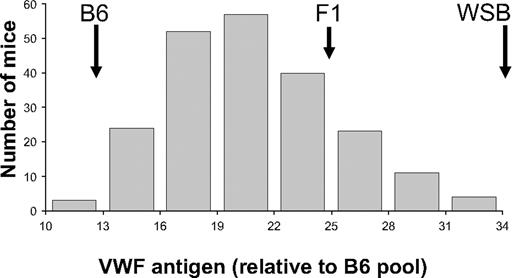
B6 was chosen to cross with WSB because it has been used as a standard strain in many previous hemostasis and other disease models, and it is also the reference strain for the mouse genome.29,30 Importantly, neither B6 nor WSB contain allelic variants associated with the previously reported modifier loci Mvwf1 and Mwf2.17,31 (B6 × WSB) F1 plasma VWF levels were intermediate between the 2 parental strains but closer to WSB (Figure 2). Therefore, an outcross/backcross approach with B6 as the backcross parent was chosen. Plasma VWF levels were determined on 207 N2 progeny generated from a (B6 × WSB) F1 backcross to B6 (Figure 2). The observed levels in N2 animals encompassed the entire phenotypic range, extending to the pure parental strains and peaking just below the level of F1 mice. Heritability of VWF levels in the (B6 × WSB) N2 population was determined to be approximately 71%. There was also a statistically significant difference in population averages among bleeds (population averages: bleed 1, 24.5; bleed 2, 18.2; bleed 3, 16.4; P < .001).
Histogram of N2 generation VWF plasma levels. Individual N2 mice were bled, and the results were averaged and normalized to an adult B6 plasma pool that was arbitrarily assigned a value of 10. The VWF antigen levels were binned and plotted as indicated on the x-axis. The locations of the parental strain and (B6 × WSB) F1 hybrid values are indicated by. These are 13.1, 24.8, and 35.3, for B6, F1, and WSB, respectively.
Histogram of N2 generation VWF plasma levels. Individual N2 mice were bled, and the results were averaged and normalized to an adult B6 plasma pool that was arbitrarily assigned a value of 10. The VWF antigen levels were binned and plotted as indicated on the x-axis. The locations of the parental strain and (B6 × WSB) F1 hybrid values are indicated by. These are 13.1, 24.8, and 35.3, for B6, F1, and WSB, respectively.
Genotyping of N2 backcross progeny identifies 3 major QTLs for VWF plasma levels
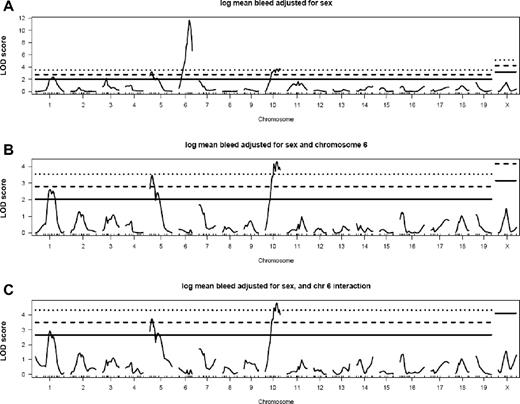
Two hundred N2 progeny were genotyped with 189 markers spaced across the genome. Additional markers were used in several subsets of mice (see “Genotyping”). Standard interval mapping at 1-centimorgan (cM) intervals using the R/qtl package27 was performed on the natural log-transformed mean VWF levels for the N2 population with adjustment for sex. Three QTLs were identified with significant linkage to VWF levels, with peak markers at D6Mit329 (Mvwf5), D10Mit269 (Mvwf6), and D5Mit66 (Mvwf7) and LOD scores of 11.65, 3.72, and 3.22, respectively (Figure 3A; Table 1).
R/qtl analysis of N2 progeny shows 3 QTLs that modify VWF plasma levels. Interval mapping was performed on natural log-transformed mean VWF levels. (A) LOD scores with adjustment for sex show 3 QTLs on chromosomes 6, 10, and 5 (Mvwf5, Mvwf6, and Mvwf7, respectively). No additional loci were identified by analysis for additive (B) or epistatic (C) effects upon adjustment for Mvwf5. Solid, dashed, and dotted lines indicate α = 0.25 (suggestive), 0.05 (significant), and 0.01 (highly significant) thresholds, respectively, obtained by permutation testing.
R/qtl analysis of N2 progeny shows 3 QTLs that modify VWF plasma levels. Interval mapping was performed on natural log-transformed mean VWF levels. (A) LOD scores with adjustment for sex show 3 QTLs on chromosomes 6, 10, and 5 (Mvwf5, Mvwf6, and Mvwf7, respectively). No additional loci were identified by analysis for additive (B) or epistatic (C) effects upon adjustment for Mvwf5. Solid, dashed, and dotted lines indicate α = 0.25 (suggestive), 0.05 (significant), and 0.01 (highly significant) thresholds, respectively, obtained by permutation testing.
To detect additional minor QTLs masked by the extremely strong effect of the chromosome 6 locus, the data were reanalyzed with the marker D6Mit366 (115.2 Mbp) as an additive or interactive covariate. The results were very similar to the initial analysis, without significant changes in the statistical significance of the LOD scores for Mvwf6 and Mvwf7 (Figure 3B-C). No additional loci, including the known modifier loci B4galnt2 (Mvwf1), Mvwf3-4,32 and the human ABO blood group ortholog, Abo (chromosomes 11, 4, 13, and 2, respectively), surpassed the 0.05 significance threshold in any of these analyses (as determined by permutation testing).
Examination of candidate genes in the peak of the Mvwf5 QTL showed that the Vwf gene itself is located in this region. (B6 × WSB) N2 mice that were BW (B6:WSB) at both the D6Mit329 and D6Mit339 markers (114.0 and 136.3 Mbp, respectively, flanking the Vwf locus) were backcrossed to B6 to the N4 and N5 generations, with selection for heterozygosity at both markers. VWF levels were measured as described above on mice that were BB (B6:B6, non-BW littermates derived during backcrossing) or BW at both markers (Figure 4 inset). There was a statistically significant increase in VWF levels in the BW mice (19.6 vs 14.6 in BB mice), confirming that Mvwf5 is a true QTL and narrowing the candidate region to 11.6 cM, which includes Vwf (Figure 4).
VWF levels in congenic mice. VWF plasma levels were determined. B6 represents C57BL/6J mice purchased from The Jackson Laboratory (n = 7). Mvwf5 BW mice were backcrossed to B6 to the N4 or N5 generation, and VWF levels on littermates that were BB (n = 18) or BW (n = 11) at both D6Mit329 and D6Mit339 were determined (see inset; numbers indicate centimorgan [cM] distances between markers and Vwf indicates the XhoI restriction fragment polymorphism described in “Methods”). *The difference between these 2 groups is statistically significant (P < .001 by t test). **The difference between these 2 groups is statistically significant (P < .004 by t test). There was no significant difference between the B6 and BB groups (P > .1 by t test). Error bars represent SD.
VWF levels in congenic mice. VWF plasma levels were determined. B6 represents C57BL/6J mice purchased from The Jackson Laboratory (n = 7). Mvwf5 BW mice were backcrossed to B6 to the N4 or N5 generation, and VWF levels on littermates that were BB (n = 18) or BW (n = 11) at both D6Mit329 and D6Mit339 were determined (see inset; numbers indicate centimorgan [cM] distances between markers and Vwf indicates the XhoI restriction fragment polymorphism described in “Methods”). *The difference between these 2 groups is statistically significant (P < .001 by t test). **The difference between these 2 groups is statistically significant (P < .004 by t test). There was no significant difference between the B6 and BB groups (P > .1 by t test). Error bars represent SD.
Strain-specific differences at the Vwf locus alter steady state Vwf mRNA levels
Sequencing of the WSB Vwf cDNA followed by comparison to published B6 sequence revealed 9 nonsynonymous SNPs, all localized to the VWF propeptide (Table 2). Eighteen synonymous SNPs were discovered, with a large block devoid of any variation at the 3′ end (Table 2). This represents a greater level of divergence between B6 and WSB, 2 strains that are thought to be primarily mus musculus domesticus in origin, versus A/J and CASA/RkJ (the strains used to identify Mvwf2-4), which are mus musculus domesticus and mus musculus castaneus, respectively.17
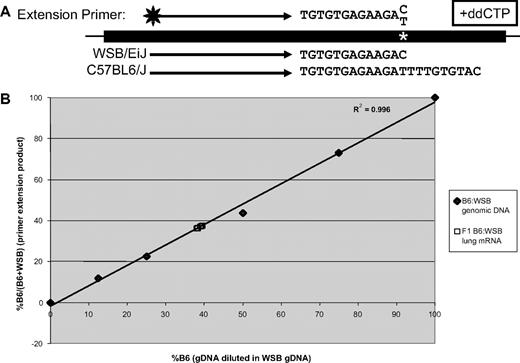
To evaluate potential cis-regulatory differences in mRNA expression or stability, lung mRNA was isolated from (B6 × WSB) F1 mice and subjected to RT-PCR for Vwf mRNA, followed by primer extension analysis that distinguishes the B6 and WSB alleles because of an exonic SNP (Figure 5A). A similar approach has been successful in distinguishing expression from maternal and paternal alleles in cases of human VWD.33 Quantitation showed that 38.7% (± 0.6%) of transcripts were derived from the B6 allele and 61.3% from the WSB locus (Figure 5B).
Allele-specific expression analysis of Vwf in (B6 × WSB) F1 mice. Adult lung cDNA was prepared from F1 mice (n = 3 males and 3 females). Polymerase chain reaction was performed with exonic primers flanking a T/C SNP, followed by primer extension with a fluorescently labeled primer and dATP, dGTP, dTTP, and ddCTP, which results in differentially sized products in each strain (A). Fluorescent primer extension products were separated, quantitated, and compared with genomic standards (B). B6 and WSB genomic DNA were mixed in various proportions to produce the standard curve. Results are expressed as a percentage of B6 transcripts from total transcripts. Linear regression was performed with Microsoft Excel.
Allele-specific expression analysis of Vwf in (B6 × WSB) F1 mice. Adult lung cDNA was prepared from F1 mice (n = 3 males and 3 females). Polymerase chain reaction was performed with exonic primers flanking a T/C SNP, followed by primer extension with a fluorescently labeled primer and dATP, dGTP, dTTP, and ddCTP, which results in differentially sized products in each strain (A). Fluorescent primer extension products were separated, quantitated, and compared with genomic standards (B). B6 and WSB genomic DNA were mixed in various proportions to produce the standard curve. Results are expressed as a percentage of B6 transcripts from total transcripts. Linear regression was performed with Microsoft Excel.
Subdivision of the N2 population into 2 groups based on genotype at Vwf (see “Methods”) resulted in an average VWF level of 18.5 in the B6:B6 group and 22.7 in the B6:WSB group. The B6:B6 group represents the contribution of 2 B6 alleles of Vwf (or 9.25 per B6 allele), whereas B6:WSB represents 1 B6 and 1 WSB allele (or 22.7 − 9.25 = 13.45 for WSB). Thus, the B6 allele contributes 41% of plasma VWF in B6:WSB heterozygotes (9.25/22.7), versus 59% for the WSB allele (13.45/22.7). The results of these calculations are indistinguishable from the mRNA data determined by primer extension (38.7% B6 and 61.3% WSB; P > .6 by χ2 analysis with the plasma values as the expected values). Therefore, we conclude that the Vwf locus contribution to VWF plasma protein levels in the B6xWSB N2 population can be fully explained by differential mRNA accumulation from the 2 alleles. Additional minor contributions from the Vwf gene to the difference in plasma VWF levels between these strains, such as biosynthesis or clearance resulting from the nonsynonymous SNPs, cannot be completely excluded but are probably small.
Mvwf6 overlaps with human VWF QTL
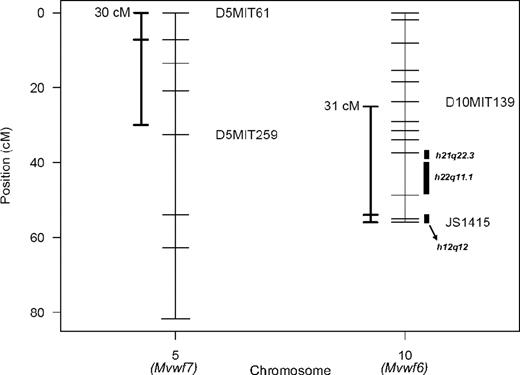
The 96.5% Bayes credible intervals were constructed for Mvwf6 and Mvwf7 (Figure 6), as previously described.25 Human orthologous regions were determined from comparative orthology maps34 and aligned to potential VWF modifier regions identified in human populations. Souto et al (from the Genetic Analysis of Idiopathic Thrombophilia or GAIT study)35 analyzed VWF levels as a quantitative trait in a group of 21 Spanish families and found potential linkage to several regions unlinked to the human VWF locus. The linkage identified in the GAIT study included 22q11.1, which displays orthology to Mvwf6 (Figure 6). Similar comparisons to linkage data from a large Amish pedigree phenotyped by ristocetin cofactor activity36 reveal orthology for Mvwf6 with human loci at 12q12 and 21q22.3 (Figure 6).
Mvwf QTL 96.5% Bayes credible intervals. Chromosomes are represented as vertical bars, and ticks represent markers used in mapping. To the left of each chromosome is the 96.5% Bayes credible interval constructed as described.25 To the right of chromosome 10 are the regions with human homology of synteny to significant regions identified in human studies 22q11.1,35 21q22.3, and 12q12.36 JS1415 is a marker derived from a B6/WSB T/G SNP downstream from the Vwf gene (128.3 Mbp) that produces an MspI restriction fragment polymorphism.
Mvwf QTL 96.5% Bayes credible intervals. Chromosomes are represented as vertical bars, and ticks represent markers used in mapping. To the left of each chromosome is the 96.5% Bayes credible interval constructed as described.25 To the right of chromosome 10 are the regions with human homology of synteny to significant regions identified in human studies 22q11.1,35 21q22.3, and 12q12.36 JS1415 is a marker derived from a B6/WSB T/G SNP downstream from the Vwf gene (128.3 Mbp) that produces an MspI restriction fragment polymorphism.
Discussion
Previous analyses of laboratory strains of mice have identified both major and minor loci regulating VWF levels.17,18,32 We now report a second strong Vwf variant associated with regulation of VWF plasma protein levels, as well as 2 additional minor loci that are unlinked to the Vwf locus. Using 2 previously unexamined strains, we found the heritability of VWF plasma levels to be 71%, similar to the value of 65% found in an (A/J × CASA/RkJ) F2 population17 and also similar to estimates of VWF level heritability in human populations.9,10 The (B6 × WSB) N2 population displayed a statistically significant decrease in VWF levels over time, falling by 49% from weaning (3 weeks) to adulthood (5-8.5 weeks). This may parallel the pattern in humans, in which plasma VWF at birth decreases by 43% to near adult levels by 6 months of age.37 In contrast, the (A/J × CASA/RkJ) F2 population17 did not show a similar age-dependent decrease in VWF. These data show strain-specific differences in the regulation of VWF levels over time in mice and raise the possibility of similar factors in humans.
Mvwf5 is a natural mouse allele altering plasma VWF because of a cis-regulatory mutation in the Vwf gene itself. This is the second example of a natural variant Vwf allele among inbred strains of mice, as Mvwf2 is due to a VWF gene-coding mutation.17 The relative elevation in steady-state levels of Vwf mRNA derived from the WSB Mvwf5 allele could be due to an alteration in transcriptional regulation, splicing, or mRNA stability, mechanisms identified in many common human genetic disorders, such as the thalassemias.38
Although most human type 1 VWD mutations identified to date affect protein function, transport, secretion, or clearance, this probably represents an ascertainment bias because most studies only examine exons, exon/intron junctions, and the proximal promoter and would miss more distant regulatory sequence changes. Consistent with this hypothesis, comprehensive exon sequencing in patients with type 1 VWD has identified mutations in only 67% of persons.12,16,39,40 Of those with identified mutations, 7% altered splice sites and 11% were localized to the promoter. Although not tested directly, both classes of mutation would be expected to alter allelic expression at the mRNA level, as shown here for murine Mvwf5. In addition, a subset of the larger class of exonic mutations could also potentially affect mRNA levels through alterations in mRNA stability. Distant regulatory mutations not covered in the human sequence analysis performed to date could also affect mRNA expression, potentially accounting for a significant percentage of the above patients for whom no mutations were identified. Recent data suggest that long-range effects of distant regulatory elements could be particularly important for VWF gene expression.41-43 Thus, a significant contribution from the large quantity of VWF intergenic and intronic sequences remains to be explored.
The presence of SNPs within the murine Vwf mRNA sequence permitted the dissection of relative mRNA expression from each Vwf allele in this and a previous study.17 An analogous approach has been used for peripheral blood platelet VWF mRNA in humans.33 Future studies applying similar RNA SNP analysis to human patients could potentially define novel subgroups of type 1 VWD.
Our findings, taken together with previous work,17,18,32 provide genetic analysis of 5 mouse strains in 3 different crosses and has identified 2 independent, naturally occurring Vwf gene mutations leading to altered plasma VWF levels. These data suggest that the equivalent of type 1 VWD is remarkably common in mice, as well as in humans. Reports of VWD in horse, cat, pig, rabbit, and dog5,44 suggest that a high prevalence of Vwf gene mutations may be common among all mammalian species. These observations, together with the highly variable levels of plasma VWF in humans45 and mice (Lemmerhirt et al17 and Johnsen et al31 ; Figure 1), might be explained by common selective pressures, potentially including interactions with infectious pathogens. Consistent with this hypothesis, analysis of B4galnt2 (Mvwf1) in wild mouse populations suggests that this locus is under selective pressure.46 These observations also suggest that the genetic regulation of plasma VWF in the inbred laboratory mouse is complex and probably involves a large number of genes. The corresponding picture in the outbred human population is probably more complex and even more difficult to approach experimentally.
The orthologous region of Mvwf6 overlaps with a significant VWF QTL identified in the GAIT study,35 as well as regions identified as potential human VWD modifiers in a large Amish pedigree.36 These human loci are distinct from those showing potential conservation of synteny with Mvwf3 and Mvwf4.32 Thus, analyses of inbred mouse strains have identified 5 potential modifier loci outside of the Vwf gene, Mvwf1,18 Mvwf3-4,32 and Mvwf6-7 (this report), 3 of which display conservation of synteny with potential human modifier loci. Although the overlapping regions are still quite large, positional cloning of these VWF modifier genes in the mouse should identify candidate genes for the modification of bleeding and thrombotic risk in humans.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the National Heart, Lung, and Blood Institute Mammalian Genotyping Service; the University of Michigan Sequencing and Genotyping Core; and Dave Siemieniak, Tricia Hayes, Diana Keung, Jennifer Myaeng, Susan Spaulding, Beverly Twiss, and Alok Swaroop for technical assistance.
This work was supported by the National Institutes of Health (R37-HL39693 and P01-HL057346, D.G.; and R01-GM074244, K.W.B.), the American Heart Association (0675025N, J.A.S.), the National Hemophilia Foundation Clinical Fellowship Program (J.A.S.), the National Institute of Child Health and Human Development (HD028820, J.A.S.), and 2 National Science Foundation Graduate Fellowship Awards (H.L.L. and A.M.). D.G. is an investigator of the Howard Hughes Medical Institute.
National Institutes of Health
Authorship
Contribution: J.A.S. participated in research design, performed the research, and wrote the paper; A.M. aided in data analysis and wrote the paper; H.L.L. participated in research design and performed the research; K.W.B. aided in data analysis and wrote the paper; and D.G. participated in research design and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for A.M. is Department of Biomedical Engineering, University of Virginia, Charlottesville, VA. The current affiliation for K.W.B. is Department of Biostatistics and Medical Informatics, University of Wisconsin, Madison, WI.
Correspondence: David Ginsburg, Howard Hughes Medical Institute, University of Michigan, 210 Washtenaw Ave, Life Sciences Institute, Rm 5036, Ann Arbor, MI 48109; e-mail: ginsburg@umich.edu.


![Figure 4. VWF levels in congenic mice. VWF plasma levels were determined. B6 represents C57BL/6J mice purchased from The Jackson Laboratory (n = 7). Mvwf5 BW mice were backcrossed to B6 to the N4 or N5 generation, and VWF levels on littermates that were BB (n = 18) or BW (n = 11) at both D6Mit329 and D6Mit339 were determined (see inset; numbers indicate centimorgan [cM] distances between markers and Vwf indicates the XhoI restriction fragment polymorphism described in “Methods”). *The difference between these 2 groups is statistically significant (P < .001 by t test). **The difference between these 2 groups is statistically significant (P < .004 by t test). There was no significant difference between the B6 and BB groups (P > .1 by t test). Error bars represent SD.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/26/10.1182_blood-2009-07-233213/4/m_zh89990944990004.jpeg?Expires=1765922781&Signature=HHeZpvTkxu-1xk3AAojb3AmJBtOYaCQ7P5mgQayKiUJW~a1HxPMVlOa5evYWv5sCdVjUpkW5BdJKYtxxrxUdErUKEGKmFrBfORdU3cdgtYWCetorcWHRM7rmTmbiAlsGBMDkxTiyh8iJjvGt5uU8I9g0Sen2e7GlK0Gvxyj5AiPCdKb-EjE4AlIuz7F4Py5mlXEjl1ByTHKvFwHYIuhIHDqGOez5eNwQR41Lk3M~YEpifp1SNQEBOCoplT4cQ4d1foNZtg196RwCq-TJcTPuQ~p3eZgJubfMdttoN1vIoogE~7PUkGB11lV8EplOzUdVmgkK9nGqaMWPRI~Imb6U6Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 4. VWF levels in congenic mice. VWF plasma levels were determined. B6 represents C57BL/6J mice purchased from The Jackson Laboratory (n = 7). Mvwf5 BW mice were backcrossed to B6 to the N4 or N5 generation, and VWF levels on littermates that were BB (n = 18) or BW (n = 11) at both D6Mit329 and D6Mit339 were determined (see inset; numbers indicate centimorgan [cM] distances between markers and Vwf indicates the XhoI restriction fragment polymorphism described in “Methods”). *The difference between these 2 groups is statistically significant (P < .001 by t test). **The difference between these 2 groups is statistically significant (P < .004 by t test). There was no significant difference between the B6 and BB groups (P > .1 by t test). Error bars represent SD.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/26/10.1182_blood-2009-07-233213/4/m_zh89990944990004.jpeg?Expires=1765922782&Signature=matqeYp6Hs0Xk0BeG4n9zAdv5pa8x8QxgK1yD27Auv8-VKHVxiubprI~PudEsjvODObAMHHQOQBmEA1rpstZI0zZ5JT0Kk2YycrHUlASO~hOB1EO4DQCDvHLuuKr9JO72C76k9899SzLchGu2PWv22c2dBQHWqLp950cGknDI2DDxOyp3Cu6-fbdcMv~MfcPZXJN4rN5Ze1Y5wbu~qfuMznCU2aEN4WV93hdOZ7qBbfsjH7iuKzm0Z0skNhitZ0FZVfP6X9lvRPJoFVteUvqcLaKtI7kj7jYqiAZumK5tWB9Grabqe79XSkcKIu9jOA0b5GlSnClRDtfqq5UU8M0vA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)